Abstract
Objective
To evaluate the feasibility of kinetic modeling-based approaches from [18F]-Flobetaben dynamic PET images as a non-invasive diagnostic method for cardiac amyloidosis (CA) and to identify the two AL- and ATTR-subtypes.
Methods and Results
Twenty-one patients with diagnoses of CA (11 patients with AL-subtype and 10 patients with ATTR-subtype of CA) and 15 Control patients with no-CA conditions underwent PET/CT imaging after [18F]Florbetaben bolus injection. A two-tissue-compartment (2TC) kinetic model was fitted to time-activity curves (TAC) obtained from left ventricle wall and left atrium cavity ROIs to estimate kinetic micro- and macro-parameters. Combinations of kinetic parameters were evaluated with the purpose of distinguishing Control subjects and CA patients, and to correctly label the last ones as AL- or ATTR-subtype. Resulting sensitivity, specificity, and accuracy for Control subjects were: 0.87, 0.9, 0.89; as far as CA patients, the sensitivity, specificity, and accuracy were respectively 0.9, 1, and 0.97 for AL-CA patients and 0.9, 0.92, 0.97 for ATTR-CA patients.
Conclusion
Pharmacokinetic analysis based on a 2TC model allows cardiac amyloidosis characterization from dynamic [18F]Florbetaben PET images. Estimated model parameters allows to not only distinguish between Control subjects and patients, but also between AL- and ATTR-amyloid patients.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Amyloidosis can involve different organs and it is characterized by the deposition within the extracellular space of protein fibrils derived by misfolded proteins.1 Cardiac involvement is a major determinant in this disease, and it is common, especially in immunoglobulin light-chain amyloidosis (AL) and in transtiretine-related amyloidosis (ATTR).2,3
AL is one of the most frequent forms of amyloidosis; in this form of amyloidosis a plasma-cell clone produces monoclonal immunoglobulins characterized by structural instability which led to fragmentation and release of free light chains that undergo to misfolding with formation of protofibrils and amyloid fibrils especially in heart and kidney. In addition to structural alteration of the tissues in which they are deposited, AL amyloid fibrils cause direct toxic damage, particularly in the heart. The incidence of this disease increases with age, doubling after the age of 65 with a mean age at diagnosis of 63 years and mild male predominance.4
The median survival of not-treated patients is about 6 months from diagnosis,5 many patients get a late diagnosis; it is estimated that in about 40% of patients the definitive diagnosis is made about 12 months after the onset of symptoms. In the absence of cardiac involvement, life expectancy can be of several years.4
ATTR is characterized by the extracellular deposition of amyloid fibrils composed by transthyretin (TTR), a protein synthesized predominantly by the liver and which physiologically binds and transports thyroid hormone thyroxine and retinol-binding protein. Native TTR circulates in plasma as a soluble tetramer; in the forms of ATTR amyloidosis the bonds between the 4 monomers of the protein are weakened allowing the monomers to separate and reassemble in an anomalous way, forming insoluble fibrils that accumulate in the extracellular matrix. At the basis of the weakening of protein bonds, there may or may not be mutations in the gene that codes for TTR which divide ATTR into two main forms: “hereditary amyloidosis related to transthyretin” when there are specific gene mutations and “senile systemic amyloidosis” when the weakening of the bonds is linked to an intrinsic characteristic of proteins due to aging. The disease incidence currently estimated is not reliable; the condition is often underdiagnosed and only with the recent use of diphosphonate scintigraphy has the diagnostic accuracy improved in this type of disease. Autopsy studies suggest that the prevalence of ATTR can reach 25% of subjects over 80 years of age and 13% among patients with heart failure with preserved ejection fraction.6 The median survival of patients with untreated hereditary disease is nearly 10 years; while the survival of patients with senile form is about 47 months. Overall, the median survival is significantly higher than that of the AL forms and this aspect is linked to the toxic effect of the light chain fibrils.
Unfortunately, early clinical symptoms of cardiac amyloidosis (CA) are uncharacteristic, and they are often confused with the signs of other mimicking diseases such as hypertensive or hypertrophic heart disease. When CA is present, it appears to have an extremely poor prognosis and becomes the main cause of morbidity and mortality. Therefore, being able to early and reliably diagnose the presence of the disease would be of the utmost importance. Furthermore, the therapies that can be used to treat the two sub-types of amyloidosis are also different: AL patients undergo chemotherapy or stem cell transplantation, while ATTR patients are subjected to small RNA-silencing molecules or stabilizers.7,8 Therefore, it would be useful to clearly differentiate between AL or ATTR sub-type.
The gold standard for definite diagnosis of CA and its sub-type is endo-myocardial biopsy with immunochemistry or mass spectroscopy.9 However, it is an invasive technique with a high rate of complications and not unlimitedly repeatable. So, alternative diagnostic tools, such as imaging technologies, could provide a valid, non-invasive, and repeatable alternative. Recently, several studies have been performed to evaluate the ability to detect CA by exploiting imaging methodologies, such as echocardiography, cardiac magnetic resonance imaging (CMR), bone-scintigraphy/SPECT, and/or PET.10,11,12,13,14,15
Promising results have been obtained from the combination of several imaging techniques and analyses.16,17,18
In literature, several studies have shown how molecular imaging after injection of bisphosphate derivative tracers labeled with the 99mTc isotope allows to identify ATTR-CA,16,19,20 as well as it can make it possible to distinguish between the CA-ATTR and CA-AL subjects.21
Recently, PET imaging with [18F]Florbetaben has been studied for CA characterization.11,12,15,22 Usually, one or two static 3D PET images are acquired, at about 5 and 20 minutes after injection, or several minutes later (from 40 minutes, to more than 1 hour), and some indexes can be extracted from the acquired images.12,18,23
Using dynamic rather than static PET protocols, a range of diagnostic indices can be computed, such as retention index (RI), target to background ratio (TBR), or kinetic parameters, which have been proven to allow distinguishing subjects with CA from Control groups.11,12,24,25 While the feasibility of [18F]Florbetaben dynamic PET imaging in diagnosing CA15,22 has already been demonstrated, indexes, or a combination of them, that could significantly distinguish between the two AL- and ATTR- sub-types of CA from PET images have not yet been identified.
