Abstract
Protein–protein interaction plays an important role in the understanding of biological processes in the body. A network of dynamic protein complexes within a cell that regulates most biological processes is known as a protein–protein interaction network (PPIN). Complex prediction from PPINs is a challenging task. Most of the previous computation approaches mine cliques, stars, linear and hybrid structures as complexes from PPINs by considering topological features and fewer of them focus on important biological information contained within protein amino acid sequence. In this study, we have computed a wide variety of topological features and integrate them with biological features computed from protein amino acid sequence such as bag of words, physicochemical and spectral domain features. We propose a new Sequential Forward Feature Selection (SFFS) algorithm, i.e., random forest-based Boruta feature selection for selecting the best features from computed large feature set. Decision tree, linear discriminant analysis and gradient boosting classifiers are used as learners. We have conducted experiments by considering two reference protein complex datasets of yeast, i.e., CYC2008 and MIPS. Human and mouse complex information is taken from CORUM 3.0 dataset. Protein interaction information is extracted from the database of interacting proteins (DIP). Our proposed SFFS, i.e., random forest-based Brouta feature selection in combination with decision trees, linear discriminant analysis and Gradient Boosting Classifiers outperforms other state of art algorithms by achieving precision, recall and F-measure rates, i.e. 94.58%, 94.92% and 94.45% for MIPS, 96.31%, 93.55% and 96.02% for CYC2008, 98.84%, 98.00%, 98.87 % for CORUM humans and 96.60%, 96.70%, 96.32% for CORUM mouse dataset complexes, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Protein–protein interaction (PPI) plays an important role in the understanding of biological functions like cellular behave in cells, illness, and health. This is still challenging to find interaction among proteins and their complexes. Identifying protein complexes (PCs) from these PPINs is an important task in biomedical natural language processing (BioNLP) field [1]. By the interaction of the protein with other proteins, most of the proteins achieve their functions. So, it is important to discover the interaction between proteins to understand the biological functions in the body. Protein complex consists of multiple proteins that (stably) interact with each other. To form PCs and to carry out their biological function, proteins interact with each other. PCs intercommunicate with each other to form protein protein interaction network (PPIN) and are significant objects to recognize the attitude of cellular functions and organization in PPIN. Therefore, correctly classifying the interaction network among proteins in an organism is a significant task as it is suitable for interpreting the molecular mechanism underlying given biological functions [2].
Although protein interactions are extraordinarily diverse, all protein interfaces share confident mutual properties. Based on these common properties, different experimental and computational techniques were developed and used to detect protein complexes in PPINs. Experimental techniques use different biochemical [3], genetic, and physical methods such as DNA [4], yeast two-hybrid (Y2H) [5], tandem affinity purification (TAP) [6], mass spectroscopy (MS) [7], protein microarrays [8] and phage display [9] for analyzing protein interactions. Other approaches focus on monitoring and illustrating specific physicochemical and biochemical properties of a PC. With the development of these throughout experimental methods, a large amount of PPI datasets has been produced. However, among the variety of experimental methods, only a small part of protein complexes has been detected due to the limitation of these methods and it is still not sufficient to provide an effective and easy method for detecting proteins in the same complex [10]. In addition, datasets from significant throughout methods frequently suffer from false positives and false negatives. Therefore, detecting PCs from PPIN data using computational approaches provides an alternative way.
There are several proteins interaction and complex databases that are curated on the consensus knowledge of experts and primary experiments such as The Biological General Repository for Interaction Datasets (BioGrid) [11], Database of interacting proteins (DIP) [12], The comprehensive resource of mammalian protein complexes (Corum) [13], Mammalian Protein–Protein Interaction Database (MIPS) [14], catalogs of yeast protein complexes (cyc2008) [15], The Molecular Interaction Database (MINT) [16], KEGG [17], STRING [18], the universal protein knowledgebase (UniProt) [19] and Reactome [20]. These databases contain high-throughput data and information about protein interactions, protein complexes of different species and amino acid sequences of different proteins.
1.1 Density-Based Unsupervised Methods
Proteins are very much interactive with each other on the same PC. In the PPI network, PCs generally correspond to fully connected subgraphs. Based on this intuition, various computational algorithms have been devised. An overview of these methods is given below.
MCODE [21] produces both small- and large-size complexes. It initially selects a vertex of high local weight and then repeating adds neighbors vertices of alike weight to grow clusters. CFinder [22] forms clusters of varying sizes. It identifies fully densed subgraphs of different least merges subgraphs and clique sizes based on their ratio of shared members. That is why each node is a member of a complete order of groups of different sizes. CFinder gives results that differ significantly with changing minimum clique size. MCL [23] uses both weighted and unweighted graphs. It detects protein complexes in these graphs by pretending random walks. It expands and inflates iteratively. DPclus [24] is a group edge tracing algorithm to detect PCs by keeping tracking of the edge of an identified group.
Limiin et al. [25] reduce the number of parameters of the algorithm in DPClus uses two topological constraints: core and attachment to detect protein complexes in PPIN. Restricted Neighbors Searching Clustering (RNSC) detects PC built on two properties, i.e., graph-theoretical and gene-ontological. Leung et al. propose a statistical framework to find out PC cores using the CORE algorithm, in which CORE ranks the forecast complexes by assigning them scores. Wu et al. presented a method to core attachment which detects PCs by identifying the central complex and part complex [2]. Similarly, Dong et al. [26] proposed a search method based on a local-structure score function that detects PCs backward and forwards.
1.2 Feature Extraction and Selection-Based Supervised Learning Methods
Methods discussed in the previous section are based on the intuition that PCs form dense subgraphs in PPINS. Despite the fact, other topological constraints also have significant importance in predicting PCs from PPINs. As proteins not only form cliques in PPINs, other shapes of PCs, i.e., linear, hybrid and stars are also found in PPINs. Various supervised learning methods have been devised that identify PCs of different structures from PPINs using a number of topological features like degree, topological coefficient, clustering coefficient, etc. Subgraph topology is important for complex prediction, but the information contained within the protein’s amino acid sequence is also very important as these sequences specify the structure of proteins. Fewer computational approaches integrate biological properties computed from protein sequences with topological properties for complex prediction tasks from PPINs. Detailed overview of these methods is given below.
