Abstract
Rhododendron delavayi is an alpine evergreen ornamental plant, but water shortage limits its growth and development in urban gardens. However, the adaptive mechanism of alpine evergreen rhododendrons to drought remains unclear. Here, a water control experiment was conducted to study the physiological and transcriptomic response of R. delavayi to drought. The drought treatment for 9 days decreased photosynthetic rate, induced accumulation of reactive oxygen species (ROS), and damaged chloroplast ultrastructure of R. delavayi. However, the photosynthetic rate quickly recovered to the level before treatment when the plants were re-watered. De novo assembly of RNA-Seq data generated 86,855 unigenes with an average length of 1870 bp. A total of 22,728 differentially expressed genes (DEGs) were identified between the control and drought plants. The expression of most DEGs related to photosynthesis were down-regulated during drought stress, and were up-regulated when the plants were re-watered, including the DEGs encoding subunits of light-harvesting chlorophyll-protein complex, photosystem II and photosystem I reaction center pigment-protein complexes, and photosynthetic electron transport. The expressions of many DEGs related to signal transduction, flavonoid biosynthesis and antioxidant activity were also significantly affected by drought stress. The results indicated that the response of R. delavayi to drought involved multiple physiological processes and metabolic pathways. Photosynthetic adjustment, ROS-scavenging system, abscisic acid and brassinosteroid signal transduction pathway may play important roles to improve drought tolerance of R. delavayi. Our findings provided valuable information for understanding the mechanisms of drought tolerance employed by Rhododendron species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought is one of the most common environmental factors limiting plant growth and productivity (Chaves et al. 2009; Khan et al. 2017). The majority of climate change scenarios predict increased incidences of drought throughout the world (Lemke et al. 2007). When experiencing a water deficit, plants can activate a series of complicated regulatory mechanisms to cope with such stress (Rasmussen et al. 2013). To understand how plants cope with drought stress, it is essential to identify the genes and metabolic pathways involved in drought tolerance.
Drought stress affects almost every aspect of plant physiology. Photosynthesis is a critical metabolic process for carbon assimilation. Drought stress may affect photosynthetic CO2 assimilation by affecting stomatal opening, diffusive resistance, or subsequent photo-oxidative damage caused by an accumulation of reactive oxygen species (ROS) (Chaves et al. 2009; Campos et al. 2014). Thus, improving photosynthetic performance under drought is a desirable goal for crop breeding. However, the genes related to photosynthesis and the metabolic pathways that are activated to protect photosynthesis in such a deficit environment have not been intensively studied (Ma et al. 2016).
Plants can optimize their morphologies, physiologies and metabolic processes to cope with drought stress. For example, drought stress destroys cell membrane integrity, accelerates the accumulation of ROS and breaks the balance of redox. Plants can reduce the risk of oxidative damage by reducing energy absorption, or activate various signal channels and antioxidases to rebuild the redox equilibrium (Saibo et al. 2009; Ranjan et al. 2012). In addition, plants can produce osmolytes such as proline and soluble sugar to lower osmotic potential, which allows plants to maintain water uptake and preserve membrane integrity. These strategies are particularly important during the later stages of stress (Yamada et al. 2005).
Transcriptome sequencing technology has been successfully used to study the molecular mechanisms of model and non-model plant species adapting to drought stress. Several metabolic, signaling pathways and thousands of genes have been identified (Joshi et al. 2016; Li et al. 2017). The molecular responses of plants to drought stress involve many genes which are related to photosynthesis, signal transduction, flavonoid biosynthesis and antioxidant activity (Nakabayashi et al. 2014; Ma et al. 2016). Identifying the differentially expressed genes (DEGs) involved in these significantly enriched metabolic pathways is important for understanding the process of drought stress response in plants. However, the physiological and molecular mechanisms of Rhododendron species in response to drought stress remain largely unclear (Fernàndez-Martínez et al. 2016).
Rhododendron, one of the most well-known alpine flowers, is quite rich in Yunnan province of Southwestern China, which harbors above 320 Rhododendron species (Fang et al. 2005). Many species or varieties in the genus Rhododendron have been used in urban landscapes. However, the drought in winter and spring, and the shortage of urban water resource limit the growth and flowering performance of these ornamental plants. Thus, the physiological response to drought stress in Rhododendron has received a few attention in recent years. A previous study has shown that some physiological parameters, such as the activity of protective enzymes and the content of malondialdehyde (MDA), can be used as the indicators to evaluate the drought resistance of Rhododendron (Huang et al. 2011). Some close correlations between drought-resistance and photosynthetic and chlorophyll fluorescence parameters have also been observed in Rhododendron (He et al. 2001). Our previous studies have found that the small stomata and large stomatal density enable the plants of R. delavayi to quickly regulate stomatal movement to respond to environmental changes, and the proportion of photosynthetic limitation caused by photo-oxidative damage is very small under severe drought stress (Cai et al. 2014; 2015). Therefore, we speculate that R. delavayi has a strong ability to scavenge ROS and avoid photo-oxidative damage. However, as a non-model plant species, the physiological and molecular adaptation of this species to drought stress is poorly understood.
In this study, we detected the variations in photosynthetic performances, biochemical substances and gene expressions of leaves under different water status to explore the mechanisms of drought tolerance involved in R. delavayi by combining with transcriptome sequencing and physiological analysis. We hypothesized that: (1) drought stress decreases the photosynthetic capacity of R. delavayi, but this species has strong resistance to photo-oxidation damage under drought stress; (2) the key genes and metabolic pathways related to photosystem II, photosystem I, signal transduction, flavonoid biosynthesis and antioxidant activity contribute to well-maintained photosynthesis during drought stress.
