Abstract
In Nokia city about 450,000 l of treated sewage water was for 2 days allowed to run into the drinking water supplies of the city due to a personal error of one employee. Within the next 5 weeks about 1,000 people sought care at the municipal health centre or regional hospital because of gastroenteritis. Here we report the results of viral analyses performed by gene amplification assays from the earliest water and sewage samples as well as from close to 300 patient samples. The contaminating treated sewage was shown to harbour several enteric viruses known to cause acute gastroenteritis. Likewise, the drinking water sample was positive for noro-, astro-, rota-, entero- and adenoviruses. Noroviruses were also found in 29.8% of stool samples from affected patients, while astro-, adeno-, rota- and enteroviruses were detected in 19.7, 18.2, 7.5 and 3.7% of the specimens, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The presence of pathogenic viruses in drinking water has emerged as a significant risk factor for water consumers. Although several viruses can cause acute gastroenteritis human noroviruses are shown to be the causative agents in most of the viral waterborne outbreaks. In addition, several other RNA viruses, like rotavirus, sapovirus, astrovirus, enterovirus, hepatitis A virus and hepatitis E virus, are capable of causing waterborne epidemics of gastroenteritis (e.g. Leclerc et al. 2002; Bosch et al. 2008). Furthermore, dsDNA virus, human adenovirus (HAdV) and especially serotypes 40 and 41 are involved in the aetiology of outbreaks (Enriquez 2002).
Human enteric viruses are non-enveloped and highly stable which means that they can survive well in the environment (Dahling and Safferman 1979; Wyn-Jones and Sellwood 2001). Noroviruses infect people in all age groups and create only short-term immunity whereas viruses like astro-, sapo-, rota- and adenovirus cause infections or symptoms mainly in children and long time immunity is developed (Koopmans et al. 2002). In non-endemic regions like Finland, hepatitis A virus infection which can be prevented by vaccination has rarely been linked to waterborne outbreaks. Hepatitis E virus has caused large waterborne outbreaks in tropical and subtropical regions (Bosch et al. 2008).
Analyses of enteric viruses are relatively simple thanks for the development of RT-PCR (Jiang et al. 1992) and real-time PCR-techniques (Kageyama et al. 2003) and the information about the presence of these viruses in the environment is growing rapidly. So far, regular monitoring of viruses in water is not mandatory and viral analyses are only performed in outbreak situations or in suspicions of an outbreak. In Finland, and elsewhere, several waterborne gastroenteritis outbreaks caused by noroviruses have been recorded (Beller et al. 1997; Maunula et al. 2005).
The most common reason for viral waterborne outbreaks has been contamination by sewage of human origin (Maunula 2007). In previous studies enteric viruses have been reported to be present both in raw and treated sewage water (van den Berg et al. 2005). In the case reported here about 450,000 l of treated sewage water was accidentally allowed to run into the drinking water supplies of a small city of 30,000 inhabitants in Southern part of Finland, resulting into faecal contamination of the household water of about 10,000 people. In microbiological studies several pathogenic bacteria and parasites were found (to be published elsewhere) in both the contaminated drinking water and the treated sewage but here we report the results of viral analyses from the early water samples and from close to 300 patient samples.
Materials and Methods
Preparation of Samples for Viral Analyses
A drinking water sample was taken from a tap of water consumer at 1st December 2007, as soon as possible after the faecal contamination of drinking water was detected. The sample was transported in a coolbox to the laboratory and concentrated by the adsorption–elution method described by Gilgen et al. (1997). Briefly, 1 l sample was run through a positively charged membrane (diameter 47 mm, pore size 45 μm; AMF-Cuno, Zetapor, Meriden, CO, USA) with or without fibreglass prefilter (Millipore, Billerica, MA, USA). Viruses were eluted in 50 mmol/l glycine buffer, pH 9.5, containing 1% beef extract and the eluate was rapidly neutralized with HCl. The volume was further reduced to about 200 μl with a microconcentrator (Vivaspin 2, Vivascience AG, Hannover, Germany or Amicon, Millipore). This sample was used for nucleic acid extraction. Nucleic acid extraction was also performed directly from the water sample or after ultrafiltration of 3 ml sample to a volume of about 200 μl by Vivaspin without any preceding filtrations.
