Abstract
Enteric viruses have been described as important contaminants in fresh and ready-to-eat foods such as sandwiches, deli meat and dairy products. This is a cross-sectional randomized survey to estimate the prevalence of norovirus and human adenovirus (HAdV) from 100 Brazilian artisanal raw milk cheese samples (Minas and Coalho) obtained from different agroindustries in four producing regions in the states of Minas Gerais and one in Piauí, respectively. From October 2017 to April 2018, norovirus genogroups I and II and HAdV were investigated in these cheese samples by RT-qPCR and qPCR, respectively. Viruses were detected in 43 samples, being 26 norovirus GI strains, 14 HAdV, and 3 both viruses. Norovirus GII strains were not detected. Viral concentrations ranged from 6.17 × 104 to 1.44 × 107 genome copies/L–1 and murine norovirus 1 used as internal process control showed 100% success rate of recovery with efficiency of 10%. There was a trend towards a higher positivity rate for both viruses in the rainy season, and HAdV were more commonly found among samples with higher fecal coliform counts. This study is a first step in assessing the risk that this contamination may pose to the consumer of raw products as well as emphasizing the need for good manufacturing practices, quality control systems in the dairy industry and markets. As a randomized survey, we established baseline figures for viruses’ prevalence in five types of ready-to-eat raw milk artisanal Brazilian cheese, to allow any monitoring trends, setting control targets and future local risk analyses studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of elution-concentration methodologies for recovery and detection of virus from food matrices has implicated enteric viruses as important etiologic agents of diseases associated with contaminated foods worldwide (Havelaar et al., 2015; Mäde et al., 2013). More recently, such methodologies were validated and standardized for norovirus and hepatis A in four food classes including drinking water, fruits and vegetables, surfaces and bivalve molluscs (ISO/TS 15,216–1:2017).
Ready-to-eat foods such as sandwiches, vegetables, deli meat and dairy products have been associated with gastrointestinal outbreaks caused by norovirus in several countries, mainly due to the inappropriate handling of those foods (Stals et al., 2011a, b). Noroviruses, family Caliciviridae, are classified into genogroups (GI-X), which GI and GII causes the majority of illnesses in humans (Chhabra et al., 2019).
Also associated with food contamination (Rodriguez-Manzano et al., 2014), human adenovirus (HAdV), Adenoviridae family, are often found in wastewater worldwide, being considered as a potential indicator of human fecal contamination (Hewitt et al., 2013). In developed countries, periodic testing of raw foods using adenovirus as a bio-indicator of quality is recommended (Lu et al., 2012).
Although Brazilian studies drew attention to the possible role of cheese as a vehicle for enteric viruses affecting humans (de Castro Carvalho et al., 2020; Melgaço et al., 2018), tests for their detection in foods are still not required in Brazil.
Raw milk cheese is the dairy product more associated with foodborne diseases (Verraes et al., 2015). In France, dairy products are responsible for 7.5% of foodborne illnesses and 2.1% of outbreaks due to viral agents (InVS, 2012).
In Brazil, the raw milk Minas artisanal cheese is the oldest and most traditional one representing the main source of revenue of several smallholders, with a production rate of about 70,000 tons per year (Emater, 2004). Artisanal Coalho cheese from Northeastern Brazil is equally important. Thus, the aim of this study was to estimate the prevalence of HAdV and norovirus GI and GII of five artisanal Brazilian cheese types, made with raw milk, using a cross-sectional randomized survey.
Methods
Design, Location of the Study and Sample Planning
From October 2017 to April 2018, 100 ready-to-eat raw milk artisanal Brazilian cheese samples, obtained from 81 different agroindustries from Minas Gerais (MG) and 19 from Piauí (PI), states of Brazil (Fig. 1) were assessed for norovirus GI and GII, and HAdV.
The inclusion criteria required the participating agroindustries to be both manufacturers from a set of single-farm milk and to be registered in the state animal health agency.
There was a list of 235 rural family based cheese-processing agroindustries registered officially in five traditional producing regions of Brazil: Canastra (MG), Serro (MG), Cerrado (MG), Triângulo Mineiro (MG) and Parnaíba (PI). Then each one was marked with a specific number, and the sample of the target population to estimate the prevalence of viruses was determined by random drawing of 100 (42.5%) agroindustry samples. The sample was chosen using the Randbetween function (Microsoft Excel). One cheese was collected by agroindustry to represent it, since each one produces cheese from a set of single-farm milk. The sample size was based on a finite population of 235, margin of error of 7.5%, confidence level of 95% and an expected proportion of 50% Viruses positive cheese.