Materials and methods
Plant materials and treatments
Rhododendron delavayi was used in our experiments. This species is an evergreen shrub or small tree with large scarlet flowers that makes it highly attractive as an ornamental plant. It is widely distributed in Southwestern China and often grows in the mixed forests, forest margins, thickets, hills, rocky slopes and open field, and shows strong drought tolerance (Zhang et al. 2002). Five-year-old plants of R. delavayi were grown in the plastic pots (one plant per pot) filled with a mixture of peat and coconut coir (v/v, 2/1) in a greenhouse. The growth conditions included air temperature of 22–28 °C (day) and 14–17 °C (night), 50–70% of relative humidity, and with a maximum light intensity of approximately 1400 μmol m−2 s−1. Drought conditions were induced by withholding normal irrigation. In total, 30 plants were used for our experiments: half of the plants were watered daily to maintain the soil water content close to field capacity as the control treatment, and watering to the other half of the plants were stopped to induce different degrees of drought stress. In this way, four experimental scenarios were designed: normal irrigation (CK), stopping irrigation for 5 days (D5), stopping irrigation for 9 days (D9), stopping irrigation for 10 days, and re-watering for 6 h (REC) after 10 days of drought. Re-watering was only carried out on 10 plants. The other 5 plants were continued under no irrigation for subsequent electron microscopic observation. All of the measurements were done at the end of each time-point.
Leaf relative water content and soil water content
The intensity of drought stress was characterized by leaf relative water content (RWC) and soil water content (SWC). The value for RWC (%) was determined as: (FW − DW)/(TW − DW) × 100, where FW, DW, and TW are leaf fresh weight, dry weight, and turgid weight, respectively. FW was determined using 4 leaves for each replication. TW was determined after immersing the fresh leaves in water for 24 h at 4 °C in the dark, and DW was determined after oven dried at 72 °C for 48 h. The value for SWC (%) was calculated as: (SFW − SDW)/SDW × 100, where SFW is soil fresh weight, and SDW is soil dry weight determined after oven dried at 105 °C for 48 h.
Gas exchange, physiological traits and chloroplast ultrastructure
Photosynthetic gas exchange was determined between 9:00 and 11:00 am using a Li-6400XT open gas exchange system (Li-Cor, Inc., Nebraska, USA). Leaf photosynthetic rate (Pn), stomatal conductance (Gs), and transpiration rate (Tr) were made on the newly matured leaves at a CO2 concentration of 400 µmol mol−1, a light intensity of 1000 µmol m−2 s−1, and 60–70% relative humidity.
Four replicates of leaf samples were harvested to monitor the changes in physiological traits under different treatment conditions. The levels of proline (Pro), MDA, superoxide dismutase (SOD), soluble sugars and chlorophyll were determined by absorption photometry using a UV-751GD spectrophotometer (Shanghai Analytical Instrument Co., China).
The mesophyll cell and chloroplast ultrastructure under normal and stress conditions were observed by transmission electron microscopy (TEM). The tissue was cut from fully expanded, mature leaves. Small pieces from the middle region of the leaves were immediately sectioned into 2 mm × 2 mm segments, and immersed in a 2.5% glutaraldehyde fixative for 48 h at 4 °C. After fixation, the samples for ultrastructural analysis were prepared for TEM as described by Irwin et al. (2009). The samples were cut using a Leica EM UC7 microtome (Leica Microsystems, Buffalo Grove, IL, USA), and were examined and photographed with a JEOL JEM-1011 transmission electron microscope (Teknolab, Indonesia).
RNA isolation, library construction and Illumina sequencing
At the end of each time-point of CK, D5, D9, and REC, the top 5 leaves of a branch were sampled from five plants, frozen in liquid nitrogen, and stored at − 80 °C prior to RNA-Seq and quantitative real-time reverse transcription, qRT-PCR analyses. The RNA of the leaves was extracted using kits from Huayueyang Biotechnology Co., Ltd., according to the manufacturer’s instructions. The 28S/23S ratios of those RNA samples ranged from 1.7 to 2.1. Their integrities were examined with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif.). The values for RIN (RNA Integrity Number) ranged from 8.1 to 8.7, with no sign of degradation. Similar quantities of total RNA isolated at each time point and belonging to the same treatment group were pooled before constructing the four libraries.
Construction of cDNA library and sequencing were performed by the Beijing Genomics Institute (BGI). The entire process followed standardized procedures and was monitored by the BGI Quality Control System. Briefly, total mRNA was isolated with Oligo (dT) cellulose, then fragmented and reverse-transcribed with random primers. Second-strand cDNA was synthesized by DNA polymerase I (New England BioLabs, Inc.) and RNase H (Invitrogen, Carlsbad, CA, USA). The paired-end cDNA libraries were constructed with insert sizes of 200 bp, and the cDNA was sequenced using an Illumina (San Diego, CA, USA) Genome Analyzer platform. Image output data from the sequencer were transformed into raw sequence data by base-calling.
