Abstract
The gene structure and sequence diversity of 5S rRNA genes were analyzed in 13 Allium species. While the lengths and sequences of the coding gene segments were conserved, the spacers were highly variable and could be characterized as either short (213–404 bp) or long (384–486 bp) spacers. The short spacers were further classified into five subtypes (SS-I to SS-V) and the long spacers into four subtypes (LS-I to LS-IV). The short spacers were more conserved than were the long spacers. There was a sequence duplication of 85 bp in SS-III that distinguished it from SS-II. The coding sequences of the 5S rRNA genes started with CGG and ended with either CCC or TCC. Both long and short spacers started with TTTT at their 5′-ends. However, the long spacers ended with a 3′-TGA sequence, whereas the short spacers terminated with various sequences, such as TTA, ATA, or TGA. GC content ranged from 27 to 41% in whole repeats, and the GC content in the long spacers was lower than in the short spacers. The nucleotide diversity in the coding regions was lower than in the spacers, and the nucleotide diversity in the coding regions did not correlate with that of the spacers. FISH analysis confirmed that each Allium species has either short spacers or long spacers. Although chromosomal locations of the 5S rRNA genes in Allium wakegi confirmed the allodiploid nature of A. cepa and A. fistulosum, spacer sequences revealed the absence of SS-II in A. cepa and in A. wakegi. The current study demonstrated that the 5S rRNA genes diverged in early stages in Allium species differentiation except of the allodiploid A. wakegi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotes, 5S rRNA forms the large subunit of the ribosome with other rRNAs and several dozenproteins. Although 5S rDNA sequences have been isolated from many species and their secondary and tertiary structures have been elucidated (Funari et al. 2000; Szymanski et al. 2002), its precise function during protein synthesis has not been fully determined. In Escherichia coli, 5S rRNA facilitates communication between different functional ribosomal domains to coordinate the various events during protein synthesis (Bogdanov et al. 1995). A similar role of 5S rRNA in eukaryotes was also described in studies with yeast, Sacchromyces cereviceae (Smith et al. 2001). Through the analysis of wild-type and mutant alleles of 5S rRNA in yeast, Kiparisov et al. (2005) showed that different regions of 5S rRNA are involved in different functional centers of the ribosome, affecting translational fidelity and ultimately gene expression.
Transcription of 5S rRNA is mediated by RNA polymerase III (pol III), but other rRNA genes are transcribed by RNA polymerase I (pol I). In fungi and protists, 5S rRNA genes are within the repeat unit of other gene families, such as histones or major rRNAs, and the transcription of 5S rRNAgene is often in the opposite direction of that of the linked gene (Drouin and Moniz 1995). RNA pol III does not bind directly to DNA, but binds instead to transcription factor IIIA (TF IIIA) that binds to internal promoters, such as Box A and Box C in humans (Nielson et al. 1993), and Box A, Box C, and Box D in Neurospora crassa (Tyler 1987).
Since the 5S rRNA genes and the major 45S rRNA genes are transcribed by different polymerases, it is not surprising that these genes are located at different chromosomal loci. In Triticeae, the major rRNA and 5S rRNA genes are closely associated on homologous chromosomes 1 and 5 (Kim et al. 1993; Kellogg and Appels 1995). In Allium species, the rRNA loci are located on different chromosomes. Lee and Seo (1997) demonstrated through in situ hybridization that the 5S rRNA gene loci are on chromosomes 9 and 15, and the 18S–26S rRNA gene loci are on chromosomes 6, 10, and 14 in Allium wakegi. In Allium victorialis, the 45S rRNA gene loci are on chromosomes 2 and 4, and a 5S rRNA gene locus is on chromosome 6 (Seo et al. 1997).
