Abstract
Pistachio is a popular snack food. Aflatoxin contamination of pistachio nuts is a serious problem for many producing countries. The development of biological control methods based on ecological parameters is an environmentally friendly approach. Thirty-eight Aspergillus flavus isolates collected from a pistachio orchard in California (CA) were analyzed for production of aflatoxin (AF), cyclopiazonic acid (CPA), vegetative compatibility groups (VCGs), and mating types. All aflatoxigenic isolates produced both AFB1 and CPA. The most toxigenic one was CA28 which produced 164 μg AFB1 per 5 ml PDA fungal culture and small sclerotia (S strain, sclertoium size less than 400 μm). The other aflatoxigenic strains produce AFB1 ranging from 1.2 μg to 80 μg per 5 ml fungal culture. Twenty-one percent of the CA isolates produced AFB1, 84% produced CPA and half formed sclerotia on at least one of three tested media. The 38 CA isolates formed 26 VCGs, 6 of which had two or more isolates and 20 contained single isolates. The S strain isolates belong to 4 different VCGs. Genomic profiling by a retrotransposon DNA probe revealed fingerprint patterns that were highly polymorphic. The predicted VCGs (Pred-VCGs) based on a similarity coefficient >80% matched the VCGs of multiple isolates determined by complementation. All isolates within a VCG had the same mating-type gene of either MAT1-1 or MAT1-2. Uncorrected and VCG-corrected MAT1-1 and MAT1-2 among the isolates were equally distributed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aspergillus flavus is a widely distributed saprophyte and an opportunistic pathogen of animals and plants. Many isolates of this fungus are able to produce hepatocarcinogenic aflatoxins (AFs) and a neurotoxin called cyclopiazonic acid (CPA). AFs often contaminate agricultural crops including corn, cotton, peanuts, and tree nuts, and cause substantial and recurrent economic loss worldwide (Amaikel and Keller 2011; Payne et al. 2006). CPA has also been found to frequently co-contaminate crops with AFs (Horn 2007). Among known mycotoxins, AFs pose the greatest threat to human and animal health. Consequently, more than 100 countries have specific regulations limiting total AFs in foodstuffs, 21 having regulations for AFs in feedstuffs (van Egmond et al. 2007). Major importers of U.S. agricultural commodities have imposed threshold levels for AFB1 below 10 μg/kg for most commodities. These restrictions have had a major impact on exportability of a number of crops including tree nuts, peanuts, corn, and cottonseed. In recent years, AF contamination has become a more critical issue for the pistachio and almond industries because of the high costs associated with rejected shipments to the European Union (EU). In the U.S., the Food and Drug Administration (FDA) has set the maximum total AF limit for tree nuts that are intended for human consumption at 20 μg/kg. The EU, a major importer of California tree nuts and dry fruits, has in the past applied tolerance levels as low as 2 μg/kg for AFB1 and 4 μg/kg total AFs (European Commission 2006). These limits have recently been somewhat relaxed, with current allowable limits of 8 μg/kg for AFB1 and 10 μg/kg total AFs [Commission Regulation (EU) No 165/2010].
A. flavus populations are genetically diverse and consist of assemblages of phylogenetically related aflatoxin- and non-aflatoxin-producing isolates (Peterson 2008). This genetic diversity is not only reflected in AF production but also in morphology and in vegetative compatibility (VC), a process that vegetatively compatible hyphae fuse to form heterokaryons. Parasexual recombination can occur between fused nuclei during segregation of whole chromosomes by mitotic recombination as in meiosis. VC is determined by a series of heterokaryon incompatibility loci whose alleles have to be identical for viable hyphal fusion (Leslie 1993). VC has been used extensively to study genetic diversity of A. flavus populations in various agricultural fields. Variability in morphology, the ability to produce AFs and other secondary metabolites, or the ability to infect and decay plants is commonly found among A. flavus isolates from different VC groups (VCGs), but little variability among isolates from the same VCG (Grubisha and Cotty 2010).