Qii et al. [27] presented a supervised graph local clustering framework. This framework predicts PCs based on topological properties including cluster size, cluster density, cluster degree, topological coefficient, cluster coefficient and biological property such as frequency of amino acids of identified complexes. Yu et al. [28] presented CDIP to discover protein complexes based on integrated properties such as graph size, graph density, degree statics, clustering, and topological coefficient statics and amino acid frequency. Amino acid frequency is measured for biological properties, and the diameter and density of the network are measured for topological constraints. Quan et al. [29] proposed that clusterEps used for the compositional score of rising pattern for measuring the likelihood of sub-graph as complex and just topological features are not enough for efficient detection of PCs . Their method uses an integrative score to measure the likelihood of a subgraph to form the protein complex. So, new protein complexes can be built from seed protein by updating the score iteratively. They used the MIPS dataset and they reported the precision, recall and F1-measure of 0.638, 0.769 and 0.696, respectively.
Jiancang Zeng et al. [30] proposed a three-step framework, in the first step from off the shell database, negative data set is collected. While in the second step for preprocessing of PPI sequence, the n-gram frequency method is used. The third step is for the connection of final features and then to find the optimal features from the computed one. Lastly, for the Random Forest classifier features are selected. The experiment is done on real data set that shows the fusion method outperformed and gives the inspiring results that are helpful for future research in protein function.
Many studies have worked for extracting the important features and reduce the size of the feature vector to improve the performance of models [31,32,33]. For extracting the important features and reducing the dimension of dataset, Quan Zou et al. [34] proposed the feature ranking method known as Max-Relevance-Max-Distance (MRMD). MRMD is used to balance the stability and accuracy of prediction task and feature ranking. This method is tested on two different datasets to prove the efficiency of big data. The first one is the benchmark dataset with high dimensionality known as image classification. The second dataset is protein protein interaction prediction data that comes from their private previous research. The proposed method maintains accuracy on both datasets proved by the experiment. Unlike other filtering and wrapping methods like information gain and mRMR, this proposed method Max-Relevance-Max-Distance (MRMD) run faster.
Shao Wu Zhang et al. [35] predicted PPI with a new technique based on amino acid distance frequency that is gathered with principal component analysis (PCA) [36] and physicochemical characteristics. First, based on four types of physicochemical property values, the twenty fundamental amino acids split into three groups. Second, the feature extraction process for distance frequency is presented for the representation of pairs of protein and also feature vectors are obtained with 4 physicochemical characteristics that are combined to create distinct set vector features. Third, the technique PCA has been used to decrease the dimension vector and adopted SVM Support Vector Machine as a classifier. The complete predicted accuracy of 4 physicochemical characteristics is 91.79%. The findings indicate that the present strategy is very capable of PPI’s prediction and can be a helpful tool in the appropriate fields.
In 2018, Aisha Sikandar et al. [37] used the topological features including Closeness Centrality, Average shortest path Length, Betweenness Centrality, Clustering Coefficient, Degree, Neighborhood Connectivity, Eccentricity, Radiality, Stress Centrality, Self-Loops Topological Coefficient, Density and Size, and 50 spectral domain biological features. They used the different versions of decision tree such as cart Cart, C4.5 and breadth first search and Cart outperforms the other models. Similarly, Misba et al. [61] worked on gene–disease association prediction using biological and topological features. They used the CORUM complexes that are used in this study for human and mouse complexes.
In 2019, Aisha Sikandar et al. [38] predicted PPI with subgraph topology and sequence entropy. Length and sequence entropy is calculated to catch the core of data comprised within the sequence of proteins. Interaction of proteins was done with one another and diverse sub-graph topologies were formed. Using a logistic tree model, the incorporation of biological characteristics was done with topological sub-graph characteristics and complexes are model. The experiment findings showed that this technique outperforms the other 4 state-of-the-art methods in terms of the number of protein complexes known to be detected. This framework also offers a perspective in future biological research and may be useful in anticipating other kinds of subgraph topologies. Similarly, in 2020, Faridoon et al. [39] proposed a supervised method to predicted protein complexes using biological and topological features. They integrated the SVM with Error-correcting output coding (ECOC) to optimize the performance based on the autodetection of multiple protein complexes idea. The features used by the comparative studies are shown in Table 1.
Most of the above-mentioned unsupervised learning methods are based on intuition that proteins combine to form dense subgraphs in PPINs, whereas supervised learning methods not only mine cliques but linear, star and hybrid structures as PCs in PPINs by computing a wide variety of topological features like degree, clustering coefficients, topological coefficients, diameter, etc. Fewer methods take into account important biological information contained within protein amino acid sequence in the form of frequency, count vectorizer, fast Fourier transform and kidera factors as biological features. These methods by incorporating biological along with topological features achieve better performance as compared to previous studies. Therefore, in this paper, we have computed a wide variety of topological features given in Table 2 and integrate them with biological features. Among the biological features, we have computed spectral domain features, physicochemical properties and bag of words features. Another limitation of recent studies is that they have not used feature selection. In recent studies, the combination of different features leads to high-dimensional data, and high-dimensional data can lead to the curse of dimensionality. We have used a sequential forward feature selection method (SFFS) i.e., random forest based Boruta feature selection [40]. In this method, the best features are fused before giving to the model for training. As not all extracted features are important, we selected the important features that lead to more accurate results.
In this paper, we used gradient boosting classifier, decision tree classifier, linear discriminant analysis to train on human, mouse and yeast complexes databases. Our methodology shows a significant improvement in protein complex prediction. Our experiments show that Linear discriminant analysis performs better on both large and small databases with the best features selected by SFFS from a list of computed biological and, topological features.