The aim of the present work is to evaluate the feasibility of a kinetic modeling-based approach applied to [18F]Florbetaben dynamic PET images to identify parameters that allow to distinguish between subjects with amyloidosis and a Control group, and among the patients with CA, that allow to separate the two AL and ATTR subgroups.
Materials and Methods
Subjects and Study Design
Twenty-one patients with systemic amyloidosis and heart involvement were included in the study, in particular 11 patients with AL, and 10 patients with ATTR types, respectively. Moreover, 15 Control-patients with the clinical suspicion of CA, which received an alternative diagnosis, such as hypertensive cardiac hypertrophy, left-ventricle hypertrophy secondary to aortic-valve stenosis, or primary hypertrophic cardiomyopathy, were included in this retrospective study.
Patients with ischemic heart disease, chronic liver disease or severe renal failure were excluded.
Diagnosis of CA was based on clinical examination, biomarkers positivity (N terminal fraction of pro-brain natriuretic peptide, high sensitivity troponin T, immunoglobulin light-chains in serum and/or in urine), electrocardiogram, echocardiography, bone-scintigraphy, cardiac magnetic resonance (CMR), and by histological evidence of amyloid deposition according to the most recent cardiological evidence and guide-lines.26,27
The study was approved by the institutional ethics committee and by the AIFA (Agenzia Italiana del Farmaco) committee; all subjects signed an informed consent form. The study complied with the Declaration of Helsinki.
Subjects’ characteristics according to the type of the cardiac disease are given in Table 1; statistical significance of the comparison between groups is also given. Left ventricle septal thickness, left ventricle posterior wall (PW) thickness, and ejection fraction (EF) value were evaluated from echocardiography.
Dynamic Cardiac PET Data Acquisition and Time-Activity-Curves Generation
PET/CT imaging was obtained using a Discovery RX VCT 64-slice tomography (GE Healthcare, Milwaukee, WI, USA). Low-dose computed tomography (CT) (tube current 30 mA, tube voltage 120 kV) was firstly performed through the heart for attenuation correction for an effective dose of 1 mSv. Then, a 3-dimensional (3D) list-mode dynamic PET acquisition was performed for about 40 minutes. The PET acquisition started at the same time as the injection of an intravenous bolus of [18F]Florbetaben (300 Mbq/1 mL) followed by a saline flush of 10 mL (1 mL s−1); the mean whole-body exposure due to the radiopharmaceutical was 5.8 mSv.
Dynamic images were reconstructed from the list-mode data using 27 (for 11 subjects) or 28 frames (for 25 subjects) with different timings: 10 × 5, 10 × 30, 7 × 300 seconds (or 8 × 300 seconds) for a total of about 40 (or 45, depending on the subject’s ability to stay in the scanner) minutes; in particular, 25 subjects were scanned 40 minutes and 11 subjects for 45 minutes. Dynamic images were reconstructed using the ordered-subset expectation maximization (OSEM) iterative technique, with 3 iterations and 21 subsets; a 128 × 128 pixels matrix × 47 slices were obtained for each time frame.
Reconstructed dynamic 3D PET images were then exported to a remote workstation to be analyzed. Emission values were converted to Standardized Uptake Values (SUV), defined as the mean voxel intensity within the volumetric region of interest (ROI) normalized by the whole-body concentration of the injected radioactivity and multiplied by body mass.
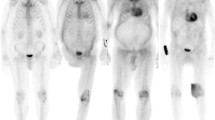
Time-activity curves (TACh) for the myocardial region were then extracted from the 4D dynamic reconstructions by manually drawing an ROI in the transaxial slice of the dynamic frame where the left-ventricular shape was best visualized; mean SUV value inside the ROI corresponds to a point in the TACh curve; similarly, plasma TACp was also obtained by manually tracing a circular ROI of about 1 cm in diameter inside the left-atrium cavity. In Figure 1, representative dynamic images with selected ROIs, and relevant TACh and TACp are shown, for a Control subject (#1), an AL patient (#4), and an ATTR patient (#5).
Given the TACh(t) from heart-tissue and TACp(t) from blood, kinetic modeling can be applied to estimate the rate parameters that characterize the tracer behavior.
In order to evaluate the reproducibility of the method, since the only variability that may arise concerns the selection of ROIs for the construction of TACs, the ROIs were selected several times; in particular, a nuclear physician (DG) with more than 10 years of experience in cardiac nuclear medicine repeated the selection twice, with about a week between the first and second measurements, in order to evaluate the intra-observer variability. Then, for evaluating the inter-observer variability, a second nuclear physician (AG, more than 10 years of experience) further performed the measurements. Both observers were blinded to each other’s results.