The suspected contamination source was treated wastewater funnelled to the drinking water distribution from the wastewater treatment plant. A treated wastewater sample was taken at 3rd December 2007 and analysed directly and in 10-fold dilutions without any filtration or concentration steps by taking a 200 μl or 140 μl to the nucleic acid extraction depending on the extraction method.
The patient stool samples originated from outbreak cases in all age groups: 24% from children were less than 10 years of age. The 10% faecal suspensions for RNA and DNA extraction were performed in phosphate-buffered saline (PBS) and the suspension was clarified by centrifugation.
Nucleic Acid Extraction
Viral RNA and DNA were extracted from all above supernatants using either the QiaAmp Viral RNA-kit (Qiagen; all RNA viruses except enteroviruses in water), the High Pure Viral Nucleic Acid Kit (Roche Molecular Biochemicals Ltd., Mannheim, Germany; adenoviruses), High Pure Viral RNA Kit (Roche; enteroviruses in water) or the E.Z.N.A.® Total RNA kit (Omega Bio-Tek Inc., Doraville, GA, USA; all RNA viruses in human faeces). All methods were carried out according to manufacturer’s instructions. Extracted nucleic acids were stored at ≤−70°C.
Gene Amplification
Samples of drinking water and wastewater as well as stool samples from patients were analysed for different enteric viruses by using the conventional or real-time methods presented in Table 1. All real-time analyses were performed by Taqman-chemistry with fluorescent probes. Real-time PCR assays were carried out in a Rotor-Gene™ 3000 real-time rotary analyser (Corbett Life Science, Sydney, Australia), LightCycler (Roche) or Mx3005P (Stratagene, USA). Conventional RT-PCR methods described by Gilgen et al. (1997) and Blomqvist et al. (1999) were used to analyse nucleic acid extracts from water and wastewater samples for rotaviruses and enteroviruses, respectively, while real-time assays (Table 1) were used to analyse nucleic acid extracts from stool samples for all the virus types. In all viral assays appropriate controls were always performed using diluted patient faecal samples containing particular viruses as a positive and nuclease-free water as a negative control in ensuring success of preparation of samples, nucleic acid extraction and gene amplification. In adenovirus assay type 40 Dugan strain (ATCC-VR-931) was used as a positive control.
The real-time assays for norovirus genogroups I and II amplified the polymerase/capsid gene junction region (Folley et al., manuscript in preparation; Loisy et al. 2005, respectively). The real-time assay for enterovirus (William Nix, CDC, personal communication) was based on primer and probe sequences derived from the highly conserved 5′-end (sense 5′-CCTGAATGCGGCTAATCC-3′, antisense 5′-TTGTCACCATWAGCAGYCA-3′, probe 5′-6FAM-CCGACTACTTTGGGWGTCCGTGT-3′-BHQ1). Briefly, 5 μl of sample RNA was added to 45 μl of a master mix (SuperScriptTM III Platinum One-Step Quantitative RT-PCR System, Invitrogen) containing 25 μl 2× Reaction Mix, 2 μl of both 10 μM primer, 1 μl of 5 μM TaqMan probe, 1 μl SS III RT/platinum Taq Mix, 1 μl of 50 mM MgSO4, 12.9 μl of PCR grade water and 0.1 μl ROX Reference Dye. The cDNA step consists of one cycle of 50°C for 30 min followed by 95°C for 5 min. PCR consists of 45 cycles of 95°C for 15 s; 58°C for 45 s and 72°C for 10 s.
