Abstract
As RNA-seq is replacing gene expression microarrays to assess genome-wide transcription abundance, gene expression Quantitative Trait Locus (eQTL) studies using RNA-seq have emerged. RNA-seq delivers two novel features that are important for eQTL studies. First, it provides information on allele-specific expression (ASE), which is not available from gene expression microarrays. Second, it generates unprecedentedly rich data to study RNA-isoform expression. In this paper, we review current methods for eQTL mapping using ASE and discuss some future directions. We also review existing works that use RNA-seq data to study RNA-isoform expression and we discuss the gaps between these works and isoform-specific eQTL mapping.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
With the completion of the human reference genome [36] and the pilot study of the 1000 Genomes Project [17], an unprecedented wealth of knowledge has been accumulated for human DNA sequence variations. In contrast, much less of this DNA-level knowledge has been translated to the understanding of human diseases. Gene expression quantitative trait locus (eQTL) mapping, which aims to dissect the genetic basis of gene expression, is one of the most promising approaches to fill this gap [11]. Many early genome-wide eQTL studies were conducted on experimental populations [7, 9, 43, 57, 67, 89]. Recently, more eQTL studies have been reported on human populations [72, 74, 75] and some of them used both DNA and RNA information to study phenotypic outcomes, such as complex diseases [18, 31, 66, 103].
RNA-seq is replacing gene expression microarrays to be the major technique for genome-wide assessment of transcript abundance. Compared with microarrays, RNA-seq provides more accurate estimates of transcript abundance for either known or unknown transcripts in a larger dynamic range, while requiring less RNA materials [90]. The central computational problems in RNA-seq include read mapping, transcriptome reconstruction (or RNA-isoform selection given exon annotations), transcript abundance estimation, and differential expression analysis. Since a number of RNA-seq protocols were developed at 2008 [10, 47, 51, 84], numerous technical improvements or computational/statistical methods have been developed for RNA-seq. We refer interested readers to Ozsolak and Milos (2010) [52] and Garber et al. (2011) [21] for recent reviews of experimental and computational methods for RNA-seq, respectively. In this review paper, we focus on the statistical/computational methods of eQTL mapping using RNA-seq.
A few pioneer studies of eQTL mapping using RNA-seq have emerged [50, 58]. These pioneer studies employed existing eQTL mapping methods that were designed for microarray data, and thus cannot fully exploit the new features in RNA-seq data. For eQTL studies, RNA-seq provides allele-specific gene expression (ASE), which is not available in microarrays, and unprecedentedly rich information for RNA-isoform expression. To the best of our knowledge, no statistical/computational method has been specifically developed for eQTL mapping using RNA-Seq, except for our recent work [77]. In the following, we will discuss the issues and potentials of eQTL mapping using ASE and isoform-specific eQTL mapping.
2 eQTL Mapping Using ASE
2.1 Introduction
In a diploid individual, each gene has two alleles: the paternal and maternal allele. The allele-specific transcript abundance is referred to as the ASE of this gene. Cis-acting regulation is due to DNA variation that directly influences the transcription process in an allele-specific manner (Fig. 1(a)). Alternatively, trans-acting regulation affects the gene expression by modifying the activity (or abundance) of the factors that regulate the gene, which leads to the same amount of expression changes for both alleles [91] (Fig. 1(b)). In this paper, we refer to an eQTL of a gene as a cis-eQTL if it alters the expression of the two alleles of this gene differently, otherwise we refer to the eQTL as a trans-eQTL. Therefore, cis- and trans-eQTL can be distinguished by ASE (Fig. 1(a),(b)) [16, 64]. In contrast, total expression of a gene cannot separate cis-eQTL and trans-eQTLs because the two types of eQTL result in similar patterns across a group of individuals (Fig. 1(c),(d)). In previous eQTL studies using microarrays, cis-eQTLs were often not distinguished from local eQTLs due to the lack of ASE. Here, we use the precise definitions of cis- and trans-eQTLs based on the ASE patterns [63]. In what follows, we introduce more details of ASE and cis-/trans-eQTL mapping using RNA-seq data.
(a) An example of a cis-eQTL in two samples. In Sample 2 where the target SNP (the SNP for which we test association) has a heterozygous genotype CG, the expressions of the two alleles are different. (b) An example of a trans-eQTL in two samples. In Sample 2 where the target SNP has a heterozygous genotype TA, the expressions of the two alleles are the same. (c) A simulated data for a cis-eQTL across 60 samples with 20 samples within each genotype class. (d) A simulated data for a trans-eQTL across 60 samples with 20 samples within each genotype class
2.1.1 ASE
In earlier studies, ASE has been assessed by quantitative genotyping following RT-PCR [12, 16, 64], which is a relatively labor-intensive low-throughput approach. Genome-wide genotyping arrays have also been used to assess ASE at predetermined polymorphic sites [23, 24, 45]. Recently, RNA-seq has been used to study the allelic imbalance of gene expression by comparing the expression of the two alleles at a single heterozygous SNP [14, 25, 48, 94]. Among these existing approaches for ASE studies, RNA-seq is the only one that provides both allelic and total expression data [55]. Previous studies have shown that allelic imbalance of gene expression is relatively common. For example, Zhang et al. [100] showed that 20 % of target polymorphic sites exhibited 1.5-fold expression difference, and Ge et al. [23] showed that 30 % of measured transcripts exhibited 1.2-fold expression difference.
Currently, ASE is often assessed by mapping the RNA-seq reads to reference genome followed by counting the number of allele-specific reads that overlap with heterozygous SNPs. Two major technical difficulties hinder accurate measurement of ASE. One is that the mapped allelic reads may be biased to the allele represented by the reference genome. The other is relative low density of heterozygous SNPs (other types of polymorphic sites) where we can assess ASE. For the former problem, one effective treatment is to remove the SNPs that tend to cause mapping bias [58]. For the latter problem, one can impute the genotypes of untyped SNPs and aggregate the information of multiple SNPs given known haplotype. While haplotypes’ information is often not available, they can be imputed (together with genotypes of untyped SNPs) using available genotype data and reference haplotypes [8, 44]. Another strategy that addresses both technical difficulties of ASE assessment is to directly map RNA-seq reads to individual-specific haploid genomes. The haploid genomes may be available for the study of experimental cross, or they can be imputed [8, 44]. The success of this strategy relies on the accuracy the haploid genomes. We are not aware of any study that has carefully compared the two strategies or mapping to reference genome or imputed haploid genomes, and it is certainly an interesting research topic. If there is no genotype data available at all, it is also possible to align RNA-seq reads to the reference genome, call genotypes, and then impute haplotypes using the genotype calls [81].