Data and Sample Collection
The cheese samples were collected for analysis in their original packaging and kept in isothermal boxes under refrigeration (< 4 ºC) during transportation to the laboratory.
Information on each agroindustry and respective samples were obtained using a structured questionnaire organized by a content group, focusing on socio-economic features, animal health, Good Agricultural Practices (GAP) and Good Handling Practices (GHP). Data about bacterial analyses of each cheese sample also were obtained from the Minas Gerais Agriculture and Livestock Institute (IMA) to assess possible associations with the presence of viruses.
Viral Concentration Method
Murine norovirus 1 (MNV-1), kindly provided by Herbert W. Virgin (Washington University School of Medicine) was spiked (10uL) in all samples as internal process control (IPC) before processing of viral concentration. It was used MNV-1 stock concentration of 1.13 × 104 genomic copies/Liter (gc/L). The cheese samples were processed using a swab and eluting in PBS buffer as described in the ISO/TS 15,216–1, 2017.
Nucleic Acids Extraction and Synthesis of Complementary DNA
Nucleic acids (RNA/DNA) were extracted from 140 µL of the concentrated samples, with a QIAamp viral RNA mini kit® (Qiagen, Valencia, USA), using an Automated System procedure following the manufacturer’s instructions QIAcube® (Qiagen, CA, USA). For RNA viruses (norovirus GI/GII and MNV-1) synthesis of complementary DNA were performed using random primers and Superscript III® Reverse Transcriptase (Invitrogen—Life Technologies, Carlsbad, CA, USA) following manufacturer instructions.
Quantitative Polymerase Chain Reaction (qPCR)
Human adenovirus (HAdV), norovirus GI and GII and MNV-1 were investigated by qPCR TaqMan® system (ABI PRISM 7500, Applied Biosystems) using a set of protocols previously described (Table 1). All samples were tested using undiluted and 1:10 diluted nucleic acid, both in duplicate, to assess and avoid possible qPCR inhibitors, with four qPCR reactions per sample. Negative and positive controls of the nucleic acid were also included in all procedures. For all determination, a specific DNA standard curve containing the target qPCR region for each of the viruses (gBlock Gene Fragment, Integrated DNA Technologies, Iowa, USA) was generated by a tenfold serial (107–100 genomic copies (gc)/µL). Samples that showed signals crossing the threshold line in both replicates until cycle threshold (Ct) value ≤ 40 presenting a characteristic sigmoidal curve were considered positives. Viral concentrations were expressed as genome copies per liter (GC/L) and calculated as the average of the duplicate data obtained, correcting for the dilution analyzed.
Molecular Characterization
Positive samples presenting Ct ≤ 35 were also tested by the conventional PCR assay (Table 1) for further Sanger sequencing. Amplicons were purified using Wizard SV Gel and PCR Clean-Up System (Promega™, Madison, WI, USA) following the manufacturer’s recommendations. Nucleotide sequencing was performed using the dideoxynucleotide chain termination method with the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit 1, v. 3.1 and the ABI Prism 3730 Genetic Analyzer (Applied Biosystems™, Foster City, CA, USA). Clustal W in Mega X (Kumar et al., 2018) was used for edition and alignment of nucleotide sequences, which were compared to those available in the National Centre for Biotechnology Information (GenBank, http://www.ncbi.nlm.nih.gov/) database using the BLAST (Basic Local Alignment Search Tool). The MEGA X (Kumar et al., 2018) was also used to build the comparative analyses, evolutionary distances and dendrogram, which were performed using Maximum Likelihood method and Tamura 3 parameter model (T92) (Tamura, 1992), Gamma Distributed (G) with 2,000 bootstraps above 70%.
Epidemiological Analyses
The prevalence of viruses-positive artisanal cheese was determined with a 95% confidence interval, and univariate analyses were performed to assess factors associated with viruses-positive artisanal cheese (outcome). Mid-p exact tests were used to assess which explanatory variables were associated with the outcome, with a 5% significance level.