A. flavus was previously thought not to have or had lost the sexual stage. In sexual heterothallic Ascomycetes, mating occurs between strains having complementary mating-type genes, that is MAT1-1 and MAT1-2. MAT1-1 encodes a protein containing a α-box domain, and MAT1-2 encodes a protein containing a high mobility group (HMG) type DNA-binding domain (Turgeon and Yoder 2000). Sexual recombination has been suggested in A. flavus (Ramirez-Prado et al. 2008), but evidence obtained cannot distinguish whether it was an ancient or is a recent activity. The genome sequence data of A. flavus NRRL3357 first indicated that A. flavus is heterothallic and only has the MAT1-1 gene (Payne et al. 2006). A single MAT1-1 or MAT1-2 gene was later reported in other A. flavus and in closely related A. parasiticus isolates (Ramirez-Prado et al. 2008). By crossing strains of opposite mating types, Horn et al. (2009) demonstrated a sexual stage in A. flavus as well as in A. parasiticus under laboratory conditions. Despite these discoveries, A. flavus is believed to reproduce clonally in agricultural and wild settings, and no evidence has demonstrated an active sexual stage of A. flavus in nature. A current biocontrol strategy to reduce AF contamination of crops in the field is to use an indigenous non-aflatoxigenic isolate from a specific VCG to competitively exclude toxigenic A. flavus strains of other VCGs (Cleveland et al. 2003). The potential of sexual reproduction between different VCGs is a concern because of perceived genetic recombination.
In this study, we analyzed 38 A. flavus isolates from a California pistachio orchard to assess their ability to produce AF, CPA and sclerotia. In addition, we grouped the isolates into VCGs by complementation and found that this classification was consistent with hybridization patterns derived from a repetitive DNA probe. Molecular characterization of mating type genes showed that MAT1-1 and MAT1-2 were equally distributed among the isolate.
Materials and methods
Isolation and characterization of A. flavus strains
Pistachio buds, flowers and fruits were collected from Wolfskill Grant Experimental Farm (University of California Davis, Winters, CA, USA). Pistachio samples were placed on salt agar (6% NaCl, 1.5% agar) supplemented with 100 mg/ml of chloramphenicol and incubated at 28°C in the dark for 14 days. Putative A. flavus growing on the plant tissues were transferred onto DRBC (dichloran rose bengal chloramphenicol) agar plates and AFPA (Aspergillus flavus and Aspergillus parasiticus agar) for the identification of A. flavus and related A. parasiticus and A. nomius due to the production of a distinctive orange pigment visible on the underside of colonies (Pitt et al. 1983). Putative isolates of A. flavus were further confirmed by conventional taxonomic systems based on colony morphology, microscopic characteristics of spores, and toxin production. Molecular characterization by comparison of gene sequences (Peterson 2008) such as ß-tubulin was performed. Purified isolates were maintained on potato dextrose agar (PDA). Fungal stock cultures were stored in silica gel maintained at 4°C and are available from the Southern Regional Research Center (SRRC) culture collection.
High performance liquid chromatography (HPLC) analysis of AFB1
AFB1 was extracted from the fungal mat and agar by methanol and analyzed by high performance liquid chromatography (HPLC) on a Agilent model 1260 ChemStation (Agilent, Palo Alto, CA, USA). HPLC was performed on a Supelcosil LC-C18 reversed-phase column (150 mm × 4.6 mm i.d., 5 μm particle size) at a flow rate of 1 ml per min. The mobile phase was methanol/acetonitrile/H2O (20:20:60). Aflatoxins were quantified by a fluorescent detector with excitation at 365 nm and emission at 455 nm.