A simple binomial test can be applied to test whether the expression of the two alleles are the same or not. However a binomial distribution cannot accommodate possible overdispersion in the data, and thus beta-binomial distribution may be preferred. Recently, Skelly et al. [71] have proposed a hierarchical Bayesian model that combines information across loci to test allelic imbalance of gene expression.
2.1.2 eQTL mapping using ASE
To the best of our knowledge, except for our recent work [77], no method has been proposed for eQTL mapping using ASE measured by multiple SNPs. In what follows, we briefly describe our eQTL mapping method using ASE by an example of a cis-eQTL for one gene in three individuals (Fig. 2). Assume that this gene has two exons and there are two exonic SNPs, one on each exon, with alleles A/T and A/G, respectively. We test the association of the gene expression with an upstream SNP (target SNP), which has two alleles, C and T. A straightforward approach is to test the association between Total Read Count (TReC) of this gene and the target SNP (Fig. 2(b)). In this example, TReC is negatively correlated with the number of T alleles of the target SNP.
Testing the association between ASE and the target SNP is less straightforward. We can consider it as a two-step procedure: (1) count the number of allele-specific reads as ASE; (2) assess the association between ASE and the target SNP (Fig. 3).
We first use the example in Fig. 2 to describe the procedure of counting allele-specific reads. An RNA-seq read is allele-specific if it can be assigned to one of the two alleles of the gene without ambiguity. As illustrated in Fig. 2(a), individuals (i) and (ii) have heterozygous genotypes for at least one exonic SNP, and thus their ASE can be measured by the RNA-seq reads that overlap with the heterozygous SNPs. Specifically, all the RNA-seq reads in individual (i) are allele-specific (Fig. 2(c)). However, for individual (ii), only the reads of the first exon are allele-specific, while the reads of the second exon do not overlap with any heterozygous SNP and hence are not allele-specific (Fig. 2(d)). Haplotype information is needed to obtain gene-level ASE by combining ASE measured at different exonic SNPs. For example, for individual (i), we count the number of allele-specific reads on the haplotype A-A and the haplotype T-G.
Next, we discuss association testing using ASE. It is important to note that the target SNP can be anywhere in the genome, and we can study the ASE association as long as the target SNP is connected with the gene of interest by contiguous haplotypes. For example, for individual (i) in Fig. 2(a), given haplotypes C-A-A and T-T-G, we can assign ASE of the gene to the two alleles of the target SNP (Fig. 2(c)). The association testing seeks to answer this question: whether one allele of the target SNP is associated with higher or lower ASE of the gene of interest. If the answer is yes, we expect ASE of one allele to be higher than the other allele when the target SNP is heterozygous, and ASE of the two alleles to be comparable when the target SNP is homozygous. For example, individual (i) has a heterozygous genotype at the target SNP, and the C-A-A allele has higher expression than the T-T-G allele. In contrast, individual (ii) has a homozygous genotype at the target SNP, and the two alleles have the same number of allele-specific reads.
Finally, we conclude this section by a real data example consisting of 65 HapMap YRI samples [58]. Figure 4(a) shows the association between TReC of the gene KLK1 (ENSG00000167748) and SNP rs1054713. There is an apparent negative correlation between TReC of KLK1 and the number of T alleles of SNP rs1054713. Figure 4(b) illustrates the association between ASE of KLK1 and the two alleles of SNP rs1054713. Denote the number of allele-specific reads pertaining to the C allele and the T allele of SNP rs1054713 by ASEc and ASEt, respectively. We are interested in whether the proportion ASEt/(ASEc + ASEt) is deviated from 0.5. The results of TReC association show that the T allele is associated with lower expression (Fig. 4(a)). If the genetic effect is allele-specific, then within one individual, the T allele should also have lower expression than the C allele; thus the proportion ASEt/(ASEc+ASEt) should be lower than 0.5. This is consistent with the observation shown in Fig. 4(b).
(a) An example of TReC association between the gene KLK1 and SNP rs1054713. The y-axis is the total number of reads mapped to the gene KLK1 and each point corresponds to one of the 65 samples. (b) An example of ASE association. The y-axis is the proportion of ASEt over all the allele-specific reads. The allele of ASEt is defined as the allele corresponding to the T allele of SNP rs1054713 when the SNP is heterozygous, and it is defined arbitrarily when the SNP is homozygous. When SNP rs1054713 is homozygous, the proportion is around 0.5; when it is heterozygous, the proportion is below 0.5, indicating that the expression from the T allele is lower than that from the C allele
2.2 Methods
Let T i and N i be respectively TReC and ASE (i.e., allele-specific read count) in sample i (1≤i≤n, where n is the number of study samples). Suppose that the target SNP has two alleles, A and B. Denote the two haplotypes of the gene of interest by H i =(H i1,H i2). Let N i1 be the number of allele-specific reads that are mapped to haplotype H i1, which implies N i1≤N i . Let G i be the genotype of the target SNP, which takes the value AA, AB or BB. Our model is based on the following factorization:

Each component is defined as follows.
-
P(T i |H i ,G i ). Given G i , the total read count T i is assumed to be independent of H i and follows a negative binomial distribution with mean μ AA , μ AB or μ BB corresponding to G i =AA, AB or BB, and a dispersion parameter ϕ. We define the association parameter β (T)≡log(μ AA /μ BB ), i.e., the log ratio of the gene expression between genotype classes AA and BB. The eQTL strength can be assessed by testing whether β (T)=0. We refer to the above model, denoted by \(P_{\beta^{(\mathrm{T})}, \phi}(T_{i} | G_{i})\), as the TReC model. The superscript (T) in β (T) indicates that the association parameter is defined in the TReC model.
-
P(N i |T i ,H i ,G i ). This part of information is irrelevant for assessing the eQTL strength, and thus can be factored out of the likelihood.
-
P(N i1|N i ,T i ,H i ,G i ). Given (N i ,H i ,G i ), the read count N i1 is assumed to be independent of T i and follows a beta-binomial distribution with a parameter π, which is the expected proportion of the allele-specific reads from haplotype H i1 over the N i allele-specific reads, and a dispersion parameter ψ. If the target SNP is homozygous in sample i, i.e., G i =AA or BB, π is fixed to be 0.5; thus the two haplotypes H i1 and H i2 can be defined arbitrarily because the likelihood remains the same if the definitions of H i1 and H i2 are flipped. The samples with homozygous genotypes at the target SNP only contribute to the estimation of the dispersion parameter ψ. If the target SNP is heterozygous, π is a free parameter, and without loss of generality we define H i1 and H i2 such that the haplotype configuration is A-H i1 and B-H i2. The eQTL strength can be assessed by testing whether π is deviated from 0.5. Following the above discussion, we have \(P(N_{i1} | N_{i},T_{i},H_{i},G_{i}) = \{P_{\pi =0.5, \psi}(N_{i1} | N_{i})\}^{I(G_{i}=AA~\text{or}~BB)} \{P_{\pi, \psi }(N_{i1} | N_{i})\}^{I(G_{i}=AB)}\), where I(.) is an indicator function. We refer to this model as the ASE model.