Results and Discussion
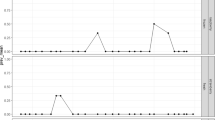
From a total of 100 samples, viral genome was detected in 43%, being 26 (26%, CI95% 17.74% to 35.73%) norovirus GI strains, 14 (14%, CI95% 7.87% to 22.37%) HAdV, and 3 (3%, CI95% 0.62%, 8.52%) both viruses. Viral concentrations ranged from 6.17 × 104 to 1.44 × 107 gc/L−1 and MNV-1, used as IPC, showed 100% success rate of recovery with efficiency of 10%. Accordingly, MNV-1 has been described as an efficient IPC (Stals et al., 2011a, b).
Although positivity results followed a strict criterion considering reproducibility of the results with replicas, the high Cts obtained did not allow the molecular characterization of all viruses detected. Out of 17 with qPCR positive results for HAdV, we were successful in the nucleotide sequencing of one (Gen Bank accession number MW713373). This exhibited high similarity with the HAdV group F (HAdV-41) strains, which is one of the most common enteric viruses involved in foodborne outbreaks (Greening & Cannon, 2016; Russell, 2009).
Concerning contamination of dairy products previously, HAdV was detected in the hands of food handlers at some farms suggesting potential source of contamination (Maunula et al., 2013).
Table 2 shows the univariate analysis for both outcomes: positivity for HAdV and norovirus. Noroviruses were more commonly associated with cheese samples with lower total coliform counts (p = 0.014); perhaps due to a confounding effect of the ‘humidity degree’ variable, since when stratified by this variable that association was no longer found (p = 0.12). Accordingly, HAdV-DNA was more commonly found in cheese samples with lower humidity degrees (bordeline p value = 0.056), but, unlikely noroviruses, this was more associated with higher faecal coliform counts (borderline p value = 0.06). This co-contamination of cheese was also found by other Brazilian studies (de Castro Carvalho et al., 2020; Melgaço et al., 2018). We found a trend towards a higher positivity rate for both viruses in the rainy season in this study, which was also showed among both water environment in Mexico (Rosiles-González et al., 2019) and human cases in Brazil (Amaral et al., 2015).
Norovirus GII strains were not detected in any cheese samples. GI strains may also be underestimated among humans and along the environment, since this genogroup has been associated with lower attack rate (Lopman et al., 2004) and less severe clinical presentation than GII-4 (Friesema et al., 2009; Huhti et al., 2011). Fankhauser et al., (2002) also showed a significant increase of GI strains in US, which was attributed to the application of newly designed primers, called region B primers, that are highly effective at detecting GI strains previously missed with region A primers. We are confident that detecting only GI among the samples tested was not the result of ineffective GII primers or probes, since GI and GII were successfully detected in one cheese (1.1%) each (Melgaço et al., 2018) in a previous study by our team using the same protocol. Thus, further research to access which noroviruses are prevalent in developing countries are needed.
The adenonoviruses, enteroviruses and hepatitis viruses (Johnson et al., 1990), and, more recently, norovirus (Langer et al., 2012), are among the major milk-associated pathogens transmitted by humans, and cheese is the main dairy product involved in foodborne outbreaks (Verraes et al., 2015). Cooling, freezing, thawing, acidification, and pasteurization, appear to be insufficient to inactivate norovirus within a food matrix, or on food surfaces (Mormann et al., 2010; Richards et al., 2012). Adenovirus also is resistant to both tertiary treatment and UV radiation (Thompson et al., 2003); however, temperatures like pasteurization were effective in inactivating this virus (Allard & Vantarakis, 2017).
Our findings of enteric virus contamination in Brazilian artisanal cheese types may be even more worrisome, since these foods are minimally processed and made from raw milk, undergoing no thermal processing, most of which marketed and consumed fresh (median of only 10 days of ripening among our sample). Thus, this study was a first step to assess the risk that such contaminations may pose to the consumer of these products, as well as to emphasize the need for good manufacturing practices, quality control systems in the dairy industry and markets.
Conclusion
HAdV and norovirus GI were found in samples of Brazilian artisanal cheese by a randomized survey. Thus, this work established representative baseline figures for virus prevalence in five types of ready-to-eat raw milk Brazilian artisanal cheese, to allow any monitoring trends, setting control targets and future local risk analysis studies.
Data Availability
All data will be available if requested to the corresponding author.