The primer and probe sequences that recognized the adenovirus (A to F) hexon gene region were according to Jothikumar et al. (2005). In the HAdV real-time Taqman assay, each PCR mix (total final volume of 25 μl) included 12.5 μl of 2× TaqMan Universal Master mix (Applied Biosystems), 0.75 μl of both 10 μM primer, 0.5 μl of 10 μM TaqMan probe, 5.5 μl of PCR-grade water and 5 μl of DNA sample or control. The real-time PCR running programme was 50°C for 2 min and 95°C for 15 min, followed by 45 cycles at 95°C for 10 s, 55°C for 30 s and 72°C for 15 s.
Molecular Typing
Norovirus genotypes were deduced after determination of the nucleic acid sequence of a PCR product from a partial viral RNA polymerase region that was amplified using primers MJV12 and RegA according to Vinje et al. (2004). In typing of adenoviruses the real-time PCR products which were produced without fluorescent probes were purified with ExoI-SAP (Fermentas) enzymatic treatment (Werle et al. 1994) and sequenced bidirectionally with the same primers used in real-time PCR. Sequences were generated by Molecular Medicine Sequencing Laboratory (National Public Health Institute, Biomedicum, Helsinki, Finland) with full service sequencing including sequencing reactions with BigDye Terminator cycle sequencing ready reaction kit v3.1 (Applied Biosystems, Foster City, CA, USA) dilution 1/18 and sequencing run with ABI3730 Automatic DNA Sequencer (Applied Biosystems). Sequence data were examined by using the Vector NTI Advance software program (Invitrogen) or by using Bioedit program (Ibis Biosciences) and the sequences compared to the sequence database in FBVE (RIVM, The Netherlands) or in GenBank by BLAST (Basic Local Alignment Search Tool). Group A rotavirus G and P types were determined based on the method first described by Gouvea et al. (1990) with modifications (Iturriza-Gomara et al. 2004).
Electron Microscopy
The 10% faecal suspensions were examined with electron microscopy for the presence of viral particles after negative staining of the sample with 2% KPT (potassium phosphotungstate), pH 6. The identification of viruses was based on typical morphology.
Results
In Nokia city about 450,000 l of treated sewage water was allowed to run into the drinking water supplies of the city due to a personal error during the 2 days in the end of November 2007 (Fig. 1). Within the following month after the accident around 1,000 people sought care at the municipal health centre and 204 visited the regional hospital in Tampere because of acute gastroenteritis. Numbers of cases of acute gastroenteritis during the epidemic in town by date of a visit at the municipal health care centre are shown in Fig. 1. In most patients the symptoms appeared within 1 week from the contamination. During the outbreak, the most typical symptoms in patients were diarrhoea and vomiting. The presence of a broad panel of enteric viruses in patients stool samples, in a tap water sample taken from the contaminated drinking water distribution network and in a sample of treated sewage suspected as the contamination source was studied by gene amplification (Table 1).
As shown in Table 2 noro-, astro-, rota-, adeno- and enteroviruses were recorded in the drinking water by PCR methods. With the exception of enteroviruses the same viruses were also present in the contaminating wastewater. The genogroup GI norovirus present in the treated sewage could not be detected in the drinking water sample. Hepatitis A virus was found in neither of the water samples. Based on ct-values obtained with real-time RT-PCR, norovirus content in the drinking water could be estimated to be over 50 PCR-units per ml and in the treated wastewater at least 10 times more.
Following the national recommendations for the epidemiological surveillance of acute gastroenteritis at outbreak situations five stool samples were collected for microbial detection on day 4 post-contamination. All of them were positive for norovirus in real time RT-PCR assay and by electron microscopy. Four samples contained both genogroups I and II norovirus, one genogroup II virus only. According to the genetic typing based on viral polymerase sequences the viruses were identified as genotypes GI.3, GI.4, GII.7 and GII.4-2006b.