The 5S rDNA is a 120-bp molecule encoded in a tandem array with a nontranscribing spacer (NTS). While the genic region is nearly invariable with 120–122 bp, the spacers are highly variable in length and sequence. The number of repeats varies from less than 1,000 to over 100,000 in plants (Cronn et al. 1996; Schneeberger et al. 1989; Sastri et al. 1992). There are two basic types of tandem arrays that have either short or long spacers (Scoles et al. 1987; Kellogg and Appels 1995; Cronn et al. 1996). Spacer length is highly variable depending on the species. The two 5S rDNA arrays likely evolved independently because the sequences of the short spacer arrays from different species are more similar than are short and long spacer sequences from the same species (Scoles et al. 1987; Dvořák et al. 1989; Cronn et al. 1996). In Triticeae species, the arrays with short spacers are on chromosome 1 and the array with long spacers is on the homologous chromosome 5 (Scoles et al. 1987). However, Shibata and Hizume (2002) showed different results in Allium species from the results of Triticeae in a study of ten taxa of Allium species. In PCR amplification using 5S rDNA universal primers, the ten taxa produced a 350 bp fragment. In addition to the 350 bp fragment, A. cepa, A. schoeoprasum, A. sativum, and A. tuberosum had another fragment in the size range of 520–620 bp fragments. Through physical mapping by fluorescence in situ hybridization (FISH) analysis, they demonstrated that the short and long units were closely associated at the interstitial region of chromosome 6. Furthermore, sequence analysis revealed that the long subunit was recently derived from the short subunit through duplication and sequence insertion in A. cepa and A. schoenoprasum. In a recent report on 5S rDNA in A. victorialis var. platyphyllum, Seo and Seo (2010) reported chromosomal distributions of the 5S rRNA gene loci with long and short spacers, in which the 5S rRNA genes were located at three distinct loci on the short arm of chromosome 6. While the short spacers of the 5S rRNA genes were present at three loci, the long spacer genes were present in only one locus proximal to the centromere.
Repeat units of tandemly repeated multigene families often undergo a homogenizing process in which sequence changes occur in a highly orchestrated manner described as concerted evolution (Arnheim 1983; Sanderson and Doyle 1992). Eickbush and Eickbush (2007) reviewed concerted evolution of rRNA genes and suggested that unequal crossing over between sister chromatids is the major driving force for concerted evolution, while gene conversion also plays a role in orchestrated gene evolution. For 5S rRNA genes, Drouin and Moniz (1995) suggested the linkage of 5S rRNA genes to other tandemly repeated gene families in fungi, protists, nematodes, and arthropods. The 5S rRNA genes have the same fate in concerted evolution as do these repeated gene families. However, Kellogg and Appels (1995) observed that the nucleotide diversity in 5S rRNA genes was not significantly different from that of the nontranscribing spacers within an array in a diploid Triticeae species. Thus, they proposed that concerted evolution was not sufficient to homogenize entire arrays. Similarly, Cronn et al. (1996) showed that 5S rDNA intralocus concerted evolutionary forces are relatively weak and the rate of interrepeat homogenization is approximately equal to the rate of speciation in diploid and polyploid cotton.
The genus Allium L., one of the largest genera among monocots, is comprised of more than 800 species, so taxonomic classifications have not been clearly defined (Harvey 1995; Friesen et al. 2006; Fritsch et al. 2010; Li et al. 2010). This study presents the 5S rRNA gene structures and sequence diversity and traces the evolution of 5S rRNA genes from 13 Allium species.
Materials and methods
Plant materials and DNA extraction
The Allium species examined were kindly provided by Dr. Kang Jung-Hoon at the Rural Development Administration of Korea. These species were chosen for their economic importance and represented species classified in the genus Allium. A list of species and their accession numbers are provided in Table 1.
Genomic DNA was extracted from young leaves using DNeasy Plant Mini Kits (Qiagen, USA) as per the manufacturer's instructions.
PCR amplification and cloning
The nucleotide sequences of the primers for 5S rDNA PCR amplification were as follows: whole repeating unit, 5S_F: GATCCCATCAGAACTCC and 5S_R: GGTGCTTTAGTGCTGGTAT; coding region, coding F: CGGGTGCGATCATACCAGCA and coding R: GGGGTGCAACACGAGGACTT; short spacers, short F: TTTGCTTCTTCTCGTTCGGA and short R: GGAAGATTACTAATGCACA; and long spacers, long F: ATCGCCTTTTCACTCATACT and long R: CGAAAGACTGAGAAAATCAA (Park et al. 2000).
PCR amplification was performed with 10 ug of template genomic DNA, 5 pM each primer, 200 uM dNTPs, and 2.5 U Taq DNA polymerase (High Fidelity, Takara, Japan). PCR reactions were amplified with 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The amplified products were checked by 1.2% agarose gel electrophoresis, ligated into pGEM-T vectors (Promega, USA), and cloned into competent DH5α E. coli cells.
Sequence analysis
Five clones from each accession were randomly picked for sequencing analysis. All sequencing reactions were performed using an ABI3730 xl DNA analyzer. The sequencing reactions and reads were performed from both directions to reduce sequencing errors. Consensus sequences were determined from multiple sequence alignments of the sequences from the five clones using ClustalW version 2. Sequence similarities, the number of variable sites, and sequence diversities (π) were analyzed using the DnaSP program version 5 (http://www.ub.edu/dnasp/). GC content was calculated using the Oligo Calculator program at http://mbcf.dfci.havard.edu/docs/oliocal.html. After duplicated clones were removed, the sequences were deposited into GenBank with the accession numbers JF496587–JF496655.