2 Methods
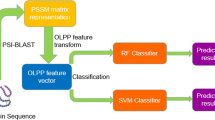
The protein–protein interaction network can be represented as graph G = E, V, where E represents the edges (interaction) between proteins, V represents the vertices (proteins). The subgraphs in PPI can be represented as Gs = Es, Vs and these subgraphs are connected to form a complete graph. To predict protein complexes, we downloaded the hand-curated yeast complexes dataset from CYC2008 [15] and MIPS [14]. Human and Mouse protein complexes dataset is taken from MIPS CORUM [13] and amino acid sequences against these genes are taken from uniport kb [19]. We downloaded the protein interaction data from DIP and created the four PPINs from which two for the yeast databases, one for humans and one for the mouse database. Then, the feature selection method is used. By considering interaction networks, we have computed 13 topological features. From proteins amino acid sequences, a total of 150 features are computed. All computed features are listed in Table 2. The extracted 163 features are passed to SFFS, i.e., random forest-based Brouta feature selection algorithm and top ranked features including Amino acid G, Physicochemical feature 6, 26, 41, 21 and 61, Density, Size, Topological Coefficient, Neighborhood connectivity, Average Shortest Path length, Closeness Centrality, Eccentricity, Radiality were selected to train the model. Figure 1 represents the flow diagram of our methodology and algorithm 1 proposed technique, respectively.

2.1 Feature Extraction
For complex detection, we have used the topological, biological features such as the spectral domain, Bag of Words and physicochemical features. For computing topological features, we have created four PPINs two for yeast by considering CYC2008 and MIPS benchmark complexes and DIP interaction data and one for humans and 1 for mouse by considering complexes from CORUM and interaction data from DIP. Complexes of size greater than 3 are considered. To compute biological features amino acid sequences of proteins in PPINs are considered. We have calculated a total of 13 topological feature and 150 biological features. The input parameter, name, and category of these features are given in Table 2.Density-based unsupervised learning methods are based on the intuition of mining dense subgraphs from PPINs as complexes and used density as the main feature. These methods are unable to mine other types of complex structures found in PPINs, i.e., star, linear, and hybrid. Therefore, feature extraction and selection-based algorithms have been devised that incorporate a variety of topological features for mining not only cliques but star, linear and hybrid complexes as well from PPINs. The detail of these methods is given in Sect. 2.2.
2.1.1 Topological Feature
Protein–protein interaction networks can be represented as graphs in which proteins are represented as nodes denoted by V and the connection between them as edges denoted by E [41]. We have computed several topological features by considering PPIN of Yeast, mouse and human detail of these features is given below:
Average shortest path Length: The average shortest path length between the vertices V and E is calculated by taking the average of shorter path length
Closeness Centrality: It is a way to identify nodes, measures the average distance of a node to all nodes, and can very easily disperse information across a network. Nodes with a high score of closeness have the shortest distances to every other node. This function is useful in mining stars and cliques. The closeness centrality is defined in the following equation.
where \(C_c(V)\) is the closeness centrality of vertex V. L(V, E) indicates the length of the shortest path between vertexes V and E. The value of closeness centrality lies between 0 and 1.
Betweenness centrality: The betweenness centrality for each node is the number of these shortest paths that traverse the node. It shows the degree to which nodes stand between each other in a PPIN, central nodes of the clique, star, and hybrid complexes have a higher betweenness centrality so this characteristic is useful in complex mining. The betweenness centrality is defined in the following equation.
where \(C_c(V)\) is betweenness centrality of vertex V, g and d are the vertices in different clusters from that of V. From vertex g to vertex d, \(\sigma\)dg represent number of shortest paths, and V represents the vertices that lies on these shortest paths, from g to d is represented by the \(\sigma\)gd (V).
Clustering coefficient: The clustering coefficient is defined in the following equation.
where \(C_d\) is the clustering coefficient of vertex d, \(k_d\) is the number of neighbors of vertex d and ed is the number of connected pairs among all the neighbors of vertex d.
Degree: The total number of vertices connected by an edge to vertex V is referred to as degree V.
Eccentricity: Vertex V’s eccentricity is the longest non infinite length of the shortest path in the network between vertex V and edge E.
Neighborhood connectivity: The number of a vertex’s neighbors is called its connectivity. The average connectivity of every neighbor of vertex V is called its neighborhood connectivity.
Radiality: Radiality is called the centrality index of a vertex V. It lies between 0 and 1. For computing radiality, the average shortest path length of a vertex V is subtracted from the connected component’s diameter plus 1 and then divides the result by the diameter of the connected component. Diameter is the number of edges in the shortest path between the furthest pair of vertices of a cluster.
SelfLoops: It is an edge that connects a vertex to itself.
Stress centrality: It is the number of shortest paths that include vertex V. A vertex got high stress value if it is traversed by the high number of shortest paths.
Topological coefficient : The topological coefficient is defined in the following equation.
where \(T_V\) is the topological coefficient of a vertex V and \(K_v\) is the neighbors of vertex V. Shared neighbors among vertex V and E are given by J (V, E). If there exists a single link among vertex m and vertex n then the value of J (V, E) is incremented by 1.
Size: Size can be defined as the number of vertices in a cluster.
Density: The density of a cluster G is defined in the following equation.
where E represents edges and V represents vertices in cluster G.
2.1.2 Biological Features
Protein structure is defined by the composition of amino acid sequences, so investigation of the amino acid sequences is enough to understand the interacting properties of proteins for performing any kind of biological function [23]. Therefore, it is important to consider these amino acid sequences to classify the protein complexes these features are very helpful to classify complexes. The following key features can be used to predict the complexes based on the information within the protein amino acid sequences.
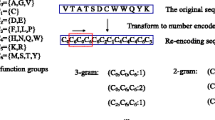
Spectral domain feature To capture the nature of data Fast Fourier Transformation (FFT) is a very useful method [42]. Previously, FFT has been used in the frequency based domain to know the patterns and in bioinformatics to classify proteins but not to classify the complexes in protein–protein interaction networks. As amino acid sequence information is enough to understand the structure of proteins, using FFT on these sequences we can extract features that can be useful in the prediction of protein complexes [36].