Thin layer chromatography (TLC) analysis of CPA
Growth medium for CPA production consisted of 2% yeast extract, 15% sucrose and 1% soytone (YESSB). The pH of YESSB was adjusted to 6.0 prior to autoclaving. Approximately 105 spores were inoculated into 1 ml YESSB in a 4-ml glass vial. Cultures were grown for 7 days at 30°C in the dark. At the end of growth, 1 ml of chloroform was added to each vial to extract metabolites. The vial was vortexed for about 30 s, sat for 2 h and vortexed again. The liquid content was then transferred to a clean microfuge tube and spun for 2 min to separate the aqueous and organic phases. Aliquots of 200 μl of the organic layer were transferred to clean microfuge tubes and air-dried. CPA detection was performed using TLC on Si250 silica gel plates (J.T. Baker, Phillipsburg, NJ, USA). Ethyl acetate/methanol/ammonium hydroxide (85:15:10) was the developing solvent system. The plates were sprayed with Ehrlich’s reagent (1 g of 4-dimethylaminobenzaldehyde dissolved in 75 ml ethanol and 25 ml concentrated HCl), and CPA appeared as a blue-purple spot.
Sclerotial formation on different growth media
Malt extract agar (MEA), Czapek yeast agar (CYA) and PDA were individually inoculated with A. flavus and incubated at 28°C in the dark for 1 month. Formation of sclerotia was confirmed visually and by using a stereoscope. The sclerotial isolates were classified according to the sclerotial size; L strain isolates produced very few sclerotia with diameter greater than 400 μm, and S strain isolates produced numerous sclerotia with diameter under 400 μm as previously reported (Cotty 1989).
Generation of nitrate non-utilizing (nit) mutants and complementation tests
Fungal spores harvested from PDA plates were suspended in 0.05% Tween 80 solution and enumerated using a hemocytometer. Approximately 106 A. flavus spores were spread onto each PDA plate supplemented with 40 g/l potassium chlorate (Fisher Scientific, Houston, TX, USA). Chlorate-resistant mutants were putative nit mutants and were purified by transferring onto fresh chlorate-PDA plates for single colony isolation. The mutants were further characterized by the growth scheme described by Cove (1976) on minimal salt media (MM) with nitrate, nitrite, ammonia or hypoxanthine as the sole nitrogen source. Three classes of nit mutants, that is niaD (nitrate non-utilizing, nitrate reductase mutant), nirA (nitrate and nitrite non-utilizing, nitrate reductase mutant), and cnx (hypoxanthine and nitrate non-utilizing permease mutant), were identified. Spores of nit mutants were inoculated on PDA and incubated at 28°C for 2 days. Agar plugs containing mycelia of each pair of nit mutants (nir A vs. niaD, nirA vs. cnx or niaD vs. cnx) were placed 1 cm apart on MM with nitrate as the nitrogen source and incubated at 28°C for 7–14 days. Dense hyphal growth and sporulation at the junction where the expansive mycelia of the paired nit mutants came in contact was indicative of genetic complementation between the paired nit mutants. The observed growth and sporulation was recognized as the interaction of vegetatively compatible isolates, which were assigned to the same VCG.
Extraction of fungal DNA
For large-scale DNA preparation used in Southern hybridization, liquid YPD broth (3 g yeast extract, 10 g peptone, 20 g dextrose/l dist. water) was used to grow submerge A. flavus cultures. Mycelial mats of A. flavus were harvested. Lyophilized mats were ground to a fine powder with mortar and pestle, resuspended in an extraction buffer [200 mmol/l Tris-HCl (pH 8.5), 250 mmol/l NaCl, 25 mmol/l EDTA, 0.5% (wt/vol) sodium dodecyl sulfate], and extracted with buffer-saturated phenol and chloroform. After centrifugation, the aqueous phase was transferred to a new tube, treated with RNase A (20 mg/ml), and extracted once with an equal volume of chloroform. The DNA was precipitated from the aqueous phase with isopropyl alcohol, rinsed once with 70% ethanol and suspended in 100 TE buffer (10 mmol/l Tris-HCl [pH 8.0], 1 mmol/l EDTA). For small-scale DNA preparation used in PCR, approximately 106 spores were inoculated into 1 ml PDB in a 2-ml microfuge tube. The tube was incubated horizontally at 30°C for 18–24 h. Harvested mycelia were processed using a Scientific Industries’ Disruptor Genie™ (Zymo Research, Orange County, CA, USA). Genomic DNA was prepared using a ZR Fungal/Bacterial DNA Kit™ (Zymo Research).