The TReC model can detect both cis- and trans-eQTLs (although it cannot distinguish cis- and trans-eQTLs), and it is more powerful than a computationally convenient approach: normal quantile transformation of the TReC data is followed by a linear regression [77]. The ASE model can only detect cis-eQTL. In the following derivation, we show that the TReC and ASE data provide consistent information for cis-eQTL mapping, and thus combining them increases the power of cis-eQTL mapping. Let

where the superscript of β (A) indicates that β (A) is the genetic effect defined in the ASE model, and μ A and μ B denote the expected number of allele-specific reads for haplotypes A-H i1 and B-H i2. Recall that β (T)≡log(μ AA /μ BB ), where μ AA and μ BB are the expected TReC when the target SNP has the genotype AA and BB, respectively. Since TReC of an individual equals to the summation of TReC on each allele, we have

Note that log(μ A /μ B ) in (1) and (2) have different meanings. In (1), the expression log(μ A /μ B ) is the log ratio of ASE from the A-H i1 allele vs. the B-H i2 allele within an individual with a heterozygous genotype at the target SNP. In contrast, log(μ A /μ B ) in (2) is the log ratio of TReC from two individuals with respective genotypes AA and BB. By the definition of cis-eQTL, the variation of gene expression abundance across individuals is due to allele-specific expression, and thus we can equate log(μ A /μ B ) in (1) and (2) for cis-eQTL but not for trans-eQTL. In other words, for cis-eQTL, we can estimate the genetic effect β based on the joint likelihood \(\mathcal{L}(\beta,\phi ,\psi)\) combining the TReC and ASE data, where
and

We refer to this joint model as the TReCASE model. We have also developed a statistical test to distinguish cis- and trans-eQTLs:
One should use the TReC model for trans-eQTL and the joint model for cis-eQTL [77]. The details of obtaining MLE from the TReC, ASE, and TReCASE models are skipped, and the interested readers are referred to Sun (2011) [77].
2.3 Implementation
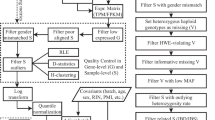
In most real data studies, the input data are RNA-seq data in the FASTA or FASTAQ format, DNA genotype data, and haplotype data from reference panels. The implementation of eQTL mapping using RNA-seq can be divided into four major steps: DNA data processing, RNA data processing, read counting, and eQTL mapping (Fig. 5).
In the step of DNA data processing, we use a phasing program, such as BEAGLE [8] or MACH [44], to impute the phase as well as to impute the genotype of a large set of SNPs that are phased against a referenced panel. It is also possible to align RNA-seq reads to the reference genome, call genotypes, and then impute haplotypes using the genotype calls [81].
The step of RNA data processing involves mapping RNA-seq reads to the genome. One can either map the reads of all the individuals to the same reference genome, or map them to the individual-specific haploid genomes that are constructed based on the phasing results. The advantages/limitations of these two approaches have been discussed in Sect. 2.1.1.
The counting step counts TReC per gene, per sample, and counts the number of allele-specific reads per allele of a gene, per sample. If there are m genes and n samples, the result of counting TReC is a matrix of size m×n, and the result of counting ASE is a matrix of size m×2n. Counting TReC is not trivial because one may prefer to count the reads that overlap and only overlap with the exonic regions of the gene of interest. Counting ASE is more complicated because one needs to compare the nucleotides in an RNA-seq read with the two alleles of any heterozygous SNP. Some Quality Control (QC) steps should be implemented. For example, the reads with mapping ambiguity or low mapping quality should be removed. While counting allele-specific reads, one should check the sequencing quality score of a read at a particular SNP. If the sequencing quality score at that particular base pair is low, the read should not be counted as allele-specific. In addition, one RNA-seq read may harbor more than one SNP and those SNPs may suggest contradicting alleles for the read, e.g., one SNP suggests this read is from paternal allele and the other SNP suggests it is from the maternal allele. Such reads should also be discarded.
Finally, in the step of eQTL mapping, the variation of TReC and/or ASE of a gene is associated with a target SNP, using the haplotype information to connect the alleles of the gene to the alleles of the target SNP. Two sets of covariates can be included in the regression model. One is the set of observed covariates, including the total number of reads per sample, batch, gender, age, etc. The other is the set of derived covariates that aim to capture unobserved batch effects. For example, one may use standardized TReCs (TReCs of all genes of a sample are normalized by the total number of reads of that sample) to estimate Principal Components (PCs) via Principal Component Analysis (PCA), and then use these PCs as derived covariates.
2.4 Discussion and Future Directions
The above discussions of eQTL mapping assume that the haplotypes are known or they are accurately estimated by a phasing program. It is reasonable to expect that the haplotypes within exonic regions of a gene can be accurately estimated. Almost 90 % of the annotated genes are shorter than 100 kb [20], in which haplotypes estimated from genotypes (i.e., phasing) are usually accurate [46]. In addition, RNA-seq assembly can fix possible switch errors from phasing. Although most existing methods for genome-wide de novo RNA-seq assembly do not produce allele-specific assembly yet [5, 69], we conjecture that reference-genome guided assembly, which is sufficient to fix switch errors from phasing, is feasible and computationally efficient. The main challenge is to infer the haplotypes connecting the target SNP and the gene body. Phasing across a long genetic distance is often inaccurate, and RNA-seq assembly cannot help if the target SNP is located in a non-exonic region, which is true in most cases. Due to this limitation, we have carried out eQTL mapping only for local SNPs within 200 kb of each gene [77]. Although recent developments render whole-genome phasing possible [19, 35, 97], these techniques are not mature enough for large-scale studies yet. Therefore, there is a pressing need to develop statistical methods for eQTL mapping using ASE that can accommodate the uncertainty of long-distance phasing.