To get the set of spectral domain feature, FFT can be applied to protein amino acid sequences. To apply the FFT on amino acid sequence, amino acid sequences will be converted in their molecular weights. FFT is calculated for every amino acid sequence using the following equation.
To compute the FFT on the amino acid sequences the length of sequences should be the same but here the length of the amino acid sequence is different therefore to make the length same the principal component analysis [43] is used to reduce the dimension of amino acid sequences. Lowest 50 coefficients of FFT of the generated spectrum by PCA.
2.1.3 Bag of Words Features
As amino acid sequences are a collection of 20 different characters, therefore, bag of word can be applied to these features. In NLP, bag of the word is a powerful technique to convert the text in features and has many applications like sentiment analysis, document classification tasks and analogical reasoning [44]. We use the count vectorizer [45, 46] at character level to convert our amino acid sequences in the feature. Count vectorizer converts the text to features by using the characters as labels and assign the counts of character in sequences to its relevant labels. Every amino acid sequence against a protein has different lengths and character counts that can distinguish proteins and used to predict protein complexes.
2.1.4 Physicochemical Feature
Analogous properties of amino acid sequences determine the physicochemical properties of proteins. These physicochemical properties can be used to predict complexes more accurately [47]. There are more than 554 physicochemical properties [48,49,50,51] but feature extraction using all these measures leads to the curse of dimensionality and higher computational cost. Fortunately, Gaurav et al. [52] summarized the rank of physicochemical properties based on the frequency counts on the whole dataset and proved that not all properties are important to predict protein protein interaction complexes. These four properties that play an important role in PPI complex detection are polarity (P), hydrophobicity (H), polarizability (Z) and Waals volume (V). We selected these four properties to extract physicochemical features from amino acid sequences. To integrate the physicochemical properties, we used PSI-blast to create PSSM matrix that defines the probability of any amino acid occurring at a specific position in a sequence and multiply these properties with that matrix by equation 7.
where S is (S = S\(^j _i\) i = 1,2...M, j = 1,2,...,20) S is PSSM query of protein sequence is a M X 20. Where M is the length of the protein sequence and 20 denotes the 20 amino sequences. Rn is the nth physicochemical property and F \(_n\)(n = 1,2,3,4) of the physicochemical property.
2.2 Feature Selection
After calculating the topological and biological features we have applied FSSF, i.e., Randomforest-based Boruta feature selection for mining best features from computed one. Boruta algorithm was designed on the idea of stoppiglia [53] that expands the information system with shadows that are artificial characteristics generated by changing the order of values in the original data and then using shadows’ significance scores to determine the meaning of the scores obtained by the real characteristics.
Boruta checks what features in an iteration have attained greater importance than the best shadow; these events are counted for each feature until their number is either substantially higher or lower than predicted at random, using a 0.01 p value cut-off by definition. If the p value for the feature is greater than the threshold, then the feature is selected else rejected. Then, the rejected features are removed from the information system. After every iteration, it shuffles the shadow features and this process is repeated until all relevant features are selected or set limit of reputation is reached. In this feature selection method, k best features are selected in the set of n features on which classification model gives the best accuracy. Let us say A is a set of all features.
\(A = {a_1,a_2,\ldots a_n}\).
Boruta takes A as the input feature set and gives optimal features set B from A. The set B is a subset of A containing the selected features by Boruta based on Random Forest.
\(B={bj|j=1,2,3\ldots ,k; b_j \subset A}.\)
2.3 Classification
After applying the feature selection, these datasets are split into training and testing datasets by 70:30 ratio, respectively. After preparing the datasets, we have train decision tree, gradient boosting, and linear discriminant analysis classifiers and during test phase, complex prediction task is achieved.We have used grid search to find the optimal parameters for the models. For the decision tree classifier, we set the n_estimator from 50 to 200, min_sample_split and min_sample_leafs from 1 to 10. The optimal parameters for decision tree classifiers are n_estimator=100, criterion= “gini”, max_depth=None, min_sample_split=2 and min_sample_leaf=1. For the GradientBoostingClassifiers optimal hyperparameters are loss=’deviance’, learning_rate=0.01, subsample=1.0,
n_estimators=100, min_samples_split=2, min_samples_leaf=1, max_depth=5. For the linear disrciminenta analysis parameters are tol=0.0001, shrinkage=None, solver=’svd’, n_components=None, priors=None, store_covariance=False. These classifiers are explained in the following section.
2.3.1 Decision tree classifier
Decision tree classifier breaks down the complexes database into smaller groups and constructs the tree at the same time for these smaller groups. The resulted tree is in the form of leaf vertices and decision vertices. The core algorithm of decision tree is ID3 that uses a greedy search approach to create the branches. It uses information Gain and Entropy to construct a tree. Information Gain is defined by equation 8.
where G(A,C) is the information gain of feature C in the whole training set A. A contains all possible features of the dataset to train. E(A) is the entropy of the whole original training set and defined in Eq. 9. While E(A, C) in entropy of C feature w.r.t overall features A.
where \(a_i\) is the proportion of A that belongs to complex i and root vertex is the vertex with the highest information gain. Then, the training dataset is divided into smaller groups and each group denotes the branch that extend the root vertex. The process is repeated and only those data points are used that complete the branch. The node in which all data points have the same label is called a pure node and this process continues until all nodes become pure.
2.3.2 Gradient Boosting Classifier
Gradien boosting classifier uses a forward stagewise strategy to build an additive model. It allows changing the learning and loss function value to optimize the performance of classifiers like Neural Networks. In each iteration of model regression trees are fit on the negative gradient of the multinomial or binomial loss function. In complex classification, it generates the multi regression trees. Tree generation for complexes classification is defined in equation 10.
where \(T_n (X)\) is a decision tree generated by gradient boosting for complexes. jn represents the number of nodes. The constructed tree divides the input features into jn disjoint portions \(S_{1n}\ldots\). \(S_{jn}\)n and predict a constant value for every portion.