Kinetic Modeling and TAC Fitting Method
Kinetic analysis of regional time-activity curves was performed using the two-tissue-compartment model, 2TC.23,28,29,30,31 According to such a model, a set of ordinary differential equations describes the time course of the tracer concentrations in the target tissue30:
where, for a 2TC model:
were \( C_{\text{f}} \left( t \right) \) and \( C_{\text{b}} \left( t \right) \) are the concentration of the free and bound compartments, respectively. From Eq. 1, K1, k2, k3, k4 are the tracer’s rate constants, i.e., the model parameters that need to be estimated by fitting operation. In this work, we considered the less complex irreversible model, described by fixing \( k_{4} = 0 \).
The total tracer concentration of a tissue region in time can be modeled as30:
where \( f_{\text{v}} \) is the fractional volume of blood in tissue, \( d_{\text{k}} \) is the radioactive decay constant, \( {\text{TAC}}_{\text{p}} \left( t \right) \) is the measured tracer concentration in plasma, \( {\text{TAC}}_{\text{wb}} \left( t \right) \) is the measured whole blood concentration, and \( {\text{IRF}}_{\text{c}} \left( t \right) \) represents the impulse response function for the \( c \)-th tissue compartment in such a way that the convolution with the arterial input function, \( {\text{TAC}}_{\text{p}} \left( t \right) \), yields its instantaneous concentration.
Parameter estimation from image-derived clinical TACs was performed by an analytic fitting method, as described in Ref. 30. The parameters estimated via model fitting were \( P = \left[ {K_{1} ,k_{2} ,k_{3} ,f_{\text{v}} } \right] \).
In Figure 2, measured and fitted curves obtained with the 2TC model are shown for (a) a Control subject (#15), (b) an AL-patient (#10), and (c) an ATTR-patient (#8).
Example curves of Control subject (#15), AL-(#10) and ATTR-(#8) patients, and relevant fitting curves. Quantitative parameters obtained by fitting analysis are shown in Table 2
ROIs selection, TACs generation extraction and kinetic data fitting were implemented in MATLAB™ (The MathWorks, Inc., Natick Massachusetts, United States), version 2019b.
Parameters and Indexes Used for Analysis
In order to quantitatively distinguish the three subject groups (Controls, AL- and ATTR-amyloidosis patients), the kinetic parameters obtained by model fitting were considered (i.e. K1, k2, k3), together with the following macro-parameters: the total volume of distribution (VT = K1/k2) and the net influx rate (Ki= K1k3/(k2 + k3)). The retention index (RI) and target to background ratio (TBR), often used in literature for characterizing the presence of amyloidosis,12,22,23,25 were also considered. The RI index was calculated as the mean SUV values between about 18 and 33 minutes after tracer injection, divided by the integral of the blood pool SUV between 0 and about 25 minutes, the midpoint of 18-33 minutes.22,25; more analytically:
The TBR was calculated as the ratio of the mean LV myocardial SUV to the blood pool SUV between about 5 and 30 minutes.23
Akaike information criterion (AIC) index29,32 was used as a model selection criterion to compare the adequacy of data fitting between the 1TC and the 2TC reversible and non-reversible model in a more quantitative manner. AIC is given by:
where N is the number of time points; \( \hat{C} \) is the model curve; p is the number of parameters to be estimated in the fitting.
Statistical Analysis Methods
Continuous variables are expressed as mean ± standard deviation or median and range, as appropriate.
The parametric hypothesis test of normality of the variables was evaluated according to the Shapiro-Willis normality test. When the parameters and indexes evaluated do not follow Normal Distribution, the Kruskal-Wallis test was used for comparisons between more than two group samples, and the Mann-Whitney test was used for comparison between two group samples. A 2-tailed p-value < 0.05 was considered significant.
Two-parameters group classification was performed by a support vector machine (SVM) classifier that automatically returns the equation of the line that separates the two groups. For estimating the quality of classification, a k-fold cross-validation was performed on the SVM training data, repeating the analysis 10 times (i.e., k = 10) and changing each time the data to be used in the train and in the validation sets.33 At each ‘fold’, the separation line’s equation is determined from the training set, and then the validation set is used to assess how well that line is able to classify new data. Eventually, the method returns 10 line equations and a goodness index of the results, i.e., the mean squared error in percent.
Statistical analysis was performed using MATLAB Statistics and Machine Learning Toolbox™ (version 2019b).
Results
To evaluate the adequacy of data fitting between the models, AIC indexes (mean ± standard deviation) were evaluated for the 1TC, 2TC irreversible, and 2TC reversible models, resulting in AIC values of 157.80 (±36.32), 169.68 (±40.24), and 172.86 (±41.03), respectively. The difference in the AIC index values between the three models resulted non-significant.
As an example of the results of the pharmacokinetic analysis, Table 2 shows the kinetic parameters estimated for three subjects whose measured TACs and fitting results are shown in Figure 2: a Control subject C, an AL-amyloidosis patient and an ATTR-amyloidosis patient.
From the Shapiro-Willis normality test, the values of the kinetic parameters resulted to follow a non-gaussian distribution, so the Kruskal-Wallis test was selected for the subsequent statistical analysis.
Intra- and inter-observer data also resulted to belong to a non-gaussian distribution. Therefore, the Mann-Whitney test was adopted for the reproducibility analysis. The intra-observer analysis gave the following p-values: 0.92 for K1, 0.89 for k2, 0.71 for k3, and 0.51 for Ki; the inter-observer analysis gave the following p values: 0.95 for K1, 0.72 for k2, 0.70 for k3, and 0.89 for Ki. Those results support the idea of the absence of a statistically significant difference between the estimated values of the model parameters, both in terms of inter- and intra-observer variability.