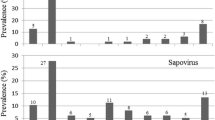
Altogether, 294 stool samples were collected from affected patients during 2–5 weeks from the beginning of the outbreak for bacterial and parasitological diagnostics. These were recovered for further virological analyses. By using gene amplification assays the samples were tested for the presence of several human enteric viruses. The results in Table 3 show that 49.7% of the specimens contain at least one virus while 50.3% of samples remained completely negative. The most common finding was norovirus; genogroups I and II were detected in 8.2% and 22.1% of samples, respectively. Other enteric RNA viruses, astrovirus, rotavirus and enterovirus, were detected in 19.7, 7.5 and 3.7% of the stool samples, respectively. Adenovirus was found in twelve samples (18.2%) out of the 66 tested. Furthermore, mixed virus infections were common with 30 double and 8 triple infections among the 294 patients.
Further genetic typing of both norovirus and adenovirus strains detected revealed genetic variation. Based on viral polymerase sequences norovirus infections were caused by genotypes GI.4, GIIb, GII.4-2006b and GII.7, the latter three of which were found in the treated sewage as well, and type GII.4 in drinking water. In cases of adenovirus positive drinking water, sewage and patient samples preliminary sequencing revealed that at least HAdV40/41 serotype was present.
Discussion
Here we report the virological results of the large waterborne outbreak in southern Finland. In the end of November the drinking water distribution network of the city in Tampere region was contaminated with treated sewage and a rapid onset of acute gastroenteritis took place in about 1,000 of 10,000 exposed people. The contaminating sewage was shown to harbour several different viruses known to cause acute gastroenteritis and they were found in patient stools as well. Accordingly, the drinking water sample was also positive for norovirus, astrovirus, rotavirus, enterovirus and adenovirus. The viral quantities can be estimated to be high, since noro-, astro-, rota- and adenoviruses were detected in drinking water and wastewater also directly without water filtration. The amounts of enterovirus and norovirus genogroup I seemed to be low or vary in concentration since they were identified either in sewage or in drinking water.
Virological analyses indicated a clear linkage between patients suffering from acute gastroenteritis and the consumption of sewage-contaminated drinking water, since all the viruses present in the drinking water were found also in patient samples. Likewise, mixed infections with two or three different enteric viruses and/or strains were common. However, based on the symptoms and virus findings in a particular patient it is difficult to make conclusions, which one of the viruses caused the disease, especially without background data from bacterial findings. Furthermore, 50.3% of the patients did not have any of the analysed viruses in their stools and they are likely to have suffered from non-viral infections at the time of sampling. The outbreak curve resembles that seen in single-source norovirus outbreaks (Hoebe et al. 2004) and is supported by the early patient sample findings.
Treated wastewater contained several norovirus genotypes, GII.4, GII.7 and GIIb that have commonly been infecting people and causing waterborne outbreaks in Finland in the recent years (Maunula and von Bonsdorff 2005). The epidemic season of 2007–2008 was about to start at the time when this drinking water accident took place and the numbers of norovirus positive cases were slightly increased in Finland. The exact prevalence of viruses and distribution of norovirus genotypes in Nokia district before the outbreak is unknown, since the sewage obtained for virus investigations was collected several days after the contamination started. Although water samples clearly contained high amounts of viruses, molecular typing was tedious mainly due to the presence of PCR inhibitors in water. The only norovirus genotype that could be determined in the drinking water was GII.4 while a broader panel of genotypes was found in sewage and affected patients. In previous waterborne viral epidemics in Finland the same single norovirus genotype has often been found in both water and patient samples (Maunula et al. 2005). Here the occurrence of multiple strains might be explained by the heavy contamination of sewage originating from a large community. Kim et al. (2005) have also reported presence of several strains of noroviruses in groundwater as well as in faecal samples from students in outbreaks of gastroenteritis in Korea.