Probe preparation and Southern hybridization
A. flavus genomic DNA was digested with PstI and fragments were separated by agarose gel electrophoresis and transferred to Nytran N membranes (Schleicher and Schuell, Keene, NH, USA) for hybridization. The pAF28, a retrotransposon DNA probe (Hua et al. 2007), was labeled by a kit of the Digoxigenin Nonradioactive Nucleic Acid Labeling and Detection System (Roche Diagnostic, Basel, Switzerland). Probe preparation and hybridization were performed as described previously (McAlpin et al. 2005b). The membranes were exposed to Kodak BioMax MR film (Eastman Kodak, Rochester, NY, USA).
DNA fingerprints and VCG prediction
Hybridization banding patterns generated by restriction fragment length polymorphisms of total genomic DNA probed with pAF28 were recorded. A phenogram based on the presence (=1) or absence (=0) of a band of a known molecular weight at a specific position was constructed using the NTSYS-pc v.2.01 (Rohlf 1997). The phenogram was generated by the unweighted pair-group arithmetic average method (UPGMA). The Dice similarity coefficient (C) value >80% was used to predict A. flavus isolates belonging to the same VCG.
PCR determination of mating types
The mating-type specific oligonucleotides primers were derived from A. flavus MAT1-1 (GenBank accession number: EU357934) and MAT1-2 (EU357936). They were MAT1-1 F: atggaaaccacagtgtctcc, MAT1-1R: tcaacgaatctagagaagtc, MAT1-2 F: atcgagaatgacgactatac and MAT1-2R: ttcttcagtagcagtcagca. PCR was performed in a Perkin Elmer GeneAmp PCR System 2400. Twenty-five pmol of each primer and about 10 ng genomic DNA were added to 25 μl Platinum Blue PCR Supermix (Invitrogen, Carlsbad, CA, USA). The mix was heated at 94°C for 5 min and then subjected to 30 cycles consisting of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 2.0 min. The expected sizes of the PCR products from MAT1-1 F/R and MAT1-2 F/R are 1.2 kb and 1.1 kb, respectively.
Results and discussion
Production of AFB1, CPA and sclerotia by California A. flavus isolates
Based on morphological characteristics and molecular analyses, we indentified 38 isolates of A. flavus from pistachio buds, flowers and fruits. Species identities based on morphological traits were consistent with those derived from the β-tubulin nucleotide sequence information (the GenBank accession numbers for CA isolates are JN394550 to JN394587). Only 21% of the A. flavus isolates produced AFB1. All aflatoxigenic isolates also produced CPA. However, 16% of the isolates did not produce detectable AFB1 and CPA (Table 1). The percentages of A. flavus isolates producing either or both mycotoxins vary greatly (Horn and Dorner 1999; Razzaghi-Abyaneh et al. 2006; Vaamonde et al. 2003). Sclerotial formation by the isolates on the three media, MEA, CYA and PDA, also differed, with PDA the most conducive followed by MEA (data not shown). Approximately 50% of the isolates produced sclerotia on all three media. This percentage of isolates capable of producing sclerotia was much lower than those reported by others. Bayman and Cotty (1991) found that all A. flavus isolates examined from Arizona fields produced sclerotia, and Shearer et al. (1992) reported that up to 92% of the A. flavus/A. parasiticus isolates from an Iowa corn field produced sclerotia. Wicklow et al. (1998) reported that 98% of the A. flavus from a cornfield in Illinois produced sclerotia. Of the 19 sclerotium-producing CA isolates, 4 were S strain and 14 were L strain.