De novo assembly, gene expression and identification of differentially expressed genes
For the assembly library, raw reads were filtered to remove those containing adapters and reads with more than 5% unknown nucleotides. Low-quality reads, i.e., those for which the percentage of low Q-value (< 10) bases was > 20%, were also removed. Clean reads were assembled de novo by SOAPaligner/SOAP2 software (Li et al. 2009). The database is available from the NCBI Short Read Archive (SRA) with an accession number PRJNA503304 (http://www.ncbi.nlm.nih.gov/sra/PRJNA503304).
Gene expression was measured in RNA-Seq analyses as the number of reads per kilobase per million mapped reads (RPKM) (Mortazavi et al. 2008). The DEGs were identified with FDR (false discovery rate) values < 0.001 and absolute values of log2 Ratio ≥ 1. Gene expression data υ were normalized to 0, log2 (υD5/υCK), log2 (υD9/υCK), and log2 (υREC/υCK). The DEGs were clustered by Short Time-series Expression Miner (STEM) (Ernst and Bar-Joseph 2006). Clustered profiles with p values < 0.05 were considered significantly expressed. The enrichment analyses of Gene Ontology (GO, http://geneontology.org/) classifications and Kyoto encyclopedia of genes and genomes (KEGG; http://www.genome.jp/kegg) pathways were conducted using the total DEGs that were detected during different periods of drought stress. GO annotation was analyzed by Blast2GO software (Götz et al. 2008). Functional classification of the unigenes was performed using WEGO software. KEGG pathway annotation was undergone by Blastall software against the KEGG database.
Validation for transcriptomics
To confirm the RNA-Seq results, a total of 10 candidate DEGs in KEGG enrichment pathway were selected for qRT-PCR. Briefly, 2 μg of total RNA was reverse-transcribed to first-strand cDNA using a ReverTra Ace qPCR RT Master Mix (TOYOBO, Japan). Afterward, qRT-PCR was performed on a Roche LightCycler 480 Real-Time PCR System (Roche, Mannheim, Germany), using a SYBR Green-based Fast qPCR kit (KAPA Biosystems, Wilmington, MA, USA). Each 20-μL reaction mixture contained 2.2 μL of diluted first-strand cDNA, 0.4 μL (200 nM) for each primer, and 10 μL of SYBR Green PCR Master Mix. Reaction conditions included an initial 95 °C for 2 min; then 40 cycles of 95 °C for 3 s and 57 °C for 30 s. Each qRT-PCR run was conducted with three independent biological replicates, with each replicate corresponding to two technical repeats. All primers were designed using PRIMER5 software (Table S1 Supplemental Materials). The relative expression values were calculated by the 2−∆∆Ct method (Livak and Schmittgen 2001).
Statistical analysis
Statistical analysis was performed with SPSS 16.0 (SPSS Inc., Chicago, USA). Difference between means were tested by one-way ANOVA and LSD multiple comparisons tests. Difference was considered significant at p values < 0.05.
Results
Variation in leaf and soil water content
The RWC of drought-stressed plants maintained the same level as the CK plants after 3 days dehydration, but decreased to 75.1% and 72.1% after 6 and 9 days dehydration, respectively. On re-watering for 6 h, the RWCs in the stressed plants recovered to the levels of the CK plants. Meanwhile, SWC declined from 147.8% at the beginning of the treatment to 25.3% after stopping irrigation for 9 days (Fig. 1).
Changes in gas exchange, physiological and ultrastructural traits
When compared with the CK plants, the values for net photosynthetic rate (Pn), stomatal conductance (gs), and transpiration rate (Tr) in R. delavayi reduced on the 5th and 9th days after stopping watering. However, the values of those parameters showed a certain degree of recovery after re-watering for 6 h (Fig. 2).
Variations in net photosynthetic rate (Pn), stomatal conductance (Gs) and transpiration rate (Tr) under different drought stresses. Values represent mean ± SE (n = 10). CK, normal irrigation; D5, stopping irrigation for 5 days; D9, stopping irrigation for 9 days; REC, re-watering for 6 h on the 10th day after stopping irrigation
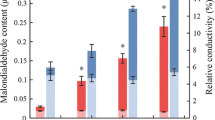
Leaf MDA content increased with the intensifying degree of drought stress. The MDA contents in the stressed plants after 6 days of stress treatment were significantly higher than that of the CK plants. After re-watering, the MDA contents decreased but were still higher than that of the CK (Fig. 3A). Similar trends were noted for SOD activity, and for the contents of proline and soluble sugars (Fig. 3B–D). Compared with the CK, the chlorophyll a and chlorophyll b contents increased significantly after stopping irrigation for 6 days, especially chlorophyll b. After re-watering, the chlorophyll a content of the stressed plants decreased to the same level as the CK plants, but the chlorophyll b content was still higher (Fig. 3E–F).
Variations in leaf malondialdehyde (MDA) content (A), SOD activity (B), proline content (C), soluble sugar content (D), chlorophyll a content per unit area (E), and chlorophyll b content per unit area (F) under different drought stresses. Day 0 corresponds to the first day of stress treatment. The arrows indicate the beginning of re-watering. Values represent mean ± SE (n = 3)
The chloroplasts in the CK plants were long and oval-shaped, and were distributed close to the edge of the plasma membrane. The chloroplast lamellae were orderly arranged and substantially parallel to the long axis of the chloroplast. The osmiophilic granules and starch grains were randomly distributed in those chloroplasts (Fig. 4A, F). After 6 days of stress treatment, the chloroplasts were slightly swollen, and the stroma lamellae were loosely arranged (Fig. 4C, H). By day 9, some chloroplasts were no longer closely attached to the plasma membrane, and plasmolysis and agglomeration were apparent, and some thylakoids had vacuolized (Fig. 4D, I). By day 12, the shape of chloroplast changed from long and oval to circular or nearly spherical, while plasmolysis and thylakoid vacuolization intensified, and the lamellae was bent to produce an overall disorganized arrangement (Fig. 4E, J).