The kinetic micro-parameters (K1, k2 and k3) and macro-parameters (Ki, VT) estimation results are shown in Figure 3 via boxplots for Control subjects (C), and patients with AL- and ATTR-amyloidosis. The Kruskal-Wallis resulting p values are also shown for each parameter.
The boxplots relevant to the RI and TBR indexes are shown in Figure 4, for C subjects and patients with AL- and ATTR-amyloidosis; for each index, the Kruskal-Wallis resulting p values are shown.
In Figures 3 and 4, it can be seen that considering only a single parameter it was not possible to classify the three groups of subjects; therefore, a combination of parameters had to be considered, i.e., the pair K1-k2 to separate the Control subjects from the CA patients and, once a diagnosis of CA was determined, to evaluate the corresponding k2-Ki parameters to classify the AL-CA patients and ATTR-CA by the SVM classifier.
Accordingly, in Figure 5, scatter plots of K1 vs. k2 (in A) for Controls and patients (both AL and ATTR sub-groups), and of k2 vs. Ki (in B) values just for AL and ATTR patients are shown.
In Figure 5A, the solid blue line and the shadowed area represent the mean line and ±std resulting from the k-fold cross-validation, respectively. The corresponding equation is: k2 = − 0.46(± 0.07)*K1 + 0.62 (± 0.06). The mean squared error in the cross-validation analysis resulted equal to 11%.
In Figure 5B, the solid red line and the shadowed area represent the mean line and ±std resulting from the k-fold cross-validation, respectively. The corresponding equation is: Ki = 9.97e−05(± 11.57e−4)*k2 + − 0.015(± 0.017). The mean squared error in the cross-validation analysis resulted equal to 9.52%
Combining the results in Figure 5, it should be possible to classify a new subject as part of the Control, AL, or ATTR group: from the combination of K1 and k2 values, it is possible to evaluate if the subject has pathology; then, from the combination of k2 and Ki values it is possible to distinguish the sub-type of CA (AL or ATTR). In Table 3 the resulting confusion matrix is shown: it describes, for each group, how many subjects were correctly estimated and how many incorrectly by the proposed method; in the latter case, it shows which group they are estimated to belong to.
To further assess the robustness of the proposed classification method, we verified whether the lines separating Controls from CA patients (in Figure 5A) and the ATTR-CA from the AL-CA patients (in Figure 5B) are also valid on the data processed by the second observer.
Sensitivity, specificity, and accuracy for each group (i.e., Controls, AL-CA and ATTR-CA) were evaluated for the data obtained from both observer #1 and observer #2, to analyze the diagnostic potential of the proposed method. Results are reported in Table 4.
Discussion
The study of cardiac amyloidosis by means of PET imaging following the injection of [18F]Florbetapir23 or [18F]Florbetaben22 is highly regarded in recent years.11 The main indexes considered to distinguish the presence or the absence of the pathology are RI and TBR, and they are derived mainly from static images generated by the mean SUV values between prefixed times, such as 10-30 minutes after injection.22,23 In Ref. 29 quantitative kinetic parameters were considered for brain amyloidosis; starting from dynamic [18F]Florbetaben PET images, kinetic models were applied for extracting quantitative parameters that lead to discrimination between β-amyloid-positive and -negative subjects. Kinetic modeling has also been used for characterizing the presence of amyloidosis in the heart: in Ref. 25 by [11C]PIB PET data were analyzed using 1-tissue and 2-tissue reversible and irreversible models.
So far, no studies are available in the literature that uses PET kinetic modeling for characterizing cardiac amyloidosis, following a [18F]Florbetaben injection. Moreover, at the moment, no accurate workflow has been suggested to distinguish whether the patient suffers from AL or ATTR sub-type of amyloidosis, in case PET images are to be used. In any case, there is currently no single diagnostic tool that allows to make a differential diagnosis between AL and ATTR alone.
Therefore, the main objective of the present work was to apply kinetic modeling to [18F]Florbetaben dynamic cardiac PET images so to identify a set of quantitative parameters capable of (i) determining the presence of amyloidosis in the heart and (ii) identifying the AL or ATTR sub-type of amyloidosis.
The two-tissue irreversible kinetic model has been adopted in this work; it implies the estimation of three kinetic parameters \( K_{1} , k_{2} , k_{3} \) plus the fractional volume of blood \( f_{\text{v}} \). The fitting results, shown as an example in Figure 2 and Table 2, demonstrate how the 2TC model provides a very good fitting on acquired data.
The boxplots shown in Figure 3 summarize the distribution of these kinetic parameters between the three groups of subjects. From a qualitative point of view, it could be stated that both \( K_{1} \) and \( k_{2} \) parameters allow to distinguish the Control group from amyloidosis patients, but they are not capable to distinguish the two AL- and ATTR-amyloidosis groups. In the chosen model, the parameter \( K_{1} \) describes the tracer flow rate from blood to the tissue; so, from the results obtained, it can be infer that this flow is higher in the Controls compared to CA patients. This can also be seen in the representative curves shown in Figure 1: the first part of the TAC curves is closely related to the speed with which the tracer passes from the blood to the cardiac tissue and, in Figure 1, it is clear that the curve relevant to the Control subject has a steeper rise compared to the other TAC curves.26
The \( k_{2} \) parameter, instead, provides information on the reversibility of the tracer from the tissue to the blood; in the event that a rather large quantity of tracer returns to the blood, the corresponding TAC curve, which is relative to the myocardial tissue after a certain time, decreases. This actually happens for the Control subjects and, albeit to a lesser extent, for the ATTR-CA patients (see examples in Figure 1 and 2). Also, from the results in Figure 3 and the example subjects in Table 2, it can be seen that both Controls and ATTR-CA patients have a value for \( k_{2} \) which is higher compared to that of AL-CA patients.