When aetiological roles of different enteric viruses were studied by analysing stool samples of 294 patients suffering from acute gastroenteritis at weeks 2–5 of the outbreak the same panel of enteric viruses (norovirus, astrovirus, rotavirus, enterovirus and adenovirus) were found than previously in the contaminated tap water. Again, the most common finding was norovirus that was found in 89 of 294 patients. In addition to single virus infections noroviruses were involved in 30 of 38 mixed virus infections. The high prevalence of norovirus is not a surprise as such since they are known to possess extremely high transmissibility and often prolonged virus shedding (Lopman et al. 2004; Atmar et al. 2008; Harris et al. 2008). Furthermore, the high number of norovirus infections may also reflect secondary transmissions. This conclusion was also further supported by the finding that the virus genotypes identified in the second week and later during the outbreak were somewhat different than those found in the first five patients. The first stool samples contained genotype GI.3 viruses, while later the most abundant genotypes were GI.4 and GII.4.
As discussed in a recent review by Koopmans (2008) new norovirus genotypes emerge through genetic mutations and recombination. The simultaneous infections with several norovirus strains that are found commonly not only in shellfish-linked outbreaks (Symes et al. 2007) but also in waterborne outbreaks offer ample opportunities for recombination to occur. Outbreaks like this where a large population has simultaneously been exposed to multitude of strains may help to find out the frequency and clarify the mechanisms of emergence of new viruses.
References
Atmar, R. L., Opekun, A. R., Gilger, M. A., Estes, M. K., Crawford, S. E., Neill, F. H., et al. (2008). Norwalk virus shedding after experimental human infection. Emerging Infectious Diseases, 14(10), 1553–1557.
Beller, M., Ellis, A., Lee, S. H., Drebot, M. A., Jenkerson, S. A., Funk, E., et al. (1997). Outbreak of viral gastroenteritis due to a contaminated well. International consequences. JAMA, 278, 563–568.
Blomqvist, S., Skyttä, A., Roivainen, M., & Hovi, T. (1999). Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. Journal of Clinical Microbiology, 37(9), 2813–2816.
Bosch, A., Guix, S., Sano, D., & Pintó, R. M. (2008). New tools for the study and direct surveillance of viral pathogens in water. Current Opinion in Biotechnology, 19, 295–301.
Costafreda, M. I., Bosch, A., & Pintó, R. M. (2006). Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Applied Environmental Microbiology, 72, 3846–3855.
Dahling, D. R., & Safferman, R. S. (1979). Survival of enteric viruses under natural conditions in a subarctic river. Applied Environmental Microbiology, 38, 1103–1110.
Enriquez, C. (2002). Adenoviruses. In G. Bitton (Ed.), Encyclopedia of environmental microbiology (Vol. 1, pp. 92–100). USA: Wiley.
Gilgen, M. D. G., Lüthy, J., & Hübner, P. (1997). Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. International Journal of Food Microbiology, 37, 189–199.
Gouvea, V., Glass, R. I., Woods, P., Taniguchi, K., Clark, H. F., Forrester, B., et al. (1990). Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. Journal of Clinical Microbiology, 28(2), 276–282.
Gutiérrez-Aguirre, I., Steyer, A., Boben, J., Gruden, K., Poljsak-Prijatelj, M., & Ravnikar, M. (2008). Sensitive detection of multiple rotavirus genotypes with a single RT-qPCR assay. Journal of Clinical Microbiology, 46(8), 2547–2554. (Epub 4 Jun 2008).
Harris, J. P., Edmunds, W. J., Pebody, R., Brown, D. W., & Lopman, B. A. (2008). Deaths from norovirus among the elderly, England and Wales. Emerging Infectious Diseases, 14(10), 1546–1552.
Hoebe, C. J., Vennema, H., de Roda Husman, A. M., & van Duynhoven, Y. T. (2004). Norovirus outbreak among primary schoolchildren who had played in a recreational water fountain. Journal of Infectious Diseases, 189(4), 699–705.
Iturriza-Gomara, M., Kang, G., & Gray, J. (2004). Rotavirus genotyping: Keeping up with an evolving population of human rotaviruses. Journal of Clinical Virology, 31, 259–265.