2.3.3 Linear Discriminant Analysis
Classification of protein complexes using the Linear discriminant analysis is done based on transform space based on some distance measures like Euclidean distances. The scatter matrix for classification is made using equation 11
where mi is training instances for each complex. xi is the mean of each complex and x is the overall mean of the feature vector.
3 Results and Discussion
In this section, we will discuss the characteristics of the datasets used in this study and evaluation measures to measure the performance of our model. We will also compare the performance and robustness of our model with state of the art studies and define the time and space complexity of our models. Furthermore, we will show the predicted protein complexes from the PPINs. The experiments were performed using Core i7 8th generation processor, 24 gigabytes of RAM and Python3.
3.1 Datasets Description and Evaluation Measures
We have used the five benchmark datasets in these experiments. To predict protein complexes we downloaded the hand-curated yeast complexes dataset from CYC2008 [15, 22] and MIPS [14] (versions used by Sikandar et al. [37, 38]). CYC2008 contains 148 complexes, 999 genes and 2475 interaction. MIPS yeast complexes dataset contains 109 complexes, 1004 genes and 2745 interactions. Human and Mouse protein complexes dataset is taken from MIPS CORUM [13]. CORUM contains overall 4274 complexes from the mammalian organism of which 67% human, 10% rat, 15% mouses and 4473 different genes information. It contains 678 mouse protein complexes and 2916 human protein complexes. Protein interaction information was taken from the DIP (database of interacting proteins) [12]. DIP contains 28850 proteins, 834 organisms, 81923 interactions overall. It contains 51221 proteins, 24918 interactions for S.cerevisiae, 5084 proteins and 9141 interactions for Homosepain, 2387 proteins and 3069 interactions for Mus Musculus. Amino acid sequences of all these proteins are taken from UniProt [19]. The summarize description of datasets is given in Table 3.
We have evaluated our performance of our models with Precision, Recall, F1-measure and Matthew Correlation coefficient measures. Accuracy measures the ratio of correctly classify complexes from the total number of complexes.
Precision measures the complex ratio of the predicted protein complex datasets corresponding to at least one of the benchmark datasets complexes. Recall measures the ratio of at least one predicted protein complex from the benchmark dataset. F1-measure is harmonic mean (HM) of recall and precision. Matthew correlation coefficient (MCC) is a correlation between the actual and predicted protein complexes and returns a value between + 1 and − 1. The coefficient of − 1 represents the total contradiction in actual and predicted complex, 0 for random prediction and +1 denotes perfect prediction. The accuracy, precision, recall, F1-measure and Matthew correlation coefficient are defined by the following equations.
where \(A_{ii}\) represents the truly predicted complexes , \(\sum _{i}A_{ji}\) represents all the truly and falsely predicted complexes and \(\sum A{ji}\) represents all the true complexes. TP represents the true-positive rate, TN represents the true-negative rate, FP represents the false-positive rate and FN represents the false-negative rate of predicted complexes.
3.2 Comparison with Other Methods
To validate our work, we compare our methodology for the yeast datasets MIPS and CYC2008 [15] by considering DIP [14] interactions with MCL [23], MCODE [21], CFINDER [22], CDIP [28], and Sikandar et al. [37, 38] in Table 1. MCL used the topological feature average degree and average density of the protein protein interaction graph. MCODE topological features used the vertex weighting and outward traversal densed region, while CFINDER used the weighted density, graph size, clustering coefficients. CDIP also used the topological features which include graph density, graph size, degree statistics, clustering coefficient, topological coefficient. Topological features give the localization information of PPIN but these experiments show that this information is not enough to detect the protein complexes sufficiently. Therefore, Sikandar et al. used the biological features along with topological feature and this novel technique show the significant improvement in the protein complex detection. We also used these biological and topological with our bag of words and physicochemical features and the combination of these features significantly improve the results. The feature difference and comparative results are shown in Tables 4, 5 and 6, Figures 2, 3 and 4 for cyc2008 and MIPS yeast benchmark sets.
These results show a significant improvement in results by using our methodology. We analyzed that results are getting better with the larger number of complexes. Our methodology is capable of handling the larger number of complexes and according to our observation, results of complex prediction from the PPINs will be more precise as the databases of interacting protein extend.
3.3 Time and Space Complexity of the Algorithm
The complexity of our models and algorithm for time and space is given in Table 7.
Here, we have represented the upper bound complexity. The complexity of these prediction models is defined in this table, while the other results are shown in Table 6. Sample space n represents the number of instances in the dataset and divided into A and B, where A represents the training examples and B represents the testing examples. The number of features is represented by the f while n\(_{\text{trees}}\) represents the number of trees generated by the algorithm to best fit the data. The prediction time complexity, for example, is calculated in the case of decision tree, gradient boosting and linear discriminant algorithms because these are the models that have been trained to predict the protein complexes and use for prediction. While in the case of random forest, complexity on the testing data is used because we have used the Random forest for the features selection method that selects the best feature best train test split methodology. The decision tree, random forest and gradient boosting classifiers use the trees for training and split until they reached the maximum depth d. The technique for detecting this split is to look for each feature (f) at the different thresholds (up to n) and the gain of knowledge (is O(n)). While in the linear discriminant analysis the question of determining the \(\beta\) weight function is determined by the equation: \(\beta\) = \((M' M)^1\) \(M' N\). The most computation-intensive aspect is to test the product M M, which takes place in operations f\(^2\) n, and then invert it, which takes place in operations f\(^3\). While most implementations tend to use a downward gradient to solve the equation scheme (M M) \(\beta\) = M N, the complexity remains the same.