Southern blot analysis
Genomic DNA (10 ug) was digested with ScaI and separated on 1% agarose gels at 50 V for 16 h. Depurination, Southern transfer to nylon membranes, and UV cross-linking were performed as per the manufacturer's instructions (GE Healthcare, USA). Probe labeling, hybridization, and signal detection were performed as per the manufacturer's instructions using an Amersham Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare).
Fluorescent in situ hybridization (FISH)
Root tips from young seedlings were pretreated with 2 mM 8-hydroxyquinoline at 18°C for 5 h, fixed in Carnoy's solution (3 EtOH:1 acetic acid, v/v), and stored at room temperature. Root tips were treated with a maceration solution containing 1%pectolyase (w/v) and 1% cellulase (w/v). Chromosome spreads were prepared with a drop of 45% acetic acid on the macerated root tips. Probe labeling and FISH procedures followed the protocol of Park et al. (2010). FISH signals were visualized using a Nikon Eclipse 80i fluorescent microscope and photographed with a Canon digital camera.
Results
Length of the 5S rRNA genes in Allium species
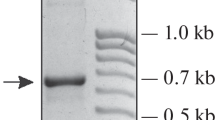
The 5S rDNA repeat was highly variable in length (333–608 bp; Table 2 and Fig. 1). Repeat lengths varied between plants of the same species, but repeats also varied within a plant due to variations in length of the NTS sequences (Table 2 and Fig. 2). There were two basic types of repeats: long spacers (384–486 bp) and short spacers (213–404 bp). While three species—A. sativum, A. tuberosum, and A. porrum—contained long spacers, the rest of the Allium species examined contained short spacers (Figs. 1 and 4). While the sequences of the long spacers were variable among the long spacers, the sequences of the short spacers were highly conserved except of the A. proliferum and A. monanthum (Fig. 2).The coding region was conserved between species in 120 bp, except for A. sativum in 120–121 bp and A. porrum in 125 bp (Fig. 3).
PCR amplified products of the 5SrRNA gene arrays in Allium species. Lanes: M: size markers, 1: A. sativum (Danyang), 2: A. sativum (Nepal), 3: A. sativum (UZB), 4–5: A. tuberosum, 6–7: A. porrum, 8–9: A. altaicum, 10–11: A. fistulosum, 12: A. proliferum, 13–14: A. wakegi, 15: A. pskemense, 16: A. vavilovii, 17: A. cepa, 18–19: A. ascalonicum, 20–21:A. monanthum, and 22: A. chinense
Intraspecific consensus sequences were determined through multiple sequence alignments. Consensus sequence comparisons revealed five length variants among the short spacers: short spacer I (SS-I) with 224–229 bp (A. wakegi-1, A. altaicum, A. fistulosum, A. chinense, and A. pskemense), SS-II with 229 bp (A. vavilovii-1, A. cepa-1, and A. ascalonicum-1), SS-III with 387 bp (A. vavilovii-2, A. cepa-2, A. wakegi-2, and A. ascalonicum-2), SS-IV with 312 bp (A. proliferum), and SS-V with 228 bp (A. monanthum; Table 2). Of the species having SS-I, A. pskemense had an extra pentanucleotide, TTTCT, to distinguish it from the other four species (Fig. 2). A. vavilovii, A. cepa, and A. ascalonicum had two types of short NTS sequences: SS-II (A. vavilovii-1, A. cepa-1, and A. ascalonicum-1) and SS-III (A. vavilovii-2, A. cepa-2, and A. ascalonicum-2). A. wakegi had SS-I (A. wakegi-1) and SS-III (A. wakegi-2). While the sequences of SS-II and SS-III were almost identical until the 140th nucleotide, the subsequent 89 bp were only 74% similar (Fig. 2). SS-III had an extra 158 bp at the 3′-end that consisted of 85 bp of duplicate sequences from the 71th–155th nucleotides (Figs. 2 and 5).
The consensus sequences of the long spacers consisted of 405–485 bp. The long spacers were classified into four types based on sequence similarities and insertion/deletion mutations: long spacer I (LS-I) with 485 bp (A. sativum Danyang-1 and A. sativum Nepal-1), LS-II with 436 bp (A. sativum Danyang-2 and A. sativum Nepal-2), LS-III with 405 bp (A. porrum), and LS-IV with 435 bp (A. sativum Uzbeckistan and A. tuberosum) (Table 2; Fig. 5).