Jiang, X., Wang, J., Graham, D. Y., & Estes, M. (1992). Detection of Norwalk virus in stool by polymerase chain reaction. Journal of Clinical Virology, 30, 2529–2534.
Jothikumar, N., Cromeans, T. L., Hill, V. R., Lu, X., Sobsey, M. D., & Erdman, D. D. (2005). Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Applied Environmental Microbiology, 71(6), 3131–3136.
Kageyama, T., Kojima, S., Shinohara, M., Uchida, K., Fukushi, S., Hoshino, F. B., et al. (2003). Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. Journal of Clinical Microbiology, 41(4), 1548–1557.
Kim, S. H., Cheon, D. S., Kim, J. H., Lee, D. H., Jheong, W. H., Heo, Y. J., et al. (2005). Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. Journal of Clinical Microbiology, 43(9), 4836–4839.
Koopmans, M. (2008). Progress in understanding norovirus epidemiology. Current Opinion in Infectious Diseases, 21, 544–552.
Koopmans, M., von Bonsdorff, C. H., Vinje, J., de Medici, D., & Monroe, S. (2002). Foodborne viruses. FEMS Microbiology Reviews, 26, 187–205.
Le Cann, P., Ranarijaona, S., Monpoeho, S., Le Guyader, F., & Ferre, V. (2004). Quantification of human astroviruses in sewage using real-time RT-PCR. Research in Microbiology, 155, 11–15.
Leclerc, H., Schwartzbrod, L., & Dei-Cas, E. (2002). Microbial agents associated with waterborne disease. Critical Reviews in Microbiology, 28, 371–409.
Loisy, F., Atmar, R. L., Guillon, P., Le Cann, P., Pommepuy, M., & Le Guyader, F. S. (2005). Real-time RT-PCR for norovirus screening in shellfish. Journal of Virological Methods, 123, 1–7.
Lopman, B., Vennema, H., Kohli, E., Pothier, P., Sanchez, A., Negredo, A., et al. (2004). Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet, 363(9410), 671–672.
Maunula, L. (2007). Waterborne norovirus outbreaks—a review. Future Virology, 2(1), 101–112.
Maunula, L., Miettinen, I. T., & von Bonsdorff, C. H. (2005). Norovirus outbreaks from drinking water. Emerging Infectious Disease, 11, 1716–1721.
Maunula, L., & von Bonsdorff, C. H. (2005). Norovirus genotypes causing gastroenteritis outbreaks in Finland 1998–2002. Journal of Clinical Virology, 34, 186–194.
Symes, S. J., Gunesekere, I. C., Marshall, J. A., & Wright, P. J. (2007). Norovirus mixed infection in an oyster-associated outbreak: An opportunity for recombination. Archives of Virology, 152(6), 1075–1086.
Van den Berg, H., Lodder, W., van der Poel, W., Vennema, H., & de Roda Husman, A. M. (2005). Genetic diversity of noroviruses in raw and treated sewage water. Research in Microbiology, 156, 532–540.
Vinje, J., Hamidjaja, R. A., & Sobsey, M. D. (2004). Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. Journal of Virological Methods, 116, 109–117.
Werle, E., Schneider, C., Renner, M., Völker, M., & Fiehn, W. (1994). Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Research, 22(20), 4354–4355.
Wyn-Jones, A., & Sellwood, J. (2001). Enteric viruses in the aquatic environment. Journal of Applied Microbiology, 91, 945–962.
Acknowledgements
The study was supported by a grant from The Ministry of Social Affairs and Health of Finland. The local health authorities are acknowledged for participating actively on solving of the outbreak.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s12560-009-9009-6
Rights and permissions
About this article
Cite this article
Maunula, L., Klemola, P., Kauppinen, A. et al. Enteric Viruses in a Large Waterborne Outbreak of Acute Gastroenteritis in Finland. Food Environ Virol 1, 31–36 (2009). https://doi.org/10.1007/s12560-008-9004-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-008-9004-3