Xiao and Scott [94] have proposed several methods for cis-eQTL mapping based on the allele-specific expression measured at a single exonic SNP from phase-unknown data: an F-test to assess whether log(N i1/N i2) has a larger variance when the target SNP is heterozygous, a t-test to assess whether the mean value of log(N i1/N i2) is deviated from 0, and a mixture-model-based test in which log(N i1/N i2) is modeled by a mixture normal distribution to account for phasing uncertainty. They found that the t-test/F-test has the highest power when the LD between the target SNP and the exonic SNP is high/low, and the mixture model approach has the highest power for moderate LD. The problem they addressed can be considered as a simplified situation of eQTL mapping using RNA-seq with a few limitations. First, they measured ASE only on a single transcribed SNP instead of across all exonic SNPs of the gene. Second, they did not borrow the information of TReC for eQTL mapping. Third, they modeled log(N i1/N i2) using normal approximation, which is less accurate than directly modeling the read counts by discrete distribution, especially for relatively lower read counts.
In addition to improving statistical power for eQTL mapping, dissecting the genetic basis of ASE can provide important insights into biology questions. For example, some recent studies have shown that cancer drivers/contributors may show imbalanced allelic expression in germline and/or tumor tissues [30, 49, 82, 101]. Such allelic imbalanced expression may be considered as biomarkers and their genetic basis may be valuable to guide personal treatments.
3 Isoform-Specific eQTL Mapping
3.1 Introduction
One important source that contributes to functional complexity of the mammalian genome is the RNA-isoforms due to alternative splicing of pre-messenger RNA [33, 36]. It has been shown that more than 90 % of human genes are alternatively spliced [54, 85], and gene expression is often differentially regulated at the isoform level in different tissues and/or at different developmental stages [85]. Previous studies have reported associations between alternative splicing events and diseases such as cystic fibrosis [22] and cancer [83, 86]. RNA-seq data provide unprecedentedly rich information to study alternative splicing events [54, 76, 85, 90]. Specifically, read depth along the gene body is informative for inferring the underlying RNA-isoforms, and reads covering exon junctions provide direct evidence of alternative splicing. Such information is also available from exon tiling arrays [95] and exon junction arrays [68], but with lower precision and limited by the probe design of the array.
There are three types of statistical/computational problems for the study of RNA-isoforms using RNA-seq data: transcriptome reconstruction, isoform abundance estimation, and differential isoform usage testing. Differential isoform usage refers to the changes of RNA-isoform expression relative to the expression of the corresponding gene. The purpose of isoform-specific eQTL mapping is to dissect the genetic basis of differential isoform usage. We also refer to isoform-specific eQTL mapping as splicing QTL mapping or sQTL mapping. Since isoform abundance cannot be directly measured, transcriptome reconstruction and abundance estimation are necessary steps of sQTL mapping, and the results of these two steps have non-negligible effect on the testing of differential isoform usage. Therefore, we review all the three topics.
3.2 Transcriptome Reconstruction
There are two types of methods for the purpose of transcriptome reconstruction: genome-independent reconstruction and genome-guided reconstruction [21]. Genome-independent reconstruction methods, such as Velvet [99], ABySS [5], and trans-ABySS [61], directly assemble the RNA-seq reads into transcripts without using a reference genome. This approach is, obviously, the only choice for organisms without a reference genome. However, when transcriptome annotation is available, the genome-guided reconstruction methods, which first map all the RNA-seq reads to the reference genome and then assemble overlapping reads into transcripts, are more accurate and computationally much more efficient. Mapping RNA-seq reads to the reference genome may involve the detection of de novo exons and exon junctions by TopHat [79], SpliceMap [3], MapSplice [87], SplitSeek [1], G-Mo.R-Se [15], QPALMA [13], or other software. Two genome-guided reconstruction methods, Cufflinks [80] and Scripture [27], have been developed. Both methods build assembly graphs (using different approaches though) in which one path in the graph corresponds to an RNA-isoform. Cufflinks reports a minimal set of isoforms by choosing a minimal set of paths while Scripture reports all compatible isoforms.
3.3 Isoform Abundance Estimation
We group the methods for isoform abundance estimation into four categories (Table 1). The methods in the first category (e.g., ALEXA-seq [26] and NEUMA [38]) estimate isoform abundance using the sequence reads that are unique to an isoform. This approach misses the information embedded in the “isoform multi-reads” [39], i.e., reads that are compatible with more than one isoform.
The methods in the other three categories use different approaches to probabilistically assign the “isoform multi-reads” to certain isoforms and then estimate isoform abundance. Methods in the second category employ a generative model to describe the stochastic process in RNA-seq experiments. The term “generative model” means that the process of generating each read is modeled so that the likelihood is a product of the likelihoods from each read. For example, following Equation (14) of Pachter (2011) [53] (with some changes of notation so that the notations are consistent in this paper), the likelihood of N single-end reads from K isoforms is

where \(\tilde{l}_{k}\) is the effective length (i.e., the number of positions where a read can start) of the kth isoform, \(\widetilde{c}_{s,k}=1\) if read s is compatible with the kth isoform and 0 otherwise, and α k is the probability of selecting a read from the kth isoform. The probability α k can be formulated as \(\alpha_{k} = \theta_{k} \tilde{l}_{k} / \sum_{k'=1}^{K} \theta_{k'} \tilde{l}_{k'}\), where θ k is the relative abundance of the kth isoform and is the parameter of interest. Extension to paired-end fragments involves modeling the distance of the two reads of a paired-end fragment. We skip the details here and refer interested readers to existing works such as Cufflinks [60, 80].
The third category includes methods that build their likelihood functions by a Poisson model [32, 59, 65]. Given a known set of isoforms, Jiang and Wong [32] modeled the fragment count of each locus (either an exon or an exon junction) by a Poisson distribution, and estimated the expression of each isoform by Maximum Likelihood Estimation (MLE). Specifically, suppose that there are K isoforms, and let N r (1≤r≤R) be the number of reads falling into the rth region of interest (e.g., an exon or exon–exon junction); the likelihood function is

where λ r is the expression rate pertaining to the rth region. Let \(\theta^{*}_{k}\) be the expression rate of the kth isoform and the parameter of interest. We define \(\lambda_{r} = l_{r} w \sum_{k=1}^{K} c_{r,k} \theta^{*}_{k}\) and \(\lambda_{r,r'} = l_{r,r'} w\sum_{k=1}^{K} c_{r,k} c_{r',k} \theta^{*}_{k}\), where w is the total number of sequence reads, l r and l r,r′ are the lengths of the rth exon and the junction of the rth and r′th exon, respectively, and c r,k =1 if the rth region is compatible with the kth isoform and 0 otherwise. Note that it is more appropriate to use the effective length instead of the actual length of exons and exon–exon junctions in the above likelihood [53]. The expression of an isoform could be zero or close to zero, which is the boundary of the parameter space and thus leads to unreliable MLE. Jiang and Wong [32] addressed this problem by importance sampling guided by MLE. Salzman et al. [65] extended the method of Jiang and Wong [32] to work with paired-end sequencing data. Richard et al. [59] developed a similar MLE approach for isoform abundance estimation of known isoforms using only the reads on exons.