This leads us to consider the fact that a 1TC (2-k parameters) model would most likely be sufficient to describe the tracer kinetics on Controls’ myocardial tissues and probably also those with ATTR-CA; this consideration is reinforced by the fact that the value of the parameter \( k_{3} \) for both the Controls and the ATTR-CAs patients is very low, as shown by the box-diagram relating to the parameter \( k_{3} \) (Figure 3) and the example subjects in Table 2. The 1TC model, however, fails to describe the behavior of the tracer for AL subjects, and in fact, the value of k3 for these patients is quite greater than 0. In fact, the parameter \( k_{3} \) in the kinetic models describes a passage/metabolization of the tracer into tissue’s compartments, irreversibly; this leads to the entrapment of the tracer in the tissue resulting in high values in the final part of the corresponding TAC curve, as it happens for AL-CA subjects (see TAC examples in Figures 1 and 2 for the AL-CA subject).
Whether or not [18F] Florbetaben is metabolized by the myocardium is not known at the moment. Senda et al.,34 have shown that in healthy subjects, [18F]Florbetaben is metabolized within minutes of injection; however, they performed the study from the plasma, and it is not known whether this metabolism occurs in an equivalent way in all the anatomical areas of the subject, or whether, for example, the heart cells metabolize it or not. From the fact that the 1TC model (i.e., with 2 k parameters) seems sufficient to describe the trend of the Control subjects, as well as ATTR-CA data, would lead to thinking that [18-F]Florbetaben is not metabolized in cardiac cells, while it would be entrapped into cardiac AL-CA deposits. Obviously, this consideration requires more in-depth studies, which go beyond the scope of this work.
Moreover, before deciding to use the irreversible 2TC model, also 2TC reversible model was tested (results not reported), which implies the estimation of the additional parameter k4, in Eq 2. The two models gave very similar results, i.e., the fitted curves were almost completely overlapping, and the fitting residuals were not significantly different in all the three groups of data.
To compare the adequacy of data fitting between the 1TC and the 2TC reversible and non-reversible model in a more quantitative manner, AIC was used as a model selection criterion. The difference in the AIC index values between the three models was non-significant. Therefore, even from more quantitative analysis, the choice of the model to use remains rather indifferent.
As it is well known, increasing the number of unknown parameters to be estimated leads to greater computational complexity and uncertainty in the estimate, so in the present work, it was decided to use the 2TC irreversible model in the statistical evaluations of the quantitative kinetic micro-parameters (\( K_{1} , k_{2} , k_{3} \)) and macro-parameters (\( K_{i} , V_{\text{T}} \)).
Micro-parameter \( k_{3} \) and macro-parameters \( K_{i} \) and \( V_{\text{T}} \) show a very high variability for AL patients and this feature is evident, even if with less entity, also for the indexes RI and TBR, shown in Figure 4. This is not new in literature: a higher variability in RI and TBR indexes for AL-patients was demonstrated in Ref. 22. Larger \( k_{3} \)value for AL patients than Controls and ATTR subjects is also reflected in the estimates of \( k_{i} \) (which is, by definition, proportional to \( k_{3} \)estimates) and \( V_{\text{T}} \) macro-parameters. The p values resulted from the Kruskal-Wallis test are < 0.05 for all the kinetic micro- and macro-parameters. The boxplots shown in Figure 4 are relevant to the RI and TBR indexes; the medians of the three groups of subjects are quite different for both the RI index and the TBR; however, the variability is such as not to allow the groups to be distinguished from each other, with the exception of Control entities and AL-CAs.
From the pairwise analysis on the Kruskal-Willis test (Figures 3 and 4), the following considerations arise: \( K_{1} \) and \( k_{2} \), and RI significantly distinguish Control subjects from amyloid patients; the macro-parameter \( {\text{K}}_{\text{i}} \) allows to significantly distinguish AL- from ATTR-patients, and Control subjects from AL, too. The ability of the RI index to distinguish also AL- from ATTR-patients confirms what was found in Ref. 12. TBR significantly distinguishes Control subjects from AL-CA and from ATTR-CA, but it is not able to distinguish AL-CA from ATTR-CA, resulting useless for diagnosis considering it as the only parameter, without information given by other indices.
Therefore, the workflow we recommend is to perform a pharmacokinetic study of myocardial TACs using an irreversible 2TC model, and to classify the subject according to the resulting estimates of the kinetic parameters, in two steps: first, considering a pair of parameters, such as \( K_{1} \) and \( k_{2} \) that is able to distinguish between Control subjects and amyloidosis patients (see Figure 5A), and then, if it results that the subject is a CA patient, considering the combination of \( k_{2} \) and \( K_{i} \), which has been proved capable of separating the AL and ATTR sub-type of CA (see Figure 5B). Such a combination of the estimated parameters not only can distinguish the patients with amyloidosis from the normal subjects but also the two types of amyloid pathology.