Figure 5 shows the sequence homologies among the different 5S rDNA NTS sequences. There were three conserved sequence blocks among the short spacers. While SS-I, -II, and -III shared the three conserved blocks, SS-IV and -V had different conserved sequences of the sequence blocks. Approximately 20 bp at the 3′-end of SS-III and –IV, high sequence similarities with the end of the long spacers, including the distinctive TAG sequences of the long spacers were seen. The 5′-end sequences were conserved among the five short spacers.
Among the four long spacers, LS-II and -III shared all the sequence conserved blocks, whereas LS-I shared almost all of the sequence conserved blocks with LS-II and -III. LS-IV was very different from the other long spacers. As mentioned above, the 3′-end 20 bp were similar to those of SS-III and SS-IV.
Comparative sequence analysis of the NTS sequences of 5S rRNA genes in Allium species
Tables 3 and 4 show the pairwise sequence similarities among the short and long spacers, respectively. The similarity among the short spacers ranged from 44.2% between A. proliferum SS-IV and A. monanthum SS-V to 99.1% among A. altaicum, A. fistulosum, and A. chinense SS-I (Table 3). The average intragroup similarity for each spacer type SS-I, -II, and -III was high, ranging between 94.1 and 97.3% (Table 4). The average similarity for SS-IV and SS-V was not valid since only a single species was represented in these spacers. In a comparison between consensus sequences, SS-I and SS-II had the highest similarity, whereas SS-III and SS-V were the least similar (Table 4). The extra 158 bp at the 3′-end of SS-III resulted in a low similarity with SS-II.
Among the long spacers, the highest similarity was observed between A. sativum Danyang and A. sativum Nepal, whereas the lowest similarity was observed between A. porrum and A. sativum Uzbeckistan (Table 3). In a comparison of the three A. sativum accessions from different geographical origins, A. sativum Uzbeckistan was the least similar to the other two A. sativum accessions: 53.0% with A. sativum Danyang and 54.5% with A. sativum Nepal. The average intragroup similarity was 79.1% among LS-1 sequences and 96.1% among LS-III sequences. A comparison of the consensus sequences indicated that LS-IV was the least similar to the other long spacers (45–46%; Table 4).
Molecular sequence variations of the 5S rRNA genes in Allium species
We sequenced 93 clones from 13 Allium species and all of the sequences were virtually different each other. The 5S rRNA coding sequences started with CGG and ended with either CCC or TCC (Fig. 3). There were 47 variable sites, 30 transition mutations, and 17 transversion mutations among the consensus sequences of the 13 Allium species. Although we analyzed a subset of clones from each species, the number of intraspecific variable sites varied from zero in A. chinense to eight in A. tuberosum (Table 2). A. vavilovii, A. monanthum, and A.proliferum each had only one variable site. The sequences toward the 3′-end were more variable than those in the 5′ regions of all 5S rRNA genes. Intragenic control regions (ICRs) (Cloix et al. 2000) were present at the 50–60th (Box A) and 80–90th nucleotides (Box C). The GC content of the coding gene was very consistent with a narrow range of 55–57%.
Both the long and short spacers started with TTTT at their 5′-end. The 3′-end sequence was TGA in the long spacers, whereas the short spacers varied and were either TTA or ATA in SS-I, -II, and -V, or TGA in SS-III and SS-IV. Both long and short spacers were highly AT-rich (Table 2). The GC content varied and ranged from 27% in A. sativum Uzbeckistan to 41% in A. vavilovii, A. cepa, and A. proliferum. The GC content of the long spacers was generally lower than that of the short spacers. There were five GC-rich regions across the short spacers, whereas GC-rich regions were present mainly at the 5′- and 3′-ends of the long spacers (Fig. 5).