The likelihoods employed by the methods in the second and third categories are different. The multinomial generative model pertains to the individual single-end read or paired-end fragment, whereas the Poisson model pertains to the read count of a region. However, the two likelihoods result in an identical estimate of isoform abundance [53], following from the equivalence between the multinomial and Poisson models [37].
The fourth category includes methods based on penalized Poisson regression [93] or penalized least squares [6, 40, 42]. These methods can simultaneously construct isoforms and estimate isoform abundance. For example, isoLasso [42] first identifies candidate isoforms for each gene using a modified connectivity-graph approach of Scripture [27]. Since Scripture reports all isoforms compatible with the observed data, it is expected that some candidate isoforms may not be expressed. Thus, one needs to simultaneously select the expressed isoforms and estimate their abundance. Towards this end, isoLasso [42] minimizes the objective function of penalized least squares

where N r is the number of sequence fragments in the rth region (e.g., exon or exon–exon junction), l r is the length of the rth region, c r,k =1 if the rth region is compatible with the kth isoform, and \(\theta^{**}_{k}\) is the expression rate of the kth isoform and is the parameter of interest. The Lasso penalty \(\lambda \sum_{k=1}^{K} | \theta^{**}_{k} |\) can penalize some of \(\theta^{**}_{k}\)’s to be 0, hence achieving the goal of isoform selection [78]. The authors of isoLasso pointed out that it is more appropriate to use the effective length instead of the actual length of exons and exon–exon junctions in their objective function.
Recent studies have shown that it is important to consider positional bias and sequence bias for the purpose of transcript abundance estimation [28, 39, 41, 60, 92]. Positional bias refers to the observation that the sequence reads are not uniformly distributed along the transcript. Sequence bias refers to the non-randomness of the sequences around the beginning and the end of each singe-end sequence read or paired-end sequence fragment; for example, reads may be more likely to start at a position of higher GC content. Methods have been developed to account for such biases for both the multinomial generative model [39, 60] and the Poisson model [41, 92]. Another approach is to reweight each sequence read by its first heptamer (seven bases), and instead of counting the number of reads mapped to a genomic region, one adds up the weight of the reads mapped to the region, and then the sums of weight are used as counts for downstream analyses [28].
3.4 Differential Isoform Usage Testing
Recall that differential isoform usage means the changes of the relative isoform expression with respect to the expression of the gene. Testing differential isoform usage is related to but different from testing differential expression. Nevertheless, some conclusions from testing differential expression are instructive for testing differential isoform usage, and are stated in this paragraph. First, for the purpose of testing differential expression, one can apply transformation such as the normal quantile transformation to read count data and then treat the transformed measurements as normally distributed random variables. Such transformation loses information, and it is more appropriate to keep the discrete feature of the RNA-seq data. Several methods have been developed for differential expression testing by modeling read counts via a discrete distribution, such as a Poisson distribution when there is no overdispersion [88], a negative binomial distribution [2, 29, 62] or a generalized Poisson distribution [73] when there is overdispersion, which is often true for expression data across biological replicates. One can also apply a two-stage approach to first test for overdispersion and then apply the appropriate modeling strategy based on the conclusion of the overdispersion test [4].
So far, only a few methods have been developed for testing differential isoform usage. Trapnell et al. [80] employed the square-root of Jensen–Shannon Divergence (JSD) as a test statistic and they derived its asymptotic distribution. Specifically, let p (1),…,p (M) be the distributions of isoform abundance under M conditions, where \(\mathbf{p}^{(m)} = (p_{1}^{(m)}, \ldots, p_{K}^{(m)})^{T}\) is a vector of length K such that \(p_{k}^{(m)}\) is the relative abundance of the kth isoform under condition m. We have \(\sum_{k=1}^{K} p_{k}^{(m)} = 1\), m=1,…,M. Then JSD is defined as

where \(H(\mathbf{p}^{(m)}) = - \sum_{k=1}^{K} p_{k}^{(m)} \log (p_{k}^{(m)})\) is the entropy across the K isoforms. The test statistic, denoted by \(f(\mathbf{p}^{(1)}, \ldots, \mathbf{p}^{(M)}) = \sqrt{JS(\mathbf {p}^{(1)},\ldots, \mathbf{p}^{(M)})}\), asymptotically follows a normal distribution with mean 0 and variance (∇f)T Σ(∇f), where (∇f) is the partial derivative of f(p (1),…,p (M)) with respect to \(p_{k}^{(m)}\), and Σ is the block-diagonal variance-covariance matrix with one block for each p (m).
Singh et al. [70] modeled the transcriptome of one condition by a splice graph, which is constructed such that one edge corresponds to a transcribed interval or a spliced site. Then they proposed a flow difference metric (FDM) to measure the isoform usage difference between two conditions by the difference between the two corresponding splice graphs. They showed that FDM is correlated with JSD and can be used as a classifier for JSD. They developed a non-parametric resampling method to obtain the null distribution of FDM under the null hypothesis of no differential isoform usage, and used this null distribution to test for differential isoform usage.
Although it is important to consider positional bias and sequence bias for isoform abundance estimation as we discussed before, it is a question of whether modeling such a bias is necessary for differential isoform usage testing. Suppose that there is a positional bias such that there is higher read depth in the 3′ end of the gene. Without modeling the positional bias, the abundance of the isoforms closer to the 3′ end of the gene may be overestimated. However, as long as such bias is consistent across all the samples, it does not lead to a false positive result for differential isoform usage testing.
3.5 Differential Isoform Expression
In addition to isoform usage testing, one can also consider differential expression of each isoform. Notably, differential isoform expression testing is different from isoform usage testing. The former produces one p-value for each isoform while the latter produces one p-value for multiple isoforms of one gene. Cufflinks [80] tests differential expression of a transcript under two conditions by assessing the following test statistic: log(FPKM 1/FPKM 2), where FPKM i is the FPKM (Fragments Per Kilo-base of the transcript and per Million RNA-seq fragments of the sample) of the transcript under condition i, and i=1 or 2. Using the conclusion Var[log(X)]≈Var(X)/E(X)2, they derived a test statistic
which follows standard normal distribution under null hypothesis of no differential expression. This testing approach did not consider the variation of FPKM estimates due to isoform selection.