From Figure 5A, it is evident that all the lines calculated by the SVM classifier and obtained from the cross-validation separate the two groups of subjects in the same way: there are always two Control subjects and two CAs that are misclassified. Instead, from Figure 5B, the lines calculated by the SVM classifier in the cross-validation classify well all the ATTR-CA patients, while there are two AL-CA patients who, depending on the line considered, can be classified incorrectly. Therefore, even if there is some variability between the lines calculated in the classification (see the shaded areas in Figure 5A and B), the areas that separate the pairs of groups are large enough to allow for classification with a low margin of error.
It is worth to note that, since the main objective of Figure 5B was to demonstrate that the simultaneous evaluation of the kinetic parameters k2-Ki allows to correctly classify ATTR-CA and AL-CA patients, in Figure 5B all CA patients were considered, i.e., even those incorrectly classified as Control subjects by Figure 5A.
From further analysis of the data incorrectly classified by Figure 5A (i.e., Controls classified as CA and, vice versa, CA classified as Controls), we saw that the two Control subjects are ultimately classified as ATTR-CA (see Table 3) and this is not surprising, given that the wash-out part of the TAC curves of the Control subjects and the ATTR-CAs are very similar (see Figures 1 and 2), and this leads to similar values of the parameters related to the wash-out, namely k2 and Ki. Furthermore, with regard to the two CA patients mistakenly classified as non-CA in Figure 5A, when represented in the k2-Ki graph (Figure 5B), they have been correctly classified, one as ATTR-CA and the other as AL-CA.
Accordingly, the results shown in Table 4 are obtained taking into account also the erroneous subjects’ classification in the first step of the method (i.e., from data of Figure 5A). However, the data obtained confirm the clinical potential of the proposed method: the accuracy is very high (≥ 0.89) for all the subgroups, while sensitivities and specificities, ranging between 0.87 and 1, confirm that the use of the method also in clinical practice could be of interest. The sensitivity, specificity, and accuracy values obtained from the data of the second observer are also very promising since they do not differ much from those obtained from the analysis of the data of the first observer. This reinforces the idea that the method will perform well even on new, unseen data.
Moreover, the reproducibility analysis by the Mann-Whitney test gave good results confirming a not significant intra- and inter-observer variability of the results.
Currently, the RI index is often used in the clinic, but it has not many advantages with respect to the proposed method as far as the acquisition time: also for RI evaluation, the exam requires the subject is in the scanner for several minutes (up to 30 minutes); moreover, although the images analyses are different, in both the methods (i.e., RI evaluation and our method), they need the ROI selection followed by a fully automatic procedure and therefore a very similar amount of task for the operator. Therefore, the method proposed here can be considered as an alternative to that suggested in the literature, even in clinical practice.
As previously stated, in compartmental kinetic models the parameter K1 is proportional to the velocity of transport of the tracer from the vessels to the tissue, and therefore it is also closely related to myocardial blood flow (MBF). Furthermore, both K1 and MBF in turn depend on the patient’s hemodynamic status; for this reason, often, to reduce this dependence a normalization of the K1 is performed with the rate-pressure product, which is closely linked to the flow at rest. In order to perform this normalization, it is also necessary to have physiological data such as the pressure and heart rate of the subject under study. In accordance with our acquisition protocols, also the heart rate and pressure were available for the subjects under study; we then analyzed our data after having normalized the parameter K1 for the rate-pressure product: the results obtained were very similar to those obtained without normalization. Certainly, to be sure that the contribution of normalization is negligible, it would be necessary to carry out a more in-depth analysis with a higher number of subjects, which is intended as a future follow-up on this work. Ultimately, the main objective of the present work was to propose a method that uses only kinetic parameters to obtain a diagnosis of CA and of its subtypes, as a proof of concept. An accurate validation analysis, with a higher number of subjects, even obtaining data from a multicentric study, is the goal of a future work.
Lastly, since a prompt diagnosis of CA is potentially life-saving, particularly in patients with a cardiac involvement of AL amyloidosis, the use of kinetic parameters as discrimination may strengthen the accuracy of [18F]-Florbetaben PET which may represent a single non-invasive diagnostic tool for AL amyloidosis, thus potentially reducing the time between symptoms onset and diagnosis.
New Knowledge Gained
Kinetic model fitting on [18F]Florbetaben dynamic PET Images allow the characterization of cardiac amyloidosis.
Study Limitations
This study has some limitations.
First of all, the number of subjects evaluated is not very high, but it is in any case the largest sample ever evaluated by means of a dynamic PET approach with extraction of the described parameters; in addition, no patient in the ATTR group had a serum or urinary monoclonal component, neither an inherited form of the disease, therefore the results described cannot be extended to this specific patient subtypes.
Other limitation is the relative low number of subjects used in the analysis. This leads to several lacks: with the available data it is not possible to accurately establish whether [18F]Florbetaben is metabolized, and in what ways, by myocardium, but only guesses are possible. It is also not possible to accurately assess how much the values of parameter K1 can be influenced by the hemodynamic state of the patient, but only indicative results can be obtained, which have shown that this dependence does not significantly change the classification between Control subjects and CAs.
Conclusion
Cardiac amyloidosis characterization from [18F]Florbetaben PET images can be performed by kinetic data fitting according to 2TC model. Estimated parameters allow not only to distinguish normal subjects from patients, but also AL- from ATTR-amyloid patients.