Changes in ultrastructure of mesophyll cells (A–E) and chloroplasts (F–J) from Rhododendron delavayi under different stages of drought treatment. CK, the control (A, F); 3d, stopping irrigation for 3 days (B, G); 6d, stopping irrigation for 6 days (C, H); 9d, stopping irrigation for 9 days (D, I); 12d, stopping irrigation for 12 days (E, J). CW, cell wall; Ch, chloroplast; IAS, intercellular air space; V, vacuole; OG, osmiophilic granules; SG, starch grains; ST, stroma thylakoid
De novo assembly and functional annotation of unigenes
The cDNA libraries from the leaves in the CK, D5, D9, and REC treatment groups produced over 4.6 G nt of clean nucleotide read data. Using a de novo assembly approach, the clean reads were assembled into 69,806, 74,468, 82,453, and 88,091 unigenes for CK, D5, D9, and REC treatments, respectively. The average unigene length was 1140 bp, and the N50 size averaged 1870 bp (Table 1). Among the 86,855 unigenes, 59.6% (51,735) were annotated by Nr database, and 40.29% (34,995) shared identity with the proteins in the Swiss-Prot database. In total, 62.95% (54,679) of the assembled unigenes were successfully annotated via KEGG, COG and GO databases (Table 2).
DEGs identification and analysis
When compared with the CK, 4503 (2804 up/1699 down), 14,430 (8329 up/6101 down), and 11,455 (7755 up/3700 down) DEGs were identified in the D5, D9, and REC treatments, respectively (Fig. 5). A total of 22,728 DEGs were detected between the stressed and CK plants (Table S2 in Supplemental Materials). Of these, 16,345 were clustered into 12 profiles which were significantly expressed (p < 0.05). The profiles included 4 down-regulated patterns (profiles 1, 2, 10, and 11) and 8 up-regulated patterns (13, 15, 16, 21, 22, 23, 24, and 25) (Fig. 6). Using GO annotation analysis, the DEGs were classified into 3 main categories including biological process, cellular component and molecular function. Under the biological process category, the part of cellular process, metabolic process, single organism process, response to stimulus were prominently represented. Under the category of cellular component, cell part and organelle represented the majorities of the category. For the molecular function category, metabolic process, catalytic activity and binding were the top abundant subcategories (Fig. 7). 43.3% (9842/22,728) of the DEGs was annotated by KEGG pathway analysis. The top 10 KEGG pathways with the highest representation of the DEGs are shown in Table 3. These DEGs were mainly located in biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, metabolism, ether lipid metabolism, flavonoid biosynthesis and plant hormone signal transduction pathway.
Short Time-series Expression Miner (STEM) cluster analysis of differentially expressed genes (DEGs) in the drought-stressed leaves of Rhododendron delavayi. 12 profile with color mark which were significantly expressed (p < 0.05), included four down-regulated patterns (profiles 1, 2, 10, and 11) and eight up-regulated patterns (profiles 13, 15, 16, 21, 22, 23, 24, and 25)
Validation of RNA-Seq
The expressions of 10 candidate DEGs in KEGG enrichment pathway were detected using qRT-PCR. In response to drought stress, the expressions of 7 genes, including flavonoid 3-galactosyl transferase (CL1213.Contig9), hydroxycinnamoyl-CoA (Unigene19117), flavonol synthase (Unigene402), photosystem I reaction center subunit XI (Unigene28281), chloroplastic-like isoform (CL1696.Contig2), chlorophyll a/b-binding protein (Unigene8237) and serine/threonine-protein kinase (CL7513.Contig1), were down-regulated, while 3 genes, including gibberellin oxidase-like (CL3049.Contig2), glutathione transferase (CL7209.Contig2) and dihydroflavonol-4-reductase (CL9210.Contig2), were up-regulated. The similar expression patterns of the candidate unigenes between qRT-PCR and RNA-Seq confirmed the high reliability of RNA-Seq data (Fig. 8).
Expression of 10 differentially expressed genes (DEGs) in Rhododendron delavayi under different drought stresses. Filled circle (left y-axis) represents relative expression of 10 DEGs using qRT-PCR, open circle (right y-axis) represents transcript abundance (RPKM) of 10 DEGs detected by RNA-seq. CK, normal irrigation; D5, stopping irrigation for 5 days; D9, stopping irrigation for 9 days; REC, re-watering for 6 h on the 10th day of stopping irrigation
Genes and pathways involved in drought-tolerance
The photosynthetic rate in R. delavayi changed obviously after stopping irrigation, and re-watering (Fig. 2). 61 DEGs were annotated by the category “photosynthesis”. Among these genes, 85.2% (52/61) genes showed down-regulated expression under drought condition. Among the down-regulated DEGs, 18 genes (one each for Lhca1, Lhca3, Lhca5, Lhcb3, Lhcb6, and Lhcb7; two each for Lhca2, Lhcb2, Lhcb4, and Lhcb5; and four for Lhcb1) encoded the subunits of the light-harvesting chlorophyll–protein (LHC) complex (Fig. 9A, Table S3); 15 genes (one each for PsbC, PsbK, PsbM, PsbO, PsbW, PsbY, and Psb28; two for PsbP; and six for PsbQ) encoded the subunits of Photosystem II (PS II) reaction center pigment-protein complexes; 11 genes (PsaD, PsaE, PsaF, PsaG, PsaK, PsaL, PsaN, and PsaO; plus three for PsaH) encoded the subunits in Photosystem I (PS I) reaction center pigment-protein complexes; 5 genes were involved in photosynthetic electron transport, and 3 genes functioned in the redox chain (Fig. 9B, Table S3). After re-watering for 6 h, most of the genes with previously decreased expression (30 out of 52) were up-regulated.