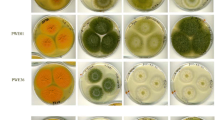
The S strain A. flavus was previously named A. flavus var. parvisclerotigenus by Saito and Tsuruta (1993), and included largely isolates of S strain that produced only AFB1 and AFB2 and also a small number of isolates of S strain that produced AFB1, AFB2, AFG1 and AFG2. The latter is commonly referred to as SBG strain. This atypical S strain has been classified to species status as A. minisclerotigenes (Pildain et al. 2008) based on the ß-tublin gene sequence and the profile of produced secondary metabolites. The S strain A. flavus of CA isolates we identified only produced AFB1 and AFB2 and therefore is not A. minisclerotigenes, which is rarely found in North America (Cotty and Cardwell 1999). Furthermore, the sequences of the AF pathway-specific regulatory aflR gene of A. flavus CA43 (GenBank accession number: AY600510) and CA44 (AY600511) are identical to that of the known S strain A. flavus such as AF70 (AY510453) and highly homologous (>99%) to many other S and L strain A. flavus isolates (data not shown). The most toxigenic one was the S strain CA28 which produced the amount of AFB1 ranging from 2- to 137-fold higher than other isolates (Table 1; Fig. 1). Only 4 L strain out of 14 CA isolates were aflatoxigenic. A few researchers have examined the relative distribution of the L and S strains in host plants, field soils and agricultural commodities. Abbas et al. (2005) reported that about 50% of the A. flavus isolates from corn, soil and peanut in Mississippi Delta produced large sclerotia and only 20% of rice isolates produced large sclerotia; S strain isolates from rice were at a higher percentage than from other sources. Horn and Dorner (1998, 1999) found that 30% of sclerotium-producing isolates of A. flavus from soil in a transect extending from eastern New Mexico through Georgia to eastern Virginia were S strain; S strain isolates were most abundant only in cotton growing regions of Texas and Louisiana. Cotty (1997) also reported that S strain A. flavus isolates were prevalent in all cotton growing areas. AF production by A. flavus differed among areas and was correlated well with S strain incidence. Barros et al. (2007) found that 27% of A. flavus sclerotial isolates collected from 30 fields of peanut growing area were S strain. The higher incidence and greater aflatoxin-producing potential of the S strain isolates (Table 1) could make the S strain A. flavus the most likely contributor to AF contamination of pistachio in California (Doster and Michailides 1994).
Classification of VCGs and VCG diversity
Of the 2,000 nit mutants generated from the 38 A. flavus isolates, 922 mutants showed low reversion frequencies on MM agar plates with nitrate. Based on the growth patterns on MM with nitrate, nitrite, ammonia or hypoxanthine as the sole nitrogen source, three types of nit mutants, niaD, nirA and cnx, were generated Cove 1976). All isolates generated niaD and nirA mutants, but only 50% of the isolates generated one or more cnx mutants. All self-paired nirD and nirA mutants were able to grow on MM nitrate medium which indicate the nit mutants are vegetatively self-compatible. The distribution of mutants was similar to that reported for the A. flavus isolates from peanut fields in Georgia (Horn and Greene 1995) and from cotton fields in Arizona (Bayman and Cotty 1991). The 38 A. flavus isolates formed 26 VCGs designated A to Z (Table 1; Fig. 2).