3.4 Robustness of our methodology
Our methodology includes the feature selection method that returns the features with their ranking and selected features with high impact that give the best results. Although biological features with the combination of topological features give better results than alone topological features but it is difficult and computationally expensive to calculate the biological features from the amino acid sequences of proteins. While sequential features and physicochemical features are easy to calculate from amino acid sequences and gives better results with the combination of the topological feature. Another limitation of biological features is that they increase the dimension of the dataset that leads to the curse of dimensionality and low the learning capability of the model. Similar to our hypothesis, our feature selection returns us the features in which no biological feature is included so we can eliminate the biological features and use sequential, physicochemical, and topological features. By using the topological, biological, sequential and physicochemical features for the CYC2008 and MIPS complexes and topological, sequential and physicochemical features for human and mouse complexes we achieve the significantly improved results and reduced computational time has shown in Tables 8, 9 and 10, and results without feature selection are shown in Table 8. Table 9 contains the results of feature selection on the whole dataset while the results of features selection just on the training dataset is shown in Table 10. These results show the robustness of our feature selection method as there is a significant difference between the result and computational time of both methods.
To show the robustness of our proposed methodology, we have compared the true positive rate of our model with other state of the art studies on eight thresholds t = 0.2, 0.3, 0.4, 0.5, 0.6; 0.7; 0.8; 0.9 in terms of the number of real complexes matched on two individual datasets. Figures 5 and 6 shows that for each value of t our proposed method matches more real complexes than other comparative methods.
3.5 Protein Complexes Prediction
We also predict the human Histone H3.1 complex, Mediator complex and Ubiquitin E3 ligase (FBXW11, SKP1A, CUL1, RBX1) and mouse Gata1–Fog1–MeCP1 complex, Ikaros and Ikaros–NuRD complex shown in Figures 7 and 8, respectively.
Predicted Human complexes using the localization and biological information (a) Histone H3.1 complex prediction based on H3C1 and H4C1 (Histone H4 protein) (b) Mediator complex prediction bases on Mediator of ribonueclic acid polymerse II transcription (MED 20) and Sterlo regularity element-binding protein (SREBF2) (c) Ubiquitin E3 ligase (FBXW11, SKP1A, CUL1, RBX1) prediction based on E3 ubiquitin protein (RBX1), S phase kinase associated protein (SKP1) and cullin 1 (CUL1)
Predicted Mouse complexes using biological and localization information (a) Gata1–Fog1–MeCP1 complex prediction based on Metastasis-associated protein (Mta 1), Histodine deacetylase 1 (Hdac1) and Histone-binding protein (Rbbp4) (b) Ikaros complex prediction based on Histone deacetylase protein (Hdac1), transcription activator BRG1 (Smarca4) and SNF/SWI subunit SMARCC1 (Smarcc1) (c) Ikaros–NuRD complex prediction based on Transcription Factor 7 like 1 (Tcf7l1)
Throughout DNA replication and possibly DNA repair, the deposition of the main histone H3 (H3.1) is combined with DNA synthesis, while the variant of Histone H3.3 acts as the substitution form for the Genetic code synthesis-independent deposition pathway [54]. The many ribonucleic acid polymerase II like the protein noncoding and coding ribonucleic acid genes need the Mediator complex to regulate their functionality. The Mediator complex is also typically targeted by DNA-binding transcription factors (TFs), sequence specific, that function in response to genetic or environmental indications to regulate gene expression systems [55]. Mediator’s main function is to relay signals from transcription activators linked to enhancer regions to the transcription machine assembled at promoters as a pre-initiation complex (PIC) for regulating transcription initiation [56]. The ubiquitin pathway is a powerful enzyme pathway that controls undesirable biosynthetic proteins for proteasome dissolution. The ubiquitin proteasome network is a master protein homeostasis regulator by means of which proteins are originally activated by E3 ligases for polyubiquitination and then degraded by the proteasome to short peptides [57].
GATA-1 is crucial to the growth of, mast cell lines, eosinophilic, erythroid megakaryocytic, and eosinophilic. It functions as a repressor and activator of various target genes, such as in erythroid cells it suppresses cell proliferation and premature hematopoietic genes while triggering erythroid genes. FOG-1 regulates GATA-1 associations with the MeCP1 complex and provides an explanation for the conflicting roles of these two erythropoiesis factors [58]. The IKAROS is a regulator of hematopoietic and its expression level influence the NuRD and the Mediator complex interact with ribonucleic acid polymerase 2 enzymes to normalize its ability to prompt protein-coding genes [59]. It Regulates the transcription expansion and suppress tumor in leukemia and mostly accompanying the poor scenario. It procedure a complex with NuRD complex and P-TEFb complex that is essential for productive transcription expansion [60]. In the predicted complexes, yellow node is the node that we used to predict the protein complex and the other nodes are true proteins in the complexes. The predicted protein complexes based on this Uniprot ID show the robustness of our model.
With the growing demand for protein protein interaction studies is presenting the computational methods for predicting the protein protein interaction and complexes. In our work, we explore the classification method for PPI combining the different features such as biological features, topological features, sequential features, and physicochemical features. In the method of feature extraction graphical features of interacting proteins, the sequence of amino acid, blosum62 and PSSM matrix representing the amino acid sequences have been proven very useful for the classification of protein complexes. Compared with earlier methods, our results have been improved using the novel features of protein and powerful classifiers. Using these feature models will perform well on other independent protein datasets. To the best of our knowledge, our method is superior, feasible, and robust.
4 Conclusion
The existing protein complex detection studies mostly used the topological features to predict the protein complexes and some recent studies have used the combination of biological and topological features. The high-dimensional data and biological feature extracted through Fast Fourier Transformation contain noise data that affect the accuracy of the model and misguide the protein complex prediction. In this study, we have computed the topological and biological features, i.e., bag of words, physicochemical and spectral domain features for mining PCs from PPINs. We have used FSSF, i.e., random forest based Boruta feature selection method to rank and select the best features that eliminate the noise from the dataset and reduce the dimension of data. Decision tree, linear discriminant analysis and gradient boosting classifiers are trained on best features selected by FSSF to predict the protein complexes. The comparison with the recent studies shows significant improvement in results. In our methodology, linear discriminant analysis classifier gives precision, recall and f-measure 94.58%, 94.92% and 94.45% for MIPS and 96.31%, 93.55% and 96.02% for CYC2008 dataset complexes, respectively. The robustness of our model is shown by the predicted complexes. In addition, our machine learning-based protein complex detection will help in future studies to predict the protein complexes based on these features much precisely. In future, we will implement the deep learning-based model to predict the protein complexes. Furthermore, we will implement the conformal prediction to mine protein complexes that will help to find the complexes with confidence.