An alternative method named BASIS (Bayesian Analysis of Splicing IsoformS) [102] directly compares RNA-isoform expression without an intermediate isoform selection step. Specifically, a hierarchical Bayesian model is employed to model the expression coverage difference at one locus between two conditions as a linear combination of the isoform expression differences plus an error term. Since the variance of the error term is dependent on the mean expression level, the error terms of all loci across the genome are grouped into 100 bins by the total coverage of the loci, and modeled separately.
3.6 Splicing QTL (sQTL) Mapping
The problem of sQTL mapping can be considered as a special case of the problem of differential isoform usage testing. To the best of our knowledge, no existing method is able to directly assess the association between the isoform usage and a quantitative covariate, which can be the additive coding of a SNP or the copy number calls at a genomic locus. The testing of differential isoform usage against a quantitative covariate is a very interesting direction for future development, not only for sQTL mapping but also for many other problems of differential isoform usage testing, for example, to assess the association between differential isoform usage and age.
The other potential research direction is to combine the eQTL mapping of total transcription abundance of a gene with the sQTL mapping of relative transcription abundance (e.g., isoform usage), because genetic variation is very likely to affect both the total expression of a gene and the relative expression of its isoforms. If this gene-level testing indicate significant differential expression, either for total expression or for isoform usage, one can further test differential expression of each isoform. We expect that this two-step approach of gene-level testing followed by isoform-level testing is more powerful than directly testing for all possible isoforms due to the reduction of the number of tests, and hence the reduced burden of multiple testing correction.
The third future direction is simultaneous allele-specific and isoform-specific eQTL mapping, which can provide unprecedented details of the genetic basis of transcription regulation. A pioneer work in this direction, a haplotype and isoform-specific expression estimation method, has been reported [81]. In fact, joint analysis of allele-specific expression and isoform-specific expression is necessary to obtain more precise conclusions. We illustrate this point by an example shown in Table 2. Suppose there is a hypothetical gene with three exons of effective length 100 bp, and to simplify the discussion, we ignore the reads overlapping with more than one exon. Here effective length of an exon is defined as the number of base pairs where an RNA-seq fragment can be sampled [80]. Further assume that this gene has two isoforms: one includes exons 1 and 3, and the other includes exons 1, 2, and 3. Isoform 1 has higher expression in paternal allele than maternal allele while isoform 2 has higher expression in maternal allele than paternal allele. If one ignores isoform expression and naively estimates FPKM at gene level, the FPKM estimates for both alleles, paternal allele, and maternal allele are 500/300=1.67,(30+30+70+70+70)/300=0.9, and (70+70+30+30+30)/300=0.77, respectively. However, given isoform configuration, the FPKM estimates at gene level for both alleles, paternal allele, and maternal allele are 500/(0.5×200+0.5×300)=2, (30+30+70+70+70)/(0.3×200+0.7×300)=1, and (70+70+30+30+30)/(0.7×200+0.3×300)=1, respectively. Therefore, ignoring isoform level expression leads to the conclusion that there is allelic imbalance of gene expression, while a more accurate explanation is that there is allele-specific isoform usage.
4 Discussion and Conclusion
Network analysis has been employed in eQTL studies to jointly mapping eQTL of multiple transcripts [56, 98]. It involves simultaneous estimation of residual covariance/precision matrix and the regression coefficient matrix. It is interesting to apply similar approaches for eQTL mapping using RNA-seq data. However, while discrete distributions such as beta-binomial or negative-binomial distributions are appropriate choices to model the RNA-seq count data for each gene, it is much more challenging to study the joint distribution of multiple genes due to the difficulty of studying multivariate beta-binomial or negative-binomial distributions. This is an interesting direction that warrants further developments of appropriate statistical methods.
We would like conclude this paper by pointing out that the developers of statistical/computational methods for eQTL mapping should not only focus on exploiting each bit of information from RNA-seq to improve statistical power. One should put even more emphasis on the scientific questions that can be answered by developing a new method. For example, using allele-specific and isoform-specific eQTL to dissect the genetic/genomic basis of complex diseases. Recent genome-wide association studies (GWAS) found that most common genetic variants can explain at most a few percents of the variance of a complex disease. This has raised some doubts on the efficacy of genetic/genomic approach for understanding complex diseases and developing treatments. eQTL studies can provide more information than GWAS because a complex disease often has tighter correlations with gene expression variations than genetic variants. This is in turn due to at least two reasons. First, by the central dogma of DNA → RNA → Protein, RNA is closer to disease than DNA in terms of signal transmission from DNA to phenotype. Second, the effects of more than one genetic variant may be accumulated on a particular transcript. On the other hand, unlike DNA data, which is stable, RNA data is noisier, e.g., RNA expression varies across tissues and development stages. RNA-seq provides more information of gene expression than expression arrays, together with more variation, e.g., the gene expression may vary in allele-specific manner or in isoform level. By combining DNA and RNA data in eQTL analysis, we may exploit both the stability of DNA data and the informativeness of RNA data for the purpose of understanding complex diseases.