KEGG map of photosynthesis-antenna proteins (A) and photosynthetic pathway (B) for Rhododendron delavayi. Analysis of DEGs in drought-treated samples compared to untreated control. The boxes with red square indicate that corresponding DEGs are down-regulated in drought-treated samples, and the boxes without red square indicate that the expression level of the corresponding genes are not changed, as determined by RNA-seq
Both hormone signal transduction and flavonoid biosynthesis were significantly enriched by KEGG analysis (Table 3). A total of 280 DEGs involved in the signal transduction pathway of plant hormone were detected. Among these DEGs, 65.0% of the DEGs (182/280) were down-regulated. Interestingly, 55.7% DEGs (156/280) were enriched in the signal transduction pathway for brassinosteroids (BRs). In this pathway, 67 and 62 DEGs encoded BAK1 and BRI1, respectively (Table 4).
Integration of KEGG and STEM cluster analysis showed that 46 DEGs belonging to flavonoid biosynthesis pathway were significantly clustered into four profiles (1, 10, 22, and 24). When the period of drought stress was prolonged, 4 genes (CL1033.Contig3_All, CL9631.Contig1_All, CL9631.Contig2_All, Unigene35052_All) encoding chalcone synthase (CHS) showed decreased transcript abundance. After the plants were re-watered, the expression of those CHS genes increased. The expression level of the genes encoding lavanone 3-hydroxylase (F3H, CL3049.Contig2_All, CL3049.Contig3_All, CL4124.Contig3_All, Unigene14035_All), dihydroflavonol 4-reductase (DFR, CL9210.Contig3_All, Unigene15174_All, Unigene8118_All), and anthocyanidin synthase (ANS, CL337.Contig2_All, Unigene2322_All, Unigene23455_All, Unigene9424_All) increased after the plants were exposed to drought stresses. Interestingly, the expressions of flavonol synthase (FLS, CL337.Contig2_All, Unigene9424_All) and DFR followed opposite trends (Fig. 10A, Table S4). This may be because they compete for the enzymes that utilized dihydroflavonols.
“Antioxidant activity” was one of the most altered GO categories (Fig. 7). We detected 61 DEGs belonging to the group for detoxification enzymes, including 5 genes encoding GSH (CL1202.Contig1_All, CL1202.Contig3_All, CL1202.Contig7_All, CL2529.Contig3_All, CL4826.Contig1_All), 2 genes encoding SOD (CL9747.Contig2_All, Unigene35071_All), 3 genes encoding APX (CL4153.Contig3_All, Unigene29990_All, Unigene29991_All), 4 genes encoding CAT (Unigene15489_All, Unigene23808_All, Unigene26028_All, Unigene6944_All), and 2 genes encoding PRX (Unigene7688_All, Unigene7689_All). Interestingly, 21 genes encoding POD were the most enriched category (Fig. 10B, Table S5).
Discussion
Drought stress can affect multiple aspects of plant physiological functions. The adaptation of plants to environmental factors is directly or indirectly related to photosynthesis. Maintenance of photosynthetic capacity under drought stress is an important feature of plants with drought tolerance (Chaves et al. 2009; Zhang et al. 2016). Down-regulation of photosynthesis under drought stress can be due to a decrease of available CO2 caused by stomatal closure or the subsequent photo-oxidative damage induced by an accumulation of ROS (Saibo et al. 2009; Campos et al. 2014). Furthermore, plants often face the cycle of drought and rehydration. The recovery efficiency and speed after rehydration are crucial to the growth and production of plants (Flexas et al. 2006). The photosynthetic rate of R. delavayi varied with water status. After stopping irrigation for 9 days, the values of Pn, Gs and Tr declined significantly, suggesting that photosynthetic capacity was depressed by drought stress. After re-watering for 6 h, the values for Pn, Gs and Tr recovered rapidly. This indicated that R. delavayi had strong self-repairing ability, and the influence of moderate drought on photosynthesis of R. delavayi was reversible.
During drought stress, some genes in the photosynthetic pathway showed decreased transcription abundance. These down-regulated genes assigned to different parts of photosynthetic process, such as LHC, PS II, PS I, cytochrome complex, photosynthetic electron transport and ATP synthase. LHC is embedded in the chloroplast thylakoid membrane and usually divided into LHCA and LHCB. The former is the primary reaction of charge separation, which is the process of converting light energy into chemical energy. The latter controls the light-harvesting function of chloroplast membranes, and plays an important role in regulating and protecting the primary photochemical process (Horton et al. 1991). The down-regulated expression of the DEGs encoding both LHCA and LHCB suggested the absorption and transformation of light energy in R. delavayi might have been limited by drought stress. The similar profile of DEGs encoding the components of the PS II and PS I protein complexes and ATP synthase suggested that the binding stabilities of PS II and PS I were affected. Meanwhile, the DEGs involved in cytochrome complex and photosynthetic electron transport was also suppressed. These indicated that the electron transport in R. delavayi was inhibited by drought. However, the transcript abundance of the DEGs encoding the components of LHC, PS II, PS I, and electron-transport chains was significantly increased after 6 h of re-watering, which resulted in the recovery of photosynthetic capacity. Thus, the photosynthesis of R. delavayi was sensitive to the change of water status, and the regulations of light energy absorption and photosynthetic electron transport were involved in the adaptation of this species to drought.