Graphic representation of the short and long spacers of the 5S rRNA genes in Allium species.The different species contained different combinations of the spacers. Long spacer (LS)-I: A. sativum (A. sativum Danyang-1 and A. sativum Nepal-1); LS-II: A. sativum (A. sativum Danyang-2 and A. sativum Nepal-2); LS-III: A. porrum; LS-IV: A.sativum and A. tuberosum. Short spacer (SS)-I: A. wakegi, A. altaicum, A. fistlosum, A. chinense, and A. pskemense; SS-II: A. vavilovii, A. cepa, and A. ascalonicum; SS-III: A. vavilovii, A. cepa, A. ascalonicum, and A. wakegi; SS-IV: A. proliferum and SS-V: A. monanthum. GC-rich regions are highlighted in yellow. Regions with sequence similarities are connected by gray. Duplicated regions in SS-III are denoted by lines with arrow heads
The NTS region revealed high intraspecific sequence variation. The number of variable sites in the NTS sequences varied from seven in A. chinense to 100 in A. porrum (Table 2). The π value, a measure of the average nucleotide differences per site between any two DNA sequences (Nei and Li 1979), was calculated for each species. As seen in the number of variable sites, π values in the coding region were lower than in the NTS sequences in all 5S rRNA genes. The nucleotide diversity in the coding region was not highly related to the nucleotide diversity in the NTS regions. For example, the coding sequence diversity was high and the NTS sequence diversity was low in A. sativum Uzbekistan, whereas A. porrum had low nucleotide diversity in the coding region and high diversity in its NTS sequence.
Chromosomal locations of the 5S rRNA genes in Allium species
FISH analysis was performed with the Allium species A. fistulosum, A. wakegi, A. cepa, and A. sativum. The Allium species with 5S rDNA short spacer repeats had hybridization signals on a pair of chromosomes in the middle of the short arm (Figs. 6a–e). The species with SS-III, which have 158 extra nucleotides at the 3′-end of the spacer, showed an additional signal at a location proximal to the centromere on the same chromosome (Fig. 6c–e). Thus, this second locus may be the site of the extra 158 bp that carry the short spacer (SS-III). As demonstrated previously (Hizume 1994; Lee et al. 1999), A. wakegi had two 5S rDNA loci: one in the short arm of a chromosome and another at the middle of the short arm of another chromosome (Fig. 6c and d).
Chromosomal locations of the 5S rRNA genes in Allium species. a A. fistulosum probed by coding region (CR; red) and short spacer (SS; green). b A. fistulosum probed by CR (red) and long spacer (LS; green; no signal). c–d A. wakegi probed by CR (red) and SS (green; no signal). e A. cepa probed by CR (red) and LS (green; no signal). f A. sativum probed by CR (green) and LS (red)
Since 5S rRNA genes with different spacers have been identified in other species, we examined the possibility that either short or long spacer types were missed in each species. The coding and spacer sequences were probed separately by labeling coding regions with biotin-dUTP (2′-deoxyuridine 5′-triphosphate) and labeling the spacers with dig-dUTP. Biotin-dUTP and dig-dUTP signals were detected by antiavidin-Cy3 (red) and antidig-FITC (green), respectively. The species with short spacers, A. fistulosum and A. wakegi, had yellow signals on the hybridization site due to the combination of coding regions of red and short spacers of green (Fig. 6a and c). When probed with coding region and long spacer sequences, the hybridization sites showed only red signals from the coding region (Fig. 6b, d, and e). Thus, these species do not carry the long spacer. A. sativum, a diploid having the long spacer, revealed three 5S rDNA loci in a chromosome in which two loci were located on the short arm and another locus was located at the middle of the long arm (Fig. 6f).
Discussion
5S rRNA genes are encoded in tandemly repeated arrays at one or more chromosomal loci in most eukaryotic species (Appel and Honeycutt 1986; Cronn et al. 1996). Each tandem array consists of 120–122 bp of coding sequence with spacers of variable lengths, and the number of tandem arrays per locus varies from a few hundred to over 100,000 (Sastri et al. 1992; Cronn et al. 1996). Usually, coding region sequences are conserved and spacer sequences are highly variable in both sequence and length. Although the chromosomal locations of the 5S rDNA loci in Allium species have been determined by cytogenetic analyses (Hizume 1994; Shibata and Hizume 2002; Seo and Seo 2010), this study combined detailed analyses of gene structures with that of sequence variations. The current study presents the sequence variation, chromosomal distribution, and relations of the 5S rRNA sequences in Allium species.
Two types of 5S rDNA repeat arrays were identified in Allium species based on spacer length variations, which is consistent with reports on other species (Scoles et al. 1987; Dvořák et al. 1989; Cronn et al. 1996). These long and short spacers were further classified into four and five subtypes, respectively, based on sequence similarities. All Allium species had 5S rDNA repeats with either short or long spacers, with slight length variations in each type as a result of sequence deletions or duplications. Because classification of the Allium species into subgenera or sections has not been firmly established, we followed the most recent classifications (Friesen et al. 2006; Fritsch et al. 2010; Li et al. 2010).