Phenogram based on cluster analysis of the fingerprints of 38 A. flavus isolates and 1 A. nomius isolate, CA45. Isolates CA47 and CA48 were collected from an almond orchid. The phenogram was generated using the NTSYS-pc v.2.01. Isolates belonging to the same vegetative compatibility group had the same DNA fingerprints
Complementation of these nit mutants revealed 65% VCG diversity, which is expressed as the ratio of VCGs to isolates examined, for the California isolates. Similarly, Horn and Greene (1995) reported 56% VCG diversity for the population from peanuts, and Papa (1986) found 69% VCG diversity for A. flavus isolates from corn kernels. However, Bayman and Cotty (1991) reported VCG diversity fluctuated from 15 to 54% in soil and cotton bolls in a single Arizona cotton field over a 3-year period. The high VCG diversity of A. flavus may be correlated with the widespread dispersal of spores from different sources into small geographic areas (Horn and Greene 1995), or with an underlying diverse A. flavus populations in the region, or due to a high mutation rate in VCG separating genes (het loci). In general, sclerotial formation, sclerotial size, and AF and CPA production are similar among isolates belonging to the same VCG. For example, the two isolates of VCG K, CA14 and CA19, are identical in these aspects. The respective multiple isolates collected that are in VCGs A, B, E, and M also exhibit identical or highly similar characteristics despite that one isolate each in VCGs E and M did not produce CPA (Table 1).
Comparison of predicted DNA fingerprint groups to VCGs determined by complementation
Fingerprints of A. flavus isolates by Southern hybridization using the pAF28 DNA as probe were highly polymorphic (Fig. 3). The phenogram (similarity index, C > 80%) established 24 predicted fingerprint groups (Fig. 2). The fingerprinting patterns were able to predict VCGs determined by complementation containing multiple isolates but not all of those containing two isolates (Table 2). Barros et al. (2007) used amplified fragment length polymorphisms (AFLP) to examine A. flavus isolates of 31 VCGs collected from a peanut field in Argentina and did not find any single AFLP pattern that is associated with a VCG containing two or more isolates. In contrast, Ehrlich et al. (2007) examined single nucleotide polymorphisms of three gene regions to infer relationships among six VCGs of A. flavus isolates collected from five regions across the southern United States and found two haplotypes for each of the genes. Although all isolates within a VCG had similar haplotypes, the six VCGs only formed four genetically distinct groups. Contrary to discriminatory power of the two methods described above, McAlpin et al. (2005b) showed that the pAF28 probe correctly assigned 75 A. flavus isolates from a peanut field in Georgia to 44 known VCGs based on a cutoff similarity coefficient of 80%; multiple strains belonging to the same VCG produced identical DNA fingerprints with rare exception. The pAF28 probe also distinguished A. flavus strains belonging to 22 VCGs characterized by Papa (1986), and VCGs containing two or more strains gave identical fingerprints from genomic DNA digested with PstI (McAlpin and Mannarelli 1995). The highly discriminatory power of pAF28 was consistently demonstrated in the analysis of 43 Aspergillus caelatus isolates of 13 VCGs collected from a Georgia peanut field (McAlpin et al., 2005). In this study, the pAF28 probe satisfactorily assigned multiple isolates belonging to the same VCGs classified by complementation. The phenogram and fingerprint patterns showed that CA10 and CA20, CA26 and CA28 formed two fingerprint groups (Fig. 2; Table 2). However, the four isolates formed four single-strain VCGs (Table 1 and 2). McAlpin et al. (2005a) reported that A. caelatus NRRL 25576 and NRRL 25577 isolated from Japan belonged to the same fingerprint group of the VCG 1 isolates from USA (similarity coefficient >80%). These two isolates were incompatible with all of the VCG 1 strains and were classified into VCG 12. Vegetative compatibility is controlled by multiple vegetative incompatibility (het) loci and heterokaryons can form only between strains with identical alleles (Leslie 1993). Variations might have occurred in the het loci of the A. caelatus during the geographic separation. These California isolates probably have mutations in some het loci making them incompatible to strains with virtually identical fingerprints.