Drought stress may stimulate ROS production (Carvalho 2008). Excess ROS induces lipid peroxidation by oxidizing unsaturated fatty acids, thereby resulting in electrolyte leakage and membrane damage (Li et al. 2015). Thus, the capacity to scavenge ROS plays an important role in improving drought tolerance (Talbi et al. 2015). MDA is a major indicator of the ROS-related oxidative damage. The level of MDA increased significantly when the stress period was prolonged, implying that drought caused oxidative damage to R. delavayi. We also observed plasmolysis and thylakoid vacuolization in the chloroplasts after stopping irrigation for 9 days. The damage to the chloroplast membrane caused by high ROS levels may reduce photosynthetic capacity. A previous study has suggested that an accumulation of ROS induces biochemical limitation of photosynthesis, which is dominant in many species under severe drought stress (Flexas et al. 2009). However, biochemical limitation has a small effect on photosynthesis in R. delavayi during drought process (Cai et al. 2015). Thus, we speculated that ROS-scavenging system plays an important role in maintaining a normal rate of photosynthesis and improving drought tolerance of R. delavayi.
Both antioxidant enzymes and non-enzymatic antioxidants are contributed to ROS-scavenging system. Here, we detected 61 DEGs associated with antioxidant activity, and most of those genes were up-regulated by drought stress. The DEGs encoding POD were the most abundant during drought process. This is consistent with the observation on Sophora moorcroftiana (Li et al. 2015). In addition, the activity of SOD increased when the stress period continued. These indicated that the activity and expression level of detoxification enzymes could be induced by drought, and could help protect tissues against oxidative damage in R. delavayi under such conditions.
Flavonoids functions in response to drought in plants (Ma et al. 2014; Nakabayashi et al. 2014; Yuan et al. 2015). Drought stress increases the expressions of several flavonoids biosynthesis genes and the accumulation of flavonoids (Yuan et al. 2015; Nakabayashi et al. 2014). Over-accumulation of flavanoids can enhance the tolerance of plants to oxidative and drought stress (Nakabayashi et al. 2014). The flavonoids are abundant in the leaves of Rhododendron (Zhang et al. 2012). Here, 46 DEGs involved in flavonoids synthesis were enriched by KEGG analysis, suggesting that the flavonoids metabolism pathway might play an important role in drought tolerance of R. delavayi. The DEGs encoding CHS, a key enzyme regulated the early steps in flavonoid biosynthesis, declined its transcript abundance in response to drought treatment. Similar result is reported in Chrysanthemum (Xu et al. 2013). However, the expression of CHS in two cultivars of Triticum aestivum is significantly induced by drought stress (Ma et al. 2014). Although this enzyme can be up-regulated by short-term drought stress (Chen et al. 2008), its activity is suppressed in the roots of Medicago sativa by long-term drought (Kang et al. 2011). F3H, DFR, and ANS are the late genes of flavanoids pathway. Their expressions in Reaumuria sooongorica are induced by drought condition (Liu et al. 2013). In this study, the transcript abundances of F3H, DFR, and ANS were up-regulated by drought stress. DFR and FLS use dihydroflavonols as their substrates. We found that the expression of DFR was up-regulated by drought stress while the expression of FLS was down-regulated. These results indicated that the synthesis of flavonol and anthocyanin competed for dihydroflavonols through a dynamic equilibrium, and that flavonoid biosynthesis transferred from flavonols to the anthocyanin pathway. Thus, a high level of anthocyanin content might improve antioxidant capacity and drought tolerance of R. delavayi.
Both abscisic acid (ABA) biosynthetic and signaling pathways play critical roles in the response to drought stress (Pizzio et al. 2013). Drought stress induces the production of endogenous ABA and subsequently up-regulate the expression of ABA-responsive genes. Dehydration-responsive genes are induced by exogenous application of ABA (Adie et al. 2007; Mehrotra et al. 2014). The core components of ABA signaling include ABA receptors (PYR/PYL), protein phosphatase 2Cs (PP2Cs), and SNF1-related kinase 2 (SnRK2). ABA combines with PYR/PYL protein and interacts with PP2Cs, inhibits the activity of PP2Cs and then relieves the inhibition of PP2Cs to SnRK2. After activation of SnRK2, downstream transcription factors or membrane proteins are phosphorylated, opening the ABA signal response (Raghavendra et al. 2010). ABRE-binging factors (ABF) subfamily transcription factors which are activated by SnRK2 can be induced by drought. The ABRE/ABF-SnRK2 pathway has a role as a positive regulator in ABA-dependent gene expression (Fujita et al. 2013). Overexpression of ABF3 gene enhances the tolerance to drought in creeping bentgrass (Choi et al. 2013). Here, we found that 1 gene encoding SnRK2 were down-regulated; 3 genes encoding PP2C and 5 genes encoding ABF were up-regulated. PP2Cs as negative regulators of ABA signaling plays a critical role to regulate stomatal movement (Yoshida et al. 2006). The downregulation of PYR/PYL and induction of PP2C genes may reduce ABA sensitivity, and then inhibits stomatal closure. R. delavayi maintained relatively high stomatal conductance during drought process. These indicated that ABA signaling pathway was involved in drought response of R. delavayi, but ABA signaling related DEGs did not promote stomatal closure in response to drought by synthesizing a large amount of ABA.