References
Ameur A, Wetterbom A, Feuk L, Gyllensten U (2010) Global and unbiased detection of splice junctions from RNA-seq data. Genome Biol 11(3):R34
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106
Au K, Jiang H, Lin L, Xing Y, Wong W (2010) Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res 38(14):4570–4578
Auer P, Doerge R (2011) A two-stage Poisson model for testing RNA-seq data. Stat Appl Genet Mol Biol 10(1):26
Birol I, Jackman S, Nielsen C, Qian J, Varhol R, Stazyk G, Morin R, Zhao Y, Hirst M, Schein J et al. (2009) De novo transcriptome assembly with ABySS. Bioinformatics 25(21):2872
Bohnert R, Rätsch G (2010) rQuant. web: a tool for RNA-seq-based transcript quantitation. Nucleic Acids Res 38(Suppl 2):W348–W351
Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296(5568):752–755
Browning S, Browning B (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81(5):1084–1097
Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW (2005) Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37(3):233–242
Cloonan N, Forrest A, Kolle G, Gardiner B, Faulkner G, Brown M, Taylor D, Steptoe A, Wani S, Bethel G et al. (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5(7):613–619
Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10(3):184–194
Cowles CR, Hirschhorn JN, Altshuler D, Lander ES (2002) Detection of regulatory variation in mouse genes. Nat Genet 32(3):432–437
De Bona F, Ossowski S, Schneeberger K, Rätsch G (2008) Optimal spliced alignments of short sequence reads. BMC Bioinform 9(Suppl 10):O7
Degner J, Marioni J, Pai A, Pickrell J, Nkadori E, Gilad Y, Pritchard J (2009) Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25(24):3207
Denoeud F, Aury J, Da Silva C, Noel B, Rogier O, Delledonne M, Morgante M, Valle G, Wincker P Scarpelli C et al. (2008) Annotating genomes with massive-scale RNA sequencing. Genome Biol 9(12):R175
Doss S, Schadt E, Drake T, Lusis A (2005) Cis-acting expression quantitative trait loci in mice. Genome Res 15(5):681
Durbin R, Altshuler D, Abecasis G, Bentley D, Chakravarti A, Clark A, Collins F, De La Vega F, Donnelly P, Egholm M et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073
Emilsson V, Thorleifsson G, Zhang B, Leonardson A, Zink F, Zhu J, Carlson S, Helgason A, Walters G, Gunnarsdottir S et al. (2008) Genetics of gene expression and its effect on disease. Nature 452(7186):423–428
Fan H, Wang J, Potanina A, Quake S (2010) Whole-genome molecular haplotyping of single cells. Nat Biotechnol 29(1):51–57
Flicek P, Amode M, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S et al. (2011) Ensembl 2011. Nucleic Acids Res 39(Suppl 1):D800
Garber M, Grabherr M, Guttman M, Trapnell C (2011) Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8(6):469–477
Garcia-Blanco M, Baraniak A, Lasda E (2004) Alternative splicing in disease and therapy. Nat Biotechnol 22(5):535–546
Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagn V, Dias J, Hoberman R, Montpetit A, Joly MM, Harvey EJ, Sinnett D, Beaulieu P, Hamon R, Graziani A, Dewar K, Harmsen E, Majewski J, Güring HH, Naumova AK, Blanchette M, Gunderson KL, Pastinen T (2009) Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet 41(11):1216–1222
Gimelbrant A, Hutchinson JN, Thompson BR, Chess A (2007) Widespread monoallelic expression on human autosomes. Science 318(5853):1136–1140
Gregg C, Zhang J, Weissbourd B, Luo S, Schroth G, Haig D, Dulac C (2010) High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329(5992):643
Griffith M, Griffith O, Mwenifumbo J, Goya R, Morrissy A, Morin R, Corbett R, Tang M, Hou Y, Pugh T et al. (2010) Alternative expression analysis by RNA sequencing. Nat Methods 7(10):843–847
Guttman M, Garber M, Levin J, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol M, Gnirke A, Nusbaum C et al. (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28(5):503–510
Hansen K, Brenner S, Dudoit S (2010) Biases in illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res 38(12):e131
Hardcastle T, Kelly K (2010) baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinform 11(1):422
Hosokawa Y, Arnold A (1998) Mechanism of cyclin d1 (ccnd1, prad1) overexpression in human cancer cells: analysis of allele-specific expression. Genes Chromosomes Cancer 22(1):66–71
Huang R, Duan S, Bleibel W, Kistner E, Zhang W, Clark T, Chen T, Schweitzer A, Blume J, Cox N et al. (2007) A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci 104(23):9758
Jiang H, Wong W (2009) Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25(8):1026
Johnson J, Castle J, Garrett-Engele P, Kan Z, Loerch P, Armour C, Santos R, Schadt E, Stoughton R, Shoemaker D (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302(5653):2141
Katz Y, Wang E, Airoldi E, Burge C (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7(12):1009–1015
Kitzman J, MacKenzie A, Adey A, Hiatt J, Patwardhan R, Sudmant P, Ng S, Alkan C, Qiu R, Eichler E et al. (2010) Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol 29(1):59–63
Lander E, Linton L, Birren B, Nusbaum C, Zody M, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al. (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Lang J (1996) On the comparison of multinomial and Poisson log-linear models. Journal of the Royal Statistical Society Series B (Methodological), 253–266
Lee S, Seo C, Lim B, Yang J, Oh J, Kim M, Lee S, Lee B, Kang C, Lee S (2011) Accurate quantification of transcriptome from RNA-seq data by effective length normalization. Nucleic Acids Res 39(2):e9
Li B, Ruotti V, Stewart R, Thomson J, Dewey C (2010) RNA-seq gene expression estimation with read mapping uncertainty. Bioinformatics 26(4):493–500
Li J, Jiang C, Hu Y, Brown B, Huang H, Bickel P (2011, in press) Sparse linear modeling of RNA-seq data for isoform discovery and abundance estimation. Proc Natl Acad Sci USA
Li J, Jiang H, Wong W (2010) Modeling non-uniformity in short-read rates in RNA-seq data. Genome Biol 11(5):R25
Li W, Feng J, Jiang T (2011) IsoLasso: a lasso regression approach to RNA-seq based transcriptome assembly. Research in Computational Molecular Biology, 168–188
Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JAG, Hazendonk E, Prins P, Plasterk RHA, Jansen RC, Breitling R, Kammenga JE (2006) Mapping determinants of gene expression plasticity by genetical genomics in C elegans. PLoS Genet 2(12):e222
Li Y, Willer C, Ding J, Scheet P, Abecasis G (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34(8):816–834
Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP (2003) Allelic variation in gene expression is common in the human genome. Genome Res 13(8):1855–1862
Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, Halperin E, Lin S, Qin Z, Munro H, Abecasis G et al. (2006) A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet 78(3):437–450
Marioni J, Mason C, Mane S, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18(9):1509–1517
McManus C, Coolon J, Duff M, Eipper-Mains J, Graveley B, Wittkopp P (2010) Regulatory divergence in drosophila revealed by mRNA-seq. Genome Res 20(6):816–825
Meyer K, Maia A, O’Reilly M, Teschendorff A, Chin S, Caldas C, Ponder B (2008) Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol 6(5):e108
Montgomery S, Sammeth M, Gutierrez-Arcelus M, Lach R, Ingle C, Nisbett J, Guigo R, Dermitzakis E (2010) Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464(7289):773–777
Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Ozsolak F, Milos P (2010) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12(2):87–98
Pachter L (2011) Models for transcript quantification from RNA-seq. arXiv:1104.3889