The low sequence similarity between the short and long spacers implies that the two 5S rRNA genes diverged during the early stages of differentiation of the Allium species, which correlates with groupings based on nuclear internal transcribed spacer of 45S rRNA genes and chloroplast sequences (Ricroch et al. 2005; Li et al. 2010; Son et al. 2010). A. altaicum is a wild species related to the cultivated bunching onion, A. fistulosum, and both species have the same 5S rRNA gene with SS-I. Both A. pskemense and A. vavilovii are considered to be the wild species related to the bulb onion, A. cepa (Harvey 1995). While A. vavilovii and A. cepa have the same 5S rRNA genes with SS-II and SS-III, the 5S rRNA gene of A. pskemense has SS-1. This result is consistent with F1 hybrid fertility analyses. The F1 hybrids from A. vavilovii and A. cepa crosses are fully fertile, but the F1 hybrids from A. pskemense and A. cepa crosses are sterile (Harvey 1995). Thus, A. pskemense may not be a direct wild relative of A. cepa, but rather is more closely related to A. altaicum and A. fistulosum.
A. wakegi (shallot or jjokpa, Korean common name) is considered to be closely related to A. fistulosum (Hizume 1994). However, the A. wakegi 5S rRNA genes have SS-I and SS-III, whereas the A. cepa 5S rRNA genes have SS-II and SS-III. The morphological phenotypes of A. wakegi are a composite of A. cepa and A. fistulosum that have underground bulbs and aboveground stems. Various cytological results, including our own, support the composite morphology. That is, A. wakegi has two chromosomes with 5S rRNA genes, one similar to A. cepa and another similar to A. fistulosum (Hizume 1994; Lee et al. 1999), supporting the allodiploid (amphihaploid) nature of A. wakegi (Tashiro 1984; Hizume 1994). Lee et al. (1999) identified the 5S rRNA gene-bearing chromosomes as chromosome 7 in A. cepa and A. fistulosum, and chromosomes 9 and 15 in A. wakegi. However, the current detailed sequence analysis showed that A. wakegi has SS-I and SS-III, but not the SS-II of A. cepa. The A. cepa accessions may carry either SS-II or SS-III, and SS-II was not identified in A. cepa in the current study. Alternatively, the 5S rRNA gene in A. wakegi may have been homogenized to SS-I by interallelic gene conversion between homologous loci (Jeffrey et al. 1994), since the SS-I and SS-II sequences are highly similar. SS-III was derived from SS-II by an 85-bp duplication. Segmental duplications in the 5S rRNA spacers occur frequently by replication slippage (Scoles et al. 1987). Three species, A. cepa, A. vavilovii, and A. ascalonicum, had both SS-II and SS-III. Since the extra 158 bp at the 3′-end of SS-III had high sequence similarities among the three species, the chromosomal loci of the 5S rRNA genes in A. vavilovii and A. ascalonicum may be the same as those of A. cepa located at two loci on the short arm of a chromosome. We examined whether SS-II and SS-III have different chromosomal sites by probing the A. cepa chromosome spread with the SS-III-specific sequence (73 bp). However, the probe size was too small to produce clear in situ hybridization signals. Thus, the possibility of different chromosomal sites for SS-II and SS-III is still unresolved.
Spacer sequences, especially the flanking regions of the coding sequences, may also have a function in proper transcription and termination of the 5S rRNA genes. As reported for other species (Scoles et al. 1987; Cronn et al. 1996; Liu et al. 2003), all of the Allium 5S rRNA spacers had polyT 5′-ends that were preceded by polyC (usually CCC) at the 3′-end of the coding region. The high GC content with only a narrow range of variation in the coding region implies that secondary structure stability may contribute to the proper functioning of the 5S rRNA in the ribosome. Hori and Osawa (1986) demonstrated that all organisms have essentially the same secondary structure, but eukaryotes, archaebacteria, and eubacteria display some different characteristics. In plants, Liu et al. (2003) reported the GC content of 5S rRNA genes and spacers in six Pinus (gymnosperm) species, in which the GC content was as high as 58.7–61.9%. In Aegilopstauchii, GC contents were 48% in the spacers and 56% in the coding region of 5S rRNA gene (Kellogg and Appels 1995). In our study, GC contents were 32–41% in the spacers, 56–58% in the coding region. Thus, the GC content of the spacers is likely related to species or genus characteristics.