References
Allard, A., & Vantarakis, A. 2017. Adenoviruses. In J. B. R. a. B. Jiménez-Cisneros (Ed.), Global Water Pathogen Project. http://www.waterpathogens.org/book/adenoviruses. https://doi.org/10.14321/waterpathogens.11
Allard, A., Albinsson, B., & Wadell, G. (2001). Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. Journal of Clinical Microbiology, 39(2), 498–505. https://doi.org/10.1128/JCM.39.2.498-505.2001
Amaral, M. S., Estevam, G. K., Penatti, M., Lafontaine, R., Lima, I. C., Spada, P. K., Gabbay, Y. B., & Matos, N. B. (2015). The prevalence of norovirus, astrovirus and adenovirus infections among hospitalised children with acute gastroenteritis in Porto Velho, state of Rondônia, western Brazilian Amazon. Memorias Do Instituto Oswaldo Cruz, 110(2), 215–221. https://doi.org/10.1590/0074-02760140381
Baert, L., Wobus, C. E., Van Coillie, E., Thackray, L. B., Debevere, J., & Uyttendaele, M. (2008). Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Applied and Environment Microbiology, 74(2), 543–546. https://doi.org/10.1128/AEM.01039-07
Chhabra, P., de Graaf, M., Parra, G. I., Chan, M. C., Green, K., Martella, V., Wang, Q., White, P. A., Katayama, K., Vennema, H., Koopmans, M. P. G., & Vinjé, J. (2019). Updated classification of norovirus genogroups and genotypes. Journal of General Virology, 100(10), 1393–1406. https://doi.org/10.1099/jgv.0.001318
de Castro Carvalho, S. V., Rogovski, P., Cadamuro, R. D., Viancelli, A., Michelon, W., Dos Reis, D. A., & Santana das Chagas, I. A., Assenço, R., da Silva Lanna, M. C., Treichel, H., & Fongaro, G. . (2020). Co-contamination of food products from family farms in an environmental disaster area in Southeast Brazil with pathogenic bacteria and enteric viruses. Archives of Virology, 165(3), 715–718. https://doi.org/10.1007/s00705-019-04501-9
Emater. 2004. Queijo Minas Artesanal: tradição e qualidade que revelam Minas. In (Vol. 80, pp. 8–9). Minas Gerais: Revista da Empresa de Assistência Técnica e Extensão Rural do Estado de Minas Gerais
Fankhauser, R. L., Monroe, S. S., Noel, J. S., Humphrey, C. D., Bresee, J. S., Parashar, U. D., Ando, T., & Glass, R. I. (2002). Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. Journal of Infectious Diseases, 186, 1–7. https://doi.org/10.1086/341085
Friesema, I. H., Vennema, H., Heijne, J. C., de Jager, C. M., Teunis, P. F., van der Linde, R., Duizer, E., & van Duynhoven, Y. T. (2009). Differences in clinical presentation between norovirus genotypes in nursing homes. Journal of Clinical Virology, 46(4), 341–344. https://doi.org/10.1016/j.jcv.2009.09.010
Greening, G. E., & Cannon, J. L. (2016). Human and Animal Viruses in Food (Including Taxonomy of Enteric Viruses). In S. Goyal & J. Cannon (Eds.), Viruses in Food. Springer, Cham. https://doi.org/10.1007/978-3-319-30723-7_2
Havelaar, A. H., Kirk, M. D., Torgerson, P. R., Gibb, H. J., Hald, T., Lake, R. J., Praet, N., Bellinger, D. C., de Silva, N. R., Gargouri, N., Speybroeck, N., Cawthorne, A., Mathers, C., Stein, C., Angulo, F. J., Devleesschauwer, B., & Group, W. H. O. F. D. B. E. R. (2015). World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Medicine, 12(12), e1001923. https://doi.org/10.1371/journal.pmed.1001923
Hernroth, B. E., Conden-Hansson, A. C., Rehnstam-Holm, A. S., Girones, R., & Allard, A. K. (2002). Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: The first Scandinavian report. Applied and Environment Microbiology, 68(9), 4523–4533. https://doi.org/10.1128/AEM.68.9.4523-4533.2002
Hewitt, J., Greening, G. E., Leonard, M., & Lewis, G. D. (2013). Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Research, 47(17), 6750–6761. https://doi.org/10.1016/j.watres.2013.09.001