Brassinosteroids are an important group of steroidal hormones that enable plants to cope with drought stress (Yuan et al. 2010; Gruszka et al. 2016). Low concentrations of BRs can promote the opening of stomata (Shakirova et al. 2016), increase the contents of proline, soluble sugars, and other osmotic substances to maintain normal leaf water potential (Zhang et al. 2008). BRs can also improve the energy metabolism balance between the chloroplasts and mitochondria, boost the initial activity of Rubisco, thereby increasing photosynthetic efficiency under water deficit condition (Li et al. 2016; Soares et al. 2016). Several components and numerous genes in the BR signaling pathways (e.g., BRI1, BAK1, BIN2, and BSU1) has been identified (Gruszka et al. 2011). BAK1 is particularly active in a BRI1-mediated BR signaling pathway, and its autophosphorylation activity is stimulated by BRI1 (Li et al. 2002). The interaction of BAK1 and BRI1 modulates BR signaling to regulate physiological reactions to drought stress (Shakirova et al. 2016). We detected 156 DEGs involved in the BR signal transduction pathway. Among these genes, 67 DEGs encode AK1 and 62 DEGs encode BRI1. This implied that the expressions of the DEGs encoding BAK1 and BRI1 in BR pathway play an important role in the response of R. delavayi to drought stress.
Conclusions
The physiological analysis suggested that drought stress depressed the photosynthetic capacity, stimulated the accumulation of ROS, and damaged the chloroplast ultrastructure of R. delavayi. However, this species showed strong resistance to photo-oxidation under drought stress, and could recover quickly from stress after re-watering. Transcriptome analysis revealed that many DEGs involved in photosynthetic pathway showed similar profile with the changes of photosynthetic during drought stress and recovery. The DEGs involved in antioxidant activity, flavonoid biosynthesis and signal transduction pathway also played important roles in maintaining photosynthesis and improving the drought tolerance of R. delavayi. Our study provided the first transcriptome profile of this species exposed to drought stress, and may improve our understanding of the adaptive mechanism of rhododendrons to drought stress.
Abbreviations
- ABA:
-
Abscisic acid
- ABF:
-
ABRE-binding factor
- ANS:
-
Anthocyanidin synthase
- APX:
-
Ascorbate peroxidase
- CAT:
-
Catalase
- CHS:
-
Chalcone synthase
- COG:
-
Clusters of orthologous groups of protein
- DEGs:
-
Differentially expressed genes
- DFR:
-
Dihydroflavonol-4-reductase
- F3H:
-
Flavanone 3-hydroxylase
- FDR:
-
False discovery rate
- FLS:
-
Flavonol synthase
- GO:
-
Gene ontology
- Gs:
-
Stomatal conductance
- GSH:
-
Glutathione peroxidase
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LHC:
-
Light-harvesting chlorophyll–protein complex
- MDA:
-
Malondialdehyde
- Nr:
-
Non-redundant protein
- OG:
-
Osmiophilic granules
- Pn :
-
Net photosynthetic rate
- POD:
-
Peroxidase
- PRX:
-
Peroxiredoxins
- PS I:
-
Photosystem I
- PS II:
-
Photosystem II
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
- STEM:
-
Short time-series expression miner
- TEM:
-
Transmission electron microscopy
- Tr :
-
Transpiration rate
References
Adie BA, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JE, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19:1665–1681. https://doi.org/10.1105/tpc.106.048041
Cai YF, Li SF, Li SF, Xie WJ, Song J (2014) How do leaf anatomies and photosynthesis of three Rhododendron species relate to their natural environments? Bot Stud 55:36–45. https://doi.org/10.1186/1999-3110-55-36
Cai YF, Wang JH, Li SF, Zhang L, Peng LC, Xie WJ, Liu FH (2015) Photosynthetic response of an alpine plant, Rhododendron delavayi Franch, to water stress and recovery: the role of mesophyll conductance. Front Plant Sci 6:1089. https://doi.org/10.3389/fpls.2015.01089
Campos H, Trejo C, Peña-Valdivia CB, García-Nava R, Conde-Martínez FV, Cruz-Ortega MR (2014) Stomatal and non-stomatal limitations of bell pepper (Capsicum annuum L.) plants under water stress and re-watering: delayed restoration of photosynthesis during recovery. Environ Exp Bot 98:56–64. https://doi.org/10.1016/j.envexpbot.2013.10.015
Carvalho MHC (2008) Drought stress and reactive oxygen species: production, scavenging and signaling. Plant Signal Behav 3:156–165. https://doi.org/10.4161/psb.3.3.5536
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560. https://doi.org/10.1093/aob/mcn125
Chen D, Liang MX, DeWald D, Weimer B, Peel MD, Bugbee B (2008) Identification of dehydration responsive genes from two non-nodulated alfalfa cultivars using Medicago truncatula microarrays. Acta Physiol Plant 30:183–199. https://doi.org/10.1007/s11738-007-0107-5