References
Peng Y, Lu Z (2017) Deep learning for extracting protein-protein interactions from biomedical literature, pp 29–38. https://doi.org/10.18653/v1/w17-2304
Qi Y, Balem F, Faloutsos C, Klein-Seetharaman J, Bar-Joseph Z (2008) Protein complex identification by supervised graph local clustering. Bioinformatics. https://doi.org/10.1093/bioinformatics/btn164
Smits AH, Vermeulen M (2016) Characterizing protein-protein interactions using mass spectrometry: challenges and opportunities. Trends Biotechnol 34(10):825–834. https://doi.org/10.1016/j.tibtech.2016.02.014
Celaj A et al (2017) Quantitative analysis of protein interaction network dynamics in yeast. Mol Syst Biol 13(7):934. https://doi.org/10.15252/msb.20177532
Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U (2009) Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci 10(6):2763–2788. https://doi.org/10.3390/ijms10062763
Puig O et al (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24(3):218–229. https://doi.org/10.1006/meth.2001.1183
George PM, Mlynash M, Adams CM, Kuo CJ, Albers GW, Olivot J-M (2015) Novel Tia biomarkers identified by mass spectrometry-based proteomics. Int J Stroke 10(8):1204–1211. https://doi.org/10.1111/ijs.12603
Templin MF, Stoll D, Schrenk M, Traub PC, Vöhringer CF, Joos TO (2002) Protein microarray technology. Drug Discov Today 7(15):815–822. https://doi.org/10.1016/S1359-6446(00)01910-2
Sidhu SS, Koide S (2007) Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol 17(4):481–487. https://doi.org/10.1016/j.sbi.2007.08.007
Shoemaker BA, Panchenko AR (2007) Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput Biol 3(3):e42. https://doi.org/10.1371/journal.pcbi.0030042
Oughtred R et al (2019) The BioGRID interaction database: 2019 update. Nucleic Acids Res 47(D1):D529–D541. https://doi.org/10.1093/nar/gky1079
Xenarios I (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30(1):303–305. https://doi.org/10.1093/nar/30.1.303
Giurgiu M et al (2019) CORUM: the comprehensive resource of mammalian protein complexes—2019. Nucleic Acids Res 47(D1):D559–D563. https://doi.org/10.1093/nar/gky973
Pagel P et al (2005) The MIPS mammalian protein-protein interaction database. Bioinformatics 21(6):832–834. https://doi.org/10.1093/bioinformatics/bti115
Pu S, Wong J, Turner B, Cho E, Wodak SJ (2009) Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res 37(3):825–831. https://doi.org/10.1093/nar/gkn1005
Licata L et al (2012) MINT, the molecular interaction database: 2012 Update. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr930
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(D1):D353–D361. https://doi.org/10.1093/nar/gkw1092
Szklarczyk D et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Bateman A et al (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45(D1):D158–D169. https://doi.org/10.1093/nar/gkw1099
Haw R, Loney F, Ong E, He Y, Wu G (2020) Perform Pathway Enrichment Analysis Using ReactomeFIViz. Humana, New York, pp 165–179. https://doi.org/10.1007/978-1-4939-9873-9_13
Bader GD, Hogue CWV (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. https://doi.org/10.1186/1471-2105-4-2
Adamcsek B, Palla G, Farkas IJ, Derényi I, Vicsek T (2006) CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics 22(8):1021–1023. https://doi.org/10.1093/bioinformatics/btl039
Wu M, Li X, Kwoh C-K, Ng S-K (2009) A core-attachment based method to detect protein complexes in PPI networks. BMC Bioinform 10(1):169. https://doi.org/10.1186/1471-2105-10-169
Li M, Chen J, Wang J, Hu B, Chen G (2008) Modifying the DPClus algorithm for identifying protein complexes based on new topological structures. BMC Bioinform 9(1):398. https://doi.org/10.1186/1471-2105-9-398
Leung HCM, Xiang Q, Yiu SM, Chin FYL (2009) Predicting protein complexes from PPI data: a core-attachment approach. J Comput Biol 16(2):133–144. https://doi.org/10.1089/cmb.2008.01TT
Dong Y, Sun Y, Qin C (2018) Predicting protein complexes using a supervised learning method combined with local structural information. PLoS One 13(3):e0194124. https://doi.org/10.1371/journal.pone.0194124
Yu Y, Lin L, Sun C, Wang X, Wang X (2011) Complex detection based on integrated properties. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), vol. 7062 LNCS, no. PART 1:121–128. https://doi.org/10.1007/978-3-642-24955-6_15
Mewes HW et al (2008) MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Res 36(SUPPL):1. https://doi.org/10.1093/nar/gkm980
Liu Q, Song J, Li J (2016) Using contrast patterns between true complexes and random subgraphs in PPI networks to predict unknown protein complexes. Sci Rep. https://doi.org/10.1038/srep21223
Zeng J, Li D, Wu Y, Zou Q, Liu X (2015) An empirical study of features fusion techniques for protein-protein interaction prediction. Curr Bioinform 11(1):4–12. https://doi.org/10.2174/1574893611666151119221435
Khan J, Bhatti MH, Khan UG, Iqbal R (2019) Multiclass EEG motor-imagery classification with sub-band common spatial patterns. Eurasip J Wirel Commun Netw 2019(1):1–9. https://doi.org/10.1186/s13638-019-1497-y
Bhatti MH et al (2019) Soft computing-based EEG classification by optimal feature selection and neural networks. IEEE Trans Ind Inform 15(10):5747–5754. https://doi.org/10.1109/TII.2019.2925624
Ahmad F, Farooq A, Ghani Khan MU, Shabbir MZ, Rabbani M, Hussain I (2020) Identification of most relevant features for classification of Francisella tularensis using machine learning. Curr Bioinform. https://doi.org/10.2174/1574893615666200219113900