Huhti, L., Szakal, E. D., Puustinen, L., Salminen, M., Huhtala, H., Valve, O., Blazevic, V., & Vesikari, T. (2011). Norovirus GII-4 causes a more severe gastroenteritis than other noroviruses in young children. Journal of Infectious Diseases, 203(10), 1442–1444. https://doi.org/10.1093/infdis/jir039
ISO 15216–1, I. 2017 Preview. Microbiology of the food chain – Horizontal method for determination of hepatitis A virus and norovirus using real-time RT-PCR – Part 1: Method for quantification. In International & O. f. Standardization (Eds.). Geneva, Switzerland
InVS. (2012). Surveillance of Toxi-Infections Alimentaires Collectives—Données de la Déclaration Obligatoire
Johnson, E. A., Nelson, J. H., & Johnson, M. (1990). Microbiological Safety of Cheese Made from Heat-Treated Milk Part II. Microbiology Journal Food Prot, 53(6), 519–540. https://doi.org/10.4315/0362-028X-53.6.519
Kageyama, T., Kojima, S., Shinohara, M., Uchida, K., Fukushi, S., Hoshino, F. B., Takeda, N., & Katayama, K. (2003). Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. Journal of Clinical Microbiology, 41(4), 1548–1557. https://doi.org/10.1128/jcm.41.4.1548-1557.2003
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. https://doi.org/10.1093/molbev/msy096
Langer, A. J., Ayers, T., Grass, J., Lynch, M., Angulo, F. J., & Mahon, B. E. (2012). Nonpasteurized dairy products, disease outbreaks, and state laws—United States, 1993–2006. Emerging Infectious Diseases, 18(3), 385–391. https://doi.org/10.3201/eid1803.111370
Lopman, B., Vennema, H., Kohli, E., Pothier, P., Sanchez, A., Negredo, A., Buesa, J., Schreier, E., Reacher, M., Brown, D., Gray, J., Iturriza, M., Gallimore, C., Bottiger, B., Hedlund, K. O., Torvén, M., von Bonsdorff, C. H., Maunula, L., Poljsak-Prijatelj, M., … Koopmans, M. (2004). Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet, 363(9410), 682–688. https://doi.org/10.1016/S0140-6736(04)15641-9
Lu, Y., Tong, H. I., Connell, C., & Wang, Z. (2012). Public Health Hotline: Enhanced Monitoring of Hawai‘i Coastal Water Quality Using Potential New Indicators. Hawaii J Med Public Health, 71(6), 163–167.
Mäde, D., Trübner, K., Neubert, E., Höhne, M., & Johne, R. (2013). Detection and typing of norovirus from frozen strawberries involved in a large-scale gastroenteritis outbreak in Germany. Food Environ Virol. https://doi.org/10.1007/s12560-013-9118-0
Maunula, L., Kaupke, A., Vasickova, P., Söderberg, K., Kozyra, I., Lazic, S., van der Poel, W. H., Bouwknegt, M., Rutjes, S., Willems, K. A., Moloney, R., D’Agostino, M., de Roda Husman, A. M., von Bonsdorff, C. H., Rzeżutka, A., Pavlik, I., Petrovic, T., & Cook, N. (2013). Tracing enteric viruses in the European berry fruit supply chain. International Journal of Food Microbiology, 167(2), 177–185. https://doi.org/10.1016/j.ijfoodmicro.2013.09.003
Melgaço, F. G., Luz, I. S., Assis, M. R. S., Caldas, M. S., Maranhão, A. G., Silva, D. A. F., Brandão, M. L. L., Medeiros, V. M., Rosas, C. O., Reis, S. M. L., & Miagostovich, M. P. (2018). Assessment of viral and bacterial contamination of fresh and ripened semi-hard cheeses. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny225
Mormann, S., Dabisch, M., & Becker, B. (2010). Effects of technological processes on the tenacity and inactivation of norovirus genogroup II in experimentally contaminated foods. Applied and Environment Microbiology, 76(2), 536–545. https://doi.org/10.1128/AEM.01797-09
Richards, G. P., Watson, M. A., Meade, G. K., Hovan, G. L., & Kingsley, D. H. (2012). Resilience of norovirus GII 4 to freezing and thawing: implications for virus infectivity. Food Environment Virology, 4(4), 192–197. https://doi.org/10.1007/s12560-012-9089-6
Rodriguez-Manzano, J., Hundesa, A., Calgua, B., Carratala, A., Maluquer de Motes, C., Rusiñol, M., Moresco, V., Ramos, A. P., Martínez-Marca, F., Calvo, M., Monte Barardi, C. R., Girones, R., & Bofill-Mas, S. (2014). Adenovirus and norovirus contaminants in commercially distributed shellfish. Food Environ Virol, 6(1), 31–41. https://doi.org/10.1007/s12560-013-9133-1