Abbreviations
- AL:
-
Immunoglobulin light-chain derived amyloidosis
- ATTR:
-
Transthyretin derived amyloidosis
- AIC:
-
Akaike information criterion
- AUC:
-
Area under curves
- CA:
-
Cardiac amyloidosis
- CMR:
-
Cardiac magnetic resonance
- EF:
-
Ejection fraction
- MBF:
-
Myocardial blood flow
- OSEM:
-
Ordered-subset expectation maximization
- PW:
-
Posterior wall
- RI:
-
Retention index
- ROI:
-
Region of interest
- SUV:
-
Standardized uptake value
- SVM:
-
Support vector machine
- TAC:
-
Time/activity curve
- TBR:
-
Target to background ratio
References
Falk RH, Dubrey SW. Amyloid heart disease. Prog Cardiovasc Dis. 2010;52:347-61.
Alexander KM, Singh A, Falk RH. Novel pharmacotherapies for cardiac amyloidosis. Pharmacol Ther. 2017;180:129-38. https://doi.org/10.1016/j.pharmthera.2017.06.011.
Falk RH. Cardiac amyloidosis: A treatable disease, often overlooked. Circulation. 2011;124:1079-85.
Merlini G, Dispenzieri A, Sanchorawala V, Schönland SO, Palladini G, Hawkins PN, Gertz MA. Systemic immunoglobulin light chain amyloidosis. Nat Rev Dis Primers. 2018;4(1):38. https://doi.org/10.1038/s41572-018-0034-3.
Wechalekar AD, Schonland SO, Kastritis E, Gillmore JD, Dimopoulos MA, Lane T, Foli A, Foard D, Milani P, Rannigan L, Hegenbart U, Hawkins PN, Merlini G, Palladini G. A European collaborative study of treatment outcomes in 346 patients with cardiac stage III AL amyloidosis. Blood. 2013;121(17):3420-7. https://doi.org/10.1182/blood-2012-12-473066.
González-López E, Gallego-Delgado M, Guzzo-Merello G, de Haro-Del Moral FJ, Cobo-Marcos M, Robles C, Bornstein B, Salas C, Lara-Pezzi E, Alonso-Pulpon L, Garcia-Pavia P. Wild-type transthyretin amyloidosis as a cause of heart failure with preserved ejection fraction. Eur Heart J. 2015;36(38):2585-94. https://doi.org/10.1093/eurheartj/ehv338.
Skinner M, Sanchorawala V, Seldin DC, Dember LM, Falk RH, Berk JL, et al. High-dose melphalan and autologous stem-cell transplantation in patients with AL amyloidosis: An 8-year study. Ann Intern Med. 2004;140:85-93.
Ruberg FL, Berk JL. Transthyretin (ATTR) cardiac amyloidosis. Circulation. 2012;126:1286-300.
Gertz MA, Lacy MQ, Dispenzieri A, Hayman SR. Amyloidosis: Diagnosis and management. Clin Lymph, Myel Leuk. 2005;6:208-19. https://doi.org/10.3816/CLM.2005.
Kim YJ, Kim HK, Sohn DW, Park JB, Grogan M, Lee SP. Contemporary imaging diagnosis of cardiac amyloidosis. J Cardiovasc Imaging. 2019;27:1. https://doi.org/10.4250/jcvi.2019.27.e9.
Kim YJ, Ha S, Kim Y. Cardiac amyloidosis imaging with amyloid positron emission tomography: A systematic review and meta-analysis. J Nucl Cardiol. 2020;27(1):123-32. https://doi.org/10.1007/s12350-018-1365-x.
Kircher M, Ihne S, Brumberg J, Morbach C, Knop S, Kortüm KM, et al. Detection of cardiac amyloidosis with 18 F-Florbetaben-PET/CT in comparison to echocardiography, cardiac MRI and DPD-scintigraphy. Eur J Nucl Med Mol Imaging. 2019;46(7):1407-16. https://doi.org/10.1007/s00259-019-04290-y.
Bhogal S, Ladia V, Sitwala P, Cook E, Bajaj K, Ramu V, et al. Cardiac amyloidosis: An updated review with emphasis on diagnosis and future directions. Curr Probl Cardiol. 2018;43(1):10-34. https://doi.org/10.1016/j.cpcardiol.2017.04.003.
Santarelli MF, Genovesi D, Positano V, Di Sarlo R, Scipioni M, Giorgetti A, Landini L, Marzullo P. Cardiac amyloidosis detection by early bisphosphonate (99mTc-HMDP) scintigraphy. J Nucl Cardiol. 2020. https://doi.org/10.1007/s12350-020-02239-5.
Genovesi D, Vergaro G, Emdin M, Giorgetti A, Marzullo P. PET-CT evaluation of amyloid systemic involvement with [18F]-florbetaben in patient with proved cardiac amyloidosis: A case report. J Nucl Cardiol. 2017;24(6):2025-9. https://doi.org/10.1007/s12350-017-0856-5.
Santarelli MF, Scipioni M, Genovesi D, Giorgetti A, Marzullo P, Landini L. Imaging techniques as an aid in the early detection of cardiac amyloidosis. Curr Pharm Des. 2020. https://doi.org/10.2174/1381612826666200813133557.
Di Bella G, Pizzino F, Minutoli F, et al. The mosaic of the cardiac amyloidosis diagnosis: Role of imaging insubtypes and stages of the disease. Eur Heart J Cardiovasc Imaging. 2014;15(12):1307-15. https://doi.org/10.1093/ehjci/jeu158.
Kyriakou P, Mouselimis D, Tsarouchas A, Rigopoulos A, Bakogiannis C, Noutsias M, Vassilikos V. Diagnosis of cardiac amyloidosis: A systematic review on the role of imaging and biomarkers. BMC Cardiovasc Disord. 2018;18(1):221.