The sequences of gene families have often been homogenized by concerted evolution via gene conversion or unequal crossing over (Eickbush and Eickbush 2007). 5S rRNA repeat arrays are a typical case of concerted evolution. However, the selection pressure of the mutations is different between coding regions and spacer sequences. From our observations of the 5S rRNA genes in Allium species, we showed that coding regions had comparatively lower genetic variation than the spacer regions, except in A. altaicum. In A. altaicum, sequence diversity in the coding region was higher than in other species, so that the π values were similar between coding and spacer regions in A. altaicum. Therefore, homogenizing forces likely allowed for similar levels of sequence polymorphisms to accumulate in both regions, but fixation of the mutations in coding regions was prohibited, as in the case of 5S rRNA genes in polyploid cotton (Cronn et al. 1996).
The sequences of and distances between the ICRs are important for transcription of the 5S rRNA genes. Alteration of the sequences or the distances could alter 5S rRNA transcription (Kellogg and Appels 1995; Cloix et al. 2000). The sequences of and distances between the 5S rRNA spacers of Allium species were similar to those of Arabidopsis thaliana (Cloix et al. 2000). However, we observed occasional sequence substitution mutations in both Box A and Box C in the Allium species. We hypothesize that these aberrant sequences carrying genes are pseudogenes. Since both wild-type and mutated copies are present in individual plants, the pseudogenes do not appear to affect the formation of ribosomes. Thus, the mutations in the control region may be fixed until purged by gene conversion from wild-type alleles.
References
Appel R, Honeycutt RL (1986) rDNA: evolution over a billion years. In: Dutta SK (ed) DNA systematic, vol II. CRC Press, Boca Raton, FL, pp 81–155
Arnheim N (1983) Concerted evolution of multigene families. In: Nei M, Koehn RK (eds) Evolution of genes and proteins. Sinauer Associates, Sunderland, MA, USA, pp 36–61
Bogdanov AA, Dontssova PA, Dokudovskaya SS, Lavrik IN (1995) Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol 73:869–876
Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillier C, Espagnol MC, Picard G, Tourmente S (2000) Analysis of the 5S RNA pool in Arabidopsis thaliana: RNAs are heterogenous and only two of the genomic 5S loci produce mature 5S RNA. Genome Res 12:132–144
Cronn RC, Zhao X, Paterson AH, Wendel JF (1996) Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J Mol Evol 42:685–705
Drouin G, Moniz de Sa M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. MolBiolEvol 12:481–493
Dvořák J, Zhang HB, Kota RS, Lassner M (1989) Organization and evolution of the 5S ribosomal RNA gene family in wheat and related species. Genome 32:1003–1016
Eickbush TH, Eickbush DG (2007) Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175:477–485
Friesen N, Fritsch R, Blattner FR (2006) Phylogeny and new intragenic classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso 22:372–395
Fritsch RM, Blattner FR, Gurushidze M (2010) New classification of Allium L. Subg. Melanocrommyum (Webb &Berthel) Rouy (Alliaceae) based on molecular and morphological characters. Phyton 49:145–220
Funari SS, Rapp G, Perbandt M, Dierks K, Vallazza M, Betzel C, Erdmann VA, Svergun DI (2000) Structure of free Thermus flavus 5S rRNA at 1.3 nm resolution from synchrotron X-ray solution scattering. J Biol Chem 275:31283–31288
Harvey MJ (1995) Onion and other cultivated alliums. Allium spp. (Liliaceae). In Evolution of crop plants, Smartt J and Simmonds NW, eds., 2nd ed., Wiley Pub.Co. New York pp. 344–350
Hizume M (1994) Allodiploid nature of Allium wakegi Araki revealed by genomic in situ hybridization and localization of 5S and 18S rDNAs. Jpn J Genet 69:407–415
Hori H, Osawa S (1986) Evolutionary change in the 5S rRNA secondary structure and a phylogenic tree of 352 5S rRNA species. Biosystems 19:163–172
Jeffrey AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JAL (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat Genet 6:136–145
Kellogg E, Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140:325–343
Kim N-S, Kuspira J, Armstrong K, Bhambhini R (1993) Genetic and cytogenetic analyses of the A genome of Triticum monococcum. VIII. Localization of rDNAs and characterization of 5S rRNA genes. Genome 36:77–86