Pan Q, Shai O, Lee L, Frey B, Blencowe B (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40(12):1413–1415
Pastinen T (2010) Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet 11(8):533–538
Peng J, Zhu J, Bergamaschi A, Han W, Noh DY, Pollack J, Wang P (2010) Regularized multivariate regression for identifying master predictors with application to integrative genomics study of breast cancer. Ann Appl Stat 4(1):53–77
Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, Hubner N, Aitman TJ (2006) Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet 2(10):e172
Pickrell J, Marioni J, Pai A, Degner J, Engelhardt B, Nkadori E, Veyrieras J, Stephens M, Gilad Y, Pritchard J (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464(7289):768–772
Richard H, Schulz M, Sultan M, Nürnberger A, Schrinner S, Balzereit D, Dagand E, Rasche A, Lehrach H, Vingron M et al. (2010) Prediction of alternative isoforms from exon expression levels in RNA-seq experiments. Nucleic Acids Res 38(10):e112
Roberts A, Trapnell C, Donaghey J, Rinn J, Pachter L et al. (2011) Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol 12(3):R22
Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman S, Mungall K, Lee S, Okada H, Qian J et al. (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7(11):909–912
Robinson M, McCarthy D, Smyth G (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Rockman M, Kruglyak L (2006) Genetics of global gene expression. Nat Rev Genet 7(11):862–872
Ronald J, Brem R, Whittle J, Kruglyak L (2005) Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet 1(2):e25
Salzman J, Jiang H, Wong W (2011) Statistical modeling of RNA-seq data. Stat Sci 26(1):62–83
Schadt E, Molony C, Chudin E, Hao K, Yang X, Lum P, Kasarskis A, Zhang B, Wang S, Suver C et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6(5):e107
Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422(6929):297–302
Shen S, Warzecha C, Carstens R, Xing Y (2010) MADS+: discovery of differential splicing events from Affymetrix exon junction array data. Bioinformatics 26(2):268
Simpson J, Wong K, Jackman S, Schein J, Jones S, Birol İ (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19(6):1117
Singh D, Orellana C, Hu Y, Jones C, Liu Y, Chiang D, Liu J, Prins J (2011) FDM: a graph-based statistical method to detect differential transcription using RNA-seq data. Bioinformatics 27(19):2633–2640
Skelly DA, Johansson M, Madeoy J, Wakefield J, Akey JM (2011) A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res 21(10):1728–1737
Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG (2007) Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 39(2):226–231
Srivastava S, Chen L (2010) A two-parameter generalized Poisson model to improve the analysis of RNA-seq data. Nucleic Acids Res 38(17):e170
Stranger B, Forrest M, Dunning M, Ingle C, Beazley C, Thorne N, Redon R, Bird C, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer S, Tavare S, Deloukas P, Hurles M, Dermitzakis E (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315:848–853
Stranger B, Nica A, Forrest M, Dimas A, Bird C, Beazley C, Ingle C, Dunning M, Flicek P, Koller D et al. (2007) Population genomics of human gene expression. Nat Genet 39(10):1217–1224
Sultan M, Schulz M, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D et al. (2008) A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321(5891):956
Sun W (2011, in press) A statistical framework for EQTL mapping using RNA-seq data. Biometrics
Tibshirani R (1996) Regression shrinkage and selection via the Lasso. J R Stat Soc B 58(1):267–288
Trapnell C, Pachter L, Salzberg S (2009) TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25(9):1105
Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, van Baren M, Salzberg S, Wold B, Pachter L (2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Turro E, Su S, Gonçalves Â, Coin L, Richardson S, Lewin A (2011) Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol 12(2):R13
Valle L, Serena-Acedo T, Liyanarachchi S, Hampel H, Comeras I, Li Z, Zeng Q, Zhang H, Pennison M, Sadim M et al. (2008) Germline allele-specific expression of tgfbr1 confers an increased risk of colorectal cancer. Science 321(5894):1361
Venables J (2004) Aberrant and alternative splicing in cancer. Cancer Res 64(21):7647
Wang E, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore S, Schroth G, Burge C (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456(7221):470–476
Wang E, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore S, Schroth G, Burge C (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456(7221):470–476
Wang G, Cooper T (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8(10):749–761
Wang K, Singh D, Zeng Z, Coleman S, Huang Y, Savich G, He X, Mieczkowski P, Grimm S, Perou C et al. (2010) Mapsplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38(18):e178
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) Degseq: an r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26(1):136–138
Wang S, Yehya N, Schadt EE, Wang H, Drake TA, Lusis AJ (2006) Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet 2(2):e15
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63
Wittkopp P, Haerum B, Clark A (2004) Evolutionary changes in cis and trans gene regulation. Nature 430(6995):85–88
Wu Z, Wang X, Zhang X (2011) Using non-uniform read distribution models to improve isoform expression inference in RNA-seq. Bioinformatics 27(4):502
Xia Z, Wen J, Chang C, Zhou X (2011) NSMAP: a method for spliced isoforms identification and quantification from RNA-seq. BMC Bioinform 12(1):162
Xiao R, Scott L (2011) Detection of cis-acting regulatory SNPs using allelic expression data. Genet Epidemiol 35:515–525
Xing Y, Stoilov P, Kapur K, Han A, Jiang H, Shen S, Black D, Wong W (2008) MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA 14(8):1470–1479
Xing Y, Yu T, Wu Y, Roy M, Kim J, Lee C (2006) An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res 34(10):3150
Yang H, Chen X, Wong W (2011) Completely phased genome sequencing through chromosome sorting. Proc Natl Acad Sci 108(1):12
Yin J, Li H (2011) A sparse conditional Gaussian graphical model for analysis of genetical genomics data. Ann Appl Stat 5(4):2630–2650
Zerbino D, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829
Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, Eggan K, Church GM (2009) Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods 6(8):613–618
Zhao Q, Kirkness E, Caballero O, Galante P, Parmigiani R, Edsall L, Kuan S, Ye Z, Levy S, Vasconcelos A et al. (2010) Systematic detection of putative tumor suppressor genes through the combined use of exome and transcriptome sequencing. Genome Biol 11(11):R114
Zheng S, Chen L (2009) A hierarchical Bayesian model for comparing transcriptomes at the individual transcript isoform level. Nucleic Acids Res 37(10):e75
Zhong H, Yang X, Kaplan L, Molony C, Schadt E (2010) Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet 86(4):581–591
Acknowledgements
We appreciate constructive comments and suggestions from an associate editor and an anonymous reviewer.
Wei Sun’s research is supported in part by the NIH Grant R01MH090936 and EPA Grant for Carolina Center for Computational Toxicology (RD-83382501). Dr. Hu’s research is supported in part by an internal grant from Emory University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, W., Hu, Y. eQTL Mapping Using RNA-seq Data. Stat Biosci 5, 198–219 (2013). https://doi.org/10.1007/s12561-012-9068-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12561-012-9068-3