Cappelli F, Gallini C, DiMario C, Costanzo EN, Vaggelli L, Tutino F, et al. Accuracy of 99mTc-hydroxymethylene diphosphonate scintigraphy for diagnosis of transthyretin cardiac amyloidosis. J Nucl Cardiol. 2019;26:497-504.
Treglia G, Glaudemans AWJM, Bertagna F, Hazenberg BPC, Erba PA, Giubbini R, et al. Diagnostic accuracy of bone scintigraphy in the assessment of cardiac transthyretin-related amyloidosis: A bivariate meta-analysis. Eur J Nuclear Med Mol Imaging. 2018;45:1945-55.
Bokhari S, Castaño A, Pozniakoff T, Deslisle S, Latif F, Maurer MS. 99mTc-pyrophosphate scintigraphy for differentiating light-chain cardiac amyloidosis from the transthyretin-related familial and senile cardiac amyloidoses. Circ Cardiovasc Imaging. 2013;6(2):195-201.
Law WP, Moore PT, Ng ACT, Wang WYS, Mollee PN. Cardiac amyloid imaging with 18F-florbetaben PET: A pilot study. J Nucl Med. 2016;57(11):1733-9. https://doi.org/10.2967/jnumed.115.169870.
Dorbala S, Vangala D, Seme J, Strader C, Bruyere JR, Di Carli MF, et al. Imaging cardiac amyloidosis: A pilot study using 18F-florbetapir positron emission tomography. Eur J Nucl Med Mol Imaging. 2014;41(9):1652-62. https://doi.org/10.1007/s00259-014-2787-6.
Osborne DR, Acuff SN, Stuckey A, Wall JS. A routine PET/CT protocol with streamlined calculations for assessing cardiac amyloidosis using 18F-florbetapir. Frontiers Cardiovasc Med. 2015;2:1-9. https://doi.org/10.3389/fcvm.2015.00023.
Kero T, Sörensen J, Antoni G, Wilking H, Carlson K, Vedin O, et al. Quantification of 11C-PIB kinetics in cardiac amyloidosis. J Nucl Cardiol. 2018. https://doi.org/10.1007/s12350-018-1349-x.
Genovesi D, Vergaro G, Giorgetti A, Marzullo P, Scipioni M, Santarelli MF, Pucci A, Buda G, Volpi E, Emdin M. [18F]-Florbetaben PET/CT for differential diagnosis among cardiac immunoglobulin light chain, transthyretin amyloidosis, and mimicking conditions. J Am Coll Cardiol Imaging. 2020. https://doi.org/10.1016/j.jcmg.2020.05.031.
Gillmore JD, et al. BCSH Committee. Guidelines on the diagnosis ad investigation of AL amyloidosis. Br J Haematol. 2015;168:2017-218.
Bentourkia M, Zaidi H. Tracer kinetic modeling in PET. PET Clin. 2007;2(2):267-77. https://doi.org/10.1016/j.cpet.2007.08.003.
Becker GA, Ichise M, Barthel H, Luthardt J, Patt M, Seese A, et al. PET quantification of 18F-florbetaben binding to amyloid deposits in human brains. J Nucl Med. 2013;54(5):723-31. https://doi.org/10.2967/jnumed.112.107185.
Scipioni M, Giorgetti A, Della Latta D, Fucci S, Positano V, Landini L, Santarelli MF. Accelerated PET kinetic maps estimation by analytic fitting method. Comput Biol Med. 2018;99:221-35.
Gunn RN, Gunn SR, Cunningham VJ. Positron emission tomography compartmental models. J Cereb Blood Flow Metab. 2001;21:635-52. https://doi.org/10.1097/00004647-200106000-00002.
Burnham KP, Anderson DR. Model selection and multimodel inference: A practical information theoretic approach. 2nd ed. New York: Springer-Verlag; 2002. p. 70-5.
Arlot Sylvain, Celisse Alain. A survey of cross-validation procedures for model selection. Stat. Surv. 2010;4:40-79. https://doi.org/10.1214/09-ss054.
Senda M, Sasaki M, Yamane T, Shimizu K, Patt M, Barthel H, et al. Ethnic comparison of pharmacokinetics of 18F-florbetaben, a PET tracer for beta-amyloid imaging, in healthy Caucasian and Japanese subjects. Eur J Nucl Med Mol Imaging. 2015;42(1):89-96. https://doi.org/10.1007/s00259-014-2890-8.
Giorgetti A, Genovesi D, Emdin M. Cardiac amyloidosis: The starched heart. J Nucl Cardiol. 2018. https://doi.org/10.1007/s12350-018-1399-0.
Gillmore JD, et al. Nonbiopsy diagnosis of cardiac transthyretine amyloidosis. Circulation. 2016;133:2404-12.
Acknowledgments
Authors would like to thank Dr. Simona Muccioli for the precious support given in the clinical data analysis.
Disclosure
No potential conflict of interest relevant to this article was reported
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors of this article have provided a PowerPoint file, available for download at SpringerLink, which summarises the contents of the paper and is free for re-use at meetings and presentations. Search for the article DOI on SpringerLink.com.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Santarelli, M.F., Genovesi, D., Scipioni, M. et al. Cardiac amyloidosis characterization by kinetic model fitting on [18F]florbetaben PET images. J. Nucl. Cardiol. 29, 1919–1932 (2022). https://doi.org/10.1007/s12350-021-02608-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12350-021-02608-8