Kiparisov S, Petrov A, Meskauskas A, Sergiev PV, Dintsova OA, Dinman JD (2005) Structural and functional analysis of 5S rRNA in Sacchromyces cereviceae. Mol Gen Genomics 274:235–247
Lee SH, Seo BB (1997) Chromosomal localization of 5S and 18S–26S rRNA genes using fluorescence in situ hybridization in Allium wakegi. Kor J Genet 19:19–26
Lee SH, Do GS, Seo BB (1999) Chromosomal localization of 5S rRNA gene loci and implications for relationships within Allium complex. Chrom Res 7:89–93
Li Q-Q, Zhou S-D, He X-J, Yu Y, Zhang Y-C, Wei X-Q (2010) Phylogeny and biogeography of Allium (Armaryllidaceae: Alliaceae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Annals Bot 106:709–733
Liu Z-L, Zhang D, Wang X-Q, Ma X-F, Wang X-R (2003) Intragenomic and intergenomic 5S rRNA sequence variation in five Asian pines. Am J Bot 90:17–24
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nielson JN, Hallenburg C, Frederiksen S, Sørensen PD, Lombolt B (1993) Transcription of human 5S rRNA genes is influenced by an upstream DNA sequence. Nucleic Acids Res 21:3631–3636
Park YK, Park KC, Kim N-S (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Mol Cells 10:18–24
Park HM, Jeon EJ, Waminal NE, Shin KS, Kweon SJ, Park BS, Suh SS, Kim HH (2010) Detection of transgenes in three genetically modified rice lines by fluorescence in situ hybridization. Genes Genom 32:527–531
Ricroch A, Yockteng R, Brown SC, Nadot S (2005) Evolution of genome size across some cultivated Allium species. Genome 48:511–520
Sanderson MJ, Doyle JJ (1992) Reconstruction of organismal and gene phylogenies from data on multigene families: concerted evolution, homoplasy, and confidence. SystBiol 41:4–17
Sastri DC, Hilu K, Appels R, Lagudah ES, Playford J, Baum BR (1992) An overview of evolution in plant 5S DNA. Pl Syst Evol 183:169–181
Schneeberger RG, Creissen GP, Cullis CA (1989) Chromosomal and molecular analysis of 5S RNA gene organization in the flax, Linum usitatissimum. Gene 83:75–84
Scoles GJ, Gill BS, Xin Z-Y, Klarke BC, McIntyre CL, Chapman C, Appels R (1987) Frequent duplication and deletion events in the 5S RNA genes and the associated spacer regions of the Triticeae. Pl Syst Evol 160:105–122
Seo JH, Seo BB (2010) Independent chromosomal localization of two different 5S rDNA of Allium victorialis var. plathphyllum by sequential fluorescence in situ hybridization in accordance with sequence polymorphism. Genes Genom 32:129–135
Seo BB, Lee SH, Mukai Y (1997) Physical mapping of 5S and 18S–26S ribosomal RNA gene familes in Allium victorialis var. platyphyllum. J Plant Biol 40:132–137
Shibata F, Hizume M (2002) Evolution of 5S rDNA units and their chromosomal locations in Allium cepa and Allium schoenoprasum revealed by microdissection and FISH. Theor Appl Genet 105:167–172
Smith MW, Meskaukas A, Wang P, Sergiev PV, Dinman JD (2001) Saturation mutagenesis of 5S rRNA in Saccharomyces cereviceae. Mol Cell Biol 21:8264–8275
Son J-H, Park K-C, Kim T-W, Park Y-J, Kang J-H, Kim N-S (2010) Sequence diversification of 45S rRNA ITS, trnH-psbA spacer, and matK genic regions in several Allium species. Genes Genom 32:165–172
Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J (2002) 5S ribosomal RNA database. Nucleic Acids Res 30:176–178
Tashiro Y (1984) Genome analysis of Allium wakegi Araki. J Jpn Hort Sci 52:399–407
Tyler BM (1987) Transcription of Neurospora crassa 5S rRNA gene requires a TATA box and three internal elements. J Mol Biol 196:801–811
Acknowledgments
This study was supported by the Bio-Green 21 Project (Grant 200804010340600080100) of the Rural Development Administration (RDA), Republic of Korea. JHS and SIL are the recipients of the BK21 Program supported by the Ministry of Education, Science and Technology (MEST), Republic of Korea. Authors express thanks to Dr. G. Fedak for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
J.-H. Son and K.-C. Park contributed equally to this work.
Rights and permissions
About this article
Cite this article
Son, JH., Park, KC., Lee, SI. et al. Sequence Variation and Comparison of the 5S rRNA Sequences in Allium Species and their Chromosomal Distribution in Four Allium Species. J. Plant Biol. 55, 15–25 (2012). https://doi.org/10.1007/s12374-011-9185-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-011-9185-4