Zou Q, Zeng J, Cao L, Ji R (2016) A novel features ranking metric with application to scalable visual and bioinformatics data classification. Neurocomputing 173:346–354. https://doi.org/10.1016/j.neucom.2014.12.123
Zhang SW, Cheng YM, Luo L, Pan Q (2011) Prediction of protein-protein interaction using distance frequency of amino acids grouped with their physicochemical properties. In: Proceedings—2011 6th International conference on bio-inspired computing: theories and applications, BIC-TA 2011, pp 70–74, https://doi.org/10.1109/BIC-TA.2011.53
Jolliffe I (2011) Principal component analysis. International encyclopedia of statistical science. Springer, Berlin, pp 1094–1096. https://doi.org/10.1007/978-3-642-04898-2_455
Sikandar A et al (2018) Decision tree based approaches for detecting protein complex in protein protein interaction network (PPI) via link and sequence analysis. IEEE Access 6:22108–22120. https://doi.org/10.1109/ACCESS.2018.2807811
Sikandar A, Anwar W, Sikandar M (2019) Combining sequence entropy and subgraph topology for complex prediction in protein protein interaction (PPI) network. Curr Bioinform 14(6):516–523. https://doi.org/10.2174/1574893614666190103100026
Faridoon A, Sikandar A, Imran M, Ghouri S, Sikandar M, Sikandar W (2020) Combining SVM and ECOC for identification of protein complexes from protein protein interaction networks by integrating amino acids’ physical properties and complex topology. Interdiscip Sci Comput Life Sci. https://doi.org/10.1007/s12539-020-00369-5
Kursa MB, Jankowski A, Rudnicki WR (2010) Boruta - a system for feature selection. Fundam Informaticae 101(4):271–285. https://doi.org/10.3233/FI-2010-288
Gursoy A, Keskin O, Nussinov R (2008) Topological properties of protein interaction networks from a structural perspective. Biochem Soc Trans 36(Pt 6):1398–403. https://doi.org/10.1042/BST0361398
Guo Y-Z et al (2006) Classifying G protein-coupled receptors and nuclear receptors on the basis of protein power spectrum from fast Fourier transform. Amino Acids 30(4):397–402. https://doi.org/10.1007/s00726-006-0332-z
Jolliffe I (2005) Principal component analysis, in encyclopedia of statistics in behavioral science. Wiley, Chichester. https://doi.org/10.1002/0470013192.bsa501
Bérard A, Servan C, Pietquin O, Besacier L (2016) MultiVec: a multilingual and multilevel representation learning toolkit for NLP. https://hal.archives-ouvertes.fr/hal-01335930/. Accessed 16 Jun 2019
Singh P (2019) Natural language processing, in machine learning with PySpark. Apress, Berkeley, pp 191–218
Kulkarni A, Shivananda A (2019) Converting text to features. Natural language processing recipes. Apress, Berkeley, pp 67–96
Li Z-W, You Z-H, Chen X, Gui J, Nie R (2016) Highly accurate prediction of protein-protein interactions via incorporating evolutionary information and physicochemical characteristics. Int J Mol Sci. https://doi.org/10.3390/ijms17091396
Kawashima S, Pokarowski P, Pokarowska M, Kolinski A, Katayama T, Kanehisa M (2008) AAindex: amino acid index database, progress report 2008. Nucleic Acids Res 36(Database issue):D202–5. https://doi.org/10.1093/nar/gkm998
Nakai K, Kidera A, Kanehisa M (2019) Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng 2(2):93–100. https://doi.org/10.1093/protein/2.2.93
Kawashima S, Kanehisa M (2000) AAindex: amino acid index database. Nucleic Acids Res 28(1):374. https://doi.org/10.1093/nar/28.1.374
Tomii K, Kanehisa M (1996) Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng 9(1):27–36. https://doi.org/10.1093/protein/9.1.27
Raicar G, Saini H, Dehzangi A, Lal S, Sharma A (2016) Improving protein fold recognition and structural class prediction accuracies using physicochemical properties of amino acids. J Theor Biol 402:117–128. https://doi.org/10.1016/J.JTBI.2016.05.002
Blei DM, Ng AY, Jordan MI (2019) Blei03a.Pdf. J Mach Learn Res 3:993–1022. http://www.jmlr.org/papers/volume3/blei03a/blei03a.pdf. Accessed 11 Nov 2003
Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116(1):51–61. https://doi.org/10.1016/S0092-8674(03)01064-X
Poss ZC, Ebmeier CC, Taatjes DJ (2013) The mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 48(6):575–608. https://doi.org/10.3109/10409238.2013.840259
Soutourina J (2018) Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19(4):262–274. https://doi.org/10.1038/nrm.2017.115
Lucas X, Ciulli A (2017) Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr Opin Struct Biol 44:101–110. https://doi.org/10.1016/j.sbi.2016.12.015
Rodriguez P et al (2005) GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24(13):2354–2366. https://doi.org/10.1038/sj.emboj.7600702
Bottardi S et al (2014) The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. PLoS Genet 10(12):e1004827. https://doi.org/10.1371/journal.pgen.1004827
Bottardi S, Mavoungou L, Milot E (2015) IKAROS: a multifunctional regulator of the polymerase II transcription cycle. Trends Genet 31(9):500–508. https://doi.org/10.1016/j.tig.2015.05.003
Sikandar M et al (2020) Analysis for disease gene association using machine learning. IEEE Access 8:160616–160626. https://doi.org/10.1109/ACCESS.2020.3020592
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Younis, H., Anwar, M.W., Khan, M.U.G. et al. A New Sequential Forward Feature Selection (SFFS) Algorithm for Mining Best Topological and Biological Features to Predict Protein Complexes from Protein–Protein Interaction Networks (PPINs). Interdiscip Sci Comput Life Sci 13, 371–388 (2021). https://doi.org/10.1007/s12539-021-00433-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00433-8