Choi YS, Kim YM, Hwang OJ, Han YJ (2013) Overexpression of Arabidopsis ABF3 gene confers enhanced tolerance to drought and heat stress in creeping bentgrass. Plant Biotechnol Rep 7:165–173. https://doi.org/10.1007/s11816-012-0245-0
Ernst J, Bar-Joseph Z (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinform 7:191. https://doi.org/10.1186/1471-2105-7-191
Fang MY, Fang RZ, He MY, Hu LZH, Yang HP (2005) Flora of China, vol 14. Science Press, Beijing, pp 260–455
Fernàndez-Martínez J, Fransi MA, Fleck I (2016) Ecophysiological responses of Betula pendula, Pinus uncinata and Rhododendron ferrugineum in the Catalan Pyrenees to low summer rainfall. Tree Physiol 36:1520–1535. https://doi.org/10.1093/treephys/tpw104
Flexas J, Bota J, Galmés J, Medrano H, Ribas-Carbó M (2006) Keeping a positive carbon balance under adverse conditions: responses of photosynthesis and respiration to water stress. Physiol Plant 127:343–352. https://doi.org/10.1111/j.1399-3054.2006.00621.x
Flexas J, Barón M, Bota J, Ducruet JM, Gallé A, Galmés J, Jiménez M, Pou A, Ribas-Carbó M, Sajnani C, Tomàs M, Medrano H (2009) Photosynthesis limitations during water stress acclimation and recovery in the drought-adapted Vitis hybrid Richter-110 (V. berlandierix V. rupestris). J Exp Bot 60:2361–2377. https://doi.org/10.1093/jxb/erp069
Fujita Y, Yoshida T, Yamaguchi Shinozaki K (2013) Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant 147:15–27. https://doi.org/10.1111/j.1399-3054.2012.01635.x
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. https://doi.org/10.1093/nar/gkn176
Gruszka D, Szarejko I, Maluszynski M (2011) Identification of barley DWARF gene involved in brassinosteroid synthesis. Plant Growth Regul 65:343–358. https://doi.org/10.1007/s10725-011-9607-9
Gruszka D, Janeczko A, Dziurka M, Pociecha E, Oklestkova J, Szarejko I (2016) Barley brassinosteroid mutants provide an insight into phytohormonal homeostasis in plant reaction to drought stress. Front Plant Sci 7:1824. https://doi.org/10.3389/fpls.2016.01824
He LS, Su JL, Liu XQ, Li C, Chen SP, Xiang LP (2001) The effects of drought stress on photosynthetic physiological characteristics of Rhododendron hybrides ‘Cosmopolitan’ simulated by PEG. J Suzhou Univ Sci Technol (Nat Sci) 28(4):62–66 (in Chinese)
Horton P, Ruban AV, Rees D, Pascal AA, Noctor G, Young AJ (1991) Control of the light-harvesting function of chloroplast membranes by aggregation of the LHCII chlorophyll–protein complex. FEBS Lett 292:1–4. https://doi.org/10.1016/0014-5793(91)80819-O
Huang CL, Chen X, Gao GL (2011) Physiological response of seedlings of three azalea species of drought stress and evaluation of drought resistance. Sci Silvae Sin 47:48–55 (in Chinese)
Irwin RB, Anciso A, Jones PJ, Glenn AL, Williams BL, Sridhar S, Arshad S (2009) Transmission electron microscopy specimen preparation perpendicular to the long axis of high aspect ratio features. J Vac Sci Technol 27:1352–1359. https://doi.org/10.1116/1.3248271
Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029. https://doi.org/10.3389/fpls.2016.01029
Kang Y, Han Y, Torres-Jerez I, Wang M, Tang Y, Monteros M (2011) System responses to long-term drought and re-watering of two contrasting alfalfa varieties. Plant J 68:871–889. https://doi.org/10.1111/j.1365-313X.2011.04738.x
Khan F, Upreti P, Singh R, Shukla PK, Shirke PA (2017) Physiological performance of two contrasting rice varieties under water stress. Physiol Mol Biol Plants 23:85–97. https://doi.org/10.1007/s12298-016-0399-2
Lemke P, Ren R, Alley I (2007) The physical science basis. Contribution of working group I to the fourth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Cambridge, pp 337–383
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222. https://doi.org/10.1016/S0092-8674(02)00812-7
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967. https://doi.org/10.1093/bioinformatics/btp336
Li H, Yao W, Fu Y, Li S, Guo Q (2015) De novo assembly and discovery of genes that are involved in drought tolerance in Tibetan Sophora moorcroftiana. PLoS ONE 10:e111054. https://doi.org/10.1371/journal.pone.0111054
Li XJ, Guo X, Zhou YH, Shi K, Zhou J, Yu JQ (2016) Overexpression of a brassinosteroid biosynthetic gene dwarf enhances photosynthetic capacity through activation of Calvin cycle enzymes in tomato. BMC Plant Biol 16:33. https://doi.org/10.1186/s12870-016-0715-6
Li YH, Zhang HN, Wu QS, Muday GK (2017) Transcriptional sequencing and analysis of major genes involved in the adventitious root formation of mango cotyledon segments. Planta 245:1193–1213. https://doi.org/10.1007/s00425-017-2677-9
Liu M, Li X, Liu Y, Cao B (2013) Regulation of flavanone 3-hydroxylase gene involved in the flavonoid biosynthesis pathway in response to UV-B radiation and drought stress in the desert