Rosiles-González, G., Ávila-Torres, G., Moreno-Valenzuela, O. A., Cháidez-Quiroz, C., Hernández-Flores, C. I., Acosta-González, G., Brown, J. K., Betancourt, W. Q., Gerba, C. P., & Hernández-Zepeda, C. (2019). Norovirus and human adenovirus occurrence and diversity in recreational water in a karst aquifer in the Yucatan Peninsula. Mexico. J Appl Microbiol, 127(4), 1255–1269. https://doi.org/10.1111/jam.14385
Russell, W. C. (2009). Adenoviruses: Update on structure and function. Journal of General Virology, 90(Pt 1), 1–20. https://doi.org/10.1099/vir.0.003087-0
Stals, A., Baert, L., De Keuckelaere, A., Van Coillie, E., & Uyttendaele, M. (2011a). Evaluation of a norovirus detection methodology for ready-to-eat foods. International Journal of Food Microbiology, 145(2–3), 420–425. https://doi.org/10.1016/j.ijfoodmicro.2011.01.013
Stals, A., Baert, L., Van Coillie, E., & Uyttendaele, M. (2011b). Evaluation of a norovirus detection methodology for soft red fruits. Food Microbiology, 28(1), 52–58. https://doi.org/10.1016/j.fm.2010.08.004
Tamura, K. (1992). Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Molecular Biology and Evolution, 9(4), 678–687. https://doi.org/10.1093/oxfordjournals.molbev.a040752
Thompson, S. S., Jackson, J. L., Suva-Castillo, M., Yanko, W. A., El Jack, Z., Kuo, J., Chen, C. L., Williams, F. P., & Schnurr, D. P. (2003). Detection of infectious human adenoviruses in tertiary-treated and ultraviolet-disinfected wastewater. Water Environment Research, 75(2), 163–170. https://doi.org/10.2175/106143003x140944
Verraes, C., Vlaemynck, G., Van Weyenberg, S., De Zutter, L., Daube, G., Sindic, M., Uyttendaele, M. M., & Herman, L. (2015). A review of the microbiological hazards of dairy products made from raw milk. International Dairy Journal, 50, 32–44. https://doi.org/10.1016/j.idairyj.2015.05.011
Acknowledgements
We thank the cheese producers who participated in this study and the IMA and Embrapa Meio Norte for carrying out the fieldwork.
Funding
This work was supported by the Minas Gerais State Agency for Research and Development (FAPEMIG) (Grant Numbers CVZ-APQ-02746–14, CVZ- APQ-03989–17 and CVZ-PPM-00526–16), Embrapa (Grant Numbers 02.13.10.007.00.00, and 12.13.10.007.00.00) and PAEF/IOC/Fiocruz.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Marcio Roberto Silva, Fernando César Ferreira, Adriana Gonçalves Maranhão, Natália Maria Lanzarini, Karina Neoob de Carvalho Castro and Marize Pereira Miagostovich. The first draft of the manuscript was written by Marcio Roberto Silva, Marize Pereira Miagostovich and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethical Approval
Ethical approval was waived by the local Ethics Committee of Oswaldo Cruz Foundation (FIOCRUZ) in view of the retrospective nature of the study. Cheese samples from selected agroindustries, already analyzed for bacteria by official laboratories, were archive stored aseptically at –20 °C and made available to this study for virus assessment. In addition, confidential information on the agroindustries, the owners and the respective samples selected was not disclosed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Preliminary results from this study were presented as a lecture entitled "Assessment of viral contamination of Brazilian artisanal raw milk cheese”, at the 30º Congresso Brasileiro de Microbiologia, 6 to 9 October, 2019, Maceió, AL, Brazil.
Rights and permissions
About this article
Cite this article
Silva, M.R., Ferreira, F.C., Maranhão, A.G. et al. Assessment of Viral Contamination of Five Brazilian Artisanal Cheese Produced from Raw Milk: a Randomized Survey. Food Environ Virol 13, 528–534 (2021). https://doi.org/10.1007/s12560-021-09491-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-021-09491-z