Distribution of mating-type genes
Horn et al. (2009) showed that sexual mating could occur between A. flavus isolates belonging to different VCGs. These findings suggest that mating type genes are not involved in incompatibility among A. flavus VCGs. The VCGs of A, B, E, and M contained multiple CA isolates, and the respective VCGs were strictly either MAT1-1 or MAT1-2 (Table 1). The MAT1-1 and MAT1-2 genes, uncorrected or VCG-corrected, distributed equally among the California isolates (Table 3). Ramirez-Prado et al (2008) reported a high frequency of MAT1-2 (80%) and an unequal VCG-corrected distribution of MAT1-1 (35%) for A. flavus strains collected from a single field in Georgia that belong to 43 VCGs. A higher frequency of MAT1-1 than MAT1-2 was also reported in A. flavus strains collected over 4 years in Arizona and Texas belonging to three common VCGs (Grubisha and Cotty 2010). A better understanding of the VCG diversity, genotypes and mycotoxin production of A. flavus isolates in the field may have implications in selecting indigenous non-aflatoxigenic A. flavus isolates for a biological control strategy to eliminate aflatoxin in food crops.
References
Abbas HK, Weaver MA, Zablotowicz RM, Horn BW, Shier WT (2005) Relationships between aflatoxin production and sclerotia formation among isolates of Aspergillus section Flavi from the Mississippi Delta. Eur J Plant Pathol 112:283–287
Amaikel S, Keller NP (2011) Aspergillus flavus. Annu Rev Phytopathol 49:107–133
Barros GG, Chiotta ML, Reynoso MM, Torres AM, Chulze SN (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909
Bayman P, Cotty PJ (1991) Vegetative compatibility and genetic diversity in the Aspergillus flavus population of a single field. Can J Bot 69:1707–1711
Cleveland TE, Dowd PF, Desjardins AE, Deepak B, Cotty PJ (2003) United States Department of Agriculture-Agricultural Research Service research on pre-harvest prevention of mycotoxins and mycotoxigenic fungi in US crops. Pest Manag Sci 59:629–642
Cotty PJ (1989) Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79:808
Cotty PJ (1997) Aflatoxin-producing potential of communities of Aspergillus section Flavi from cotton producing areas in the United States. Mycol Res 101:698–704
Cotty PJ, Cardwell KF (1999) Divergence of West African and North American communities of Aspergillus section Flavi. Appl Environ Microbiol 65:2264–2266
Cove DJ (1976) Cholorate toxicity in Aspergillus nidulans: the selection and characterisation of chlorate resistant mutants. Heredity 36:191–203
Doster MA, Michailides TJ (1994) Aspergillus molds and aflatoxins in pistachio nuts in California. Phytopathology 84:583–590
Ehrlich KC, Montalbano BG, Cotty PJ (2007) Analysis of single nucleotide polymorphisms in three genes shows evidence for genetic isolation of certain Aspergillus flavus vegetative compatibility groups. FEMS Microbiol Lett 268:231–236
European Commission 2006. The Rapid Alert System for Food and Feed (RASFF) Annual Report 2005. http://ec.europa.eu/food/food/rapidalert/report2005_en.pdf.
Grubisha LC, Cotty PJ (2010) Genetic isolation among sympatric vegetative compatibility groups of the aflatoxin–producing fungus Aspergillus flavus. Mol Ecol 19:269–280
Horn BW (2007) Biodiversity of Aspergillus section Flavi in the United States: a review. Food Addit Contam 24:1088–1101
Horn BW, Dorner JW (1998) Soil populations of Aspergillus species from section Flavi along a transect through peanut-growing regions of the United States. Mycologia 90:767–776
Horn BW, Dorner JW (1999) Regional differences in production of Aflatoxin B1 and cyclopiazonic acid by soil isolates of Aspergillus flavus along a transect within the United States. Appl Environ Microbiol 65:1444–1449
Horn BW, Greene RL (1995) Vegetative compatibility within populations of Aspergillus flavus, A. parasiticus, and A. tamarii from a peanut field. Mycologia 87:324–332
Horn BW, Moore GG, Carbone I (2009) Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429
Hua S-ST, Tarun AS, Pandey SN, Chang L, Chang P-K (2007) Characterization of AFLAV, a Tf1/Sushi retrotransposon from Aspergillus flavus. Mycopathologia 163:97–104
Leslie JF (1993) Fungal vegetative compatibility. Annu Rev Phytopathol 31:127–150
McAlpin C, Mannarelli B (1995) Construction and characterization of a DNA probe for distinguishing strains of Aspergillus flavus. Appl Environ Microbiol 61:1068–1072
McAlpin CE, Horn BW, Wicklow DT (2005a) DNA fingerprinting analysis of vegetative compatibility groups in Aspergillus caelatus. Mycologia 97:70–76
McAlpin CE, Wicklow DT, Horn BW (2005b) DNA fingerprinting analysis of vegetative compatibility groups in in Aspergillus flavus from a peanut field in Georgia. Plant Dis 86:254–258
Papa KE (1986) Heterokaryon incompatibility in Aspergillus flavus. Mycologia 78:98–101
Payne GA et al (2006) Whole genome comparison of Aspergillus flavus and A. oryzae. Med Mycol 44:9–11
Peterson SW (2008) Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226
Pildain MB, Frisvad JC, Vaamonde G, Cabral D, Varga J, Samson RA (2008) Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int J Syst Evol Microbiol 58:725–735
Pitt JI, Hocking AD, Glenn DR (1983) An improved medium for the detection of Aspergillus flavus and A. parasiticus. J Appl Bacteriol 54:109–114
Ramirez-Prado JH, Moore GG, Horn BW, Carbone I (2008) Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet Biol 45:1292–1299
Razzaghi-Abyaneh M, Shams-Ghahfarokhi M, Allameh A, Kazeroon-Shiri A, Ranjbar-Bahadori S, Mirzahoseini H, Rezaee M-B (2006) A survey on distribution of Aspergillus section Flavi in corn field soils in Iran: population patterns based on aflatoxins, cyclopiazonic acid and sclerotia production. Mycopathologia 161:183–192
Rohlf FJ (1997) NTSYS-pc numerical taxonomy and multivariate analysis system. Exeter Software, Setauket, NY
Saito M, Tsuruta O (1993) A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. Proc Jap Assoc Mycotoxicol 37:31–36
Shearer JF, Sweets LE, Baker NK, Tiffany LH (1992) A study of Aspergillus flavus/Aspergillus parasiticus in Iowa crop fields: 1988–1990. Plant Dis 76:19–22
Turgeon BG, Yoder OC (2000) Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol 31:1–5
Vaamonde G, Patriarca A, Fernández Pinto V, Comerio R, Degrossi C (2003) Variability of aflatoxin and cyclopiazonic acid production by Aspergillus section flavi from different substrates in Argentina. Int J Food Microbiol 88:79–84
van Egmond HP, Schothorst RC, Jonker MA (2007) Regulations relating to mycotoxins in food perspective in a global and European context. Anal Bioanal Chem 389:147–157
Wicklow DT, McAlpin CE, Platis CE (1998) Characterization of the Aspergillus flavus population within an Illinois maize field. Mycol Res 102:263–268
Acknowledgments
This work is supported by in-house research program funds from U.S. Department of Agriculture, Agricultural Research Services. We thank K. Ehrlich, T. McKeon, X. He for comments; C.E. Platis, S. Hong, L.T. Fang, S. Kwong, E. Scharfenstein, Bertram Lee and L. Scharfenstein for technical assistance.
Conflict of interest
The authors declare that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hua, S.S.T., McAlpin, C.E., Chang, PK. et al. Characterization of aflatoxigenic and non-aflatoxigenic Aspergillus flavus isolates from pistachio. Mycotoxin Res 28, 67–75 (2012). https://doi.org/10.1007/s12550-011-0117-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12550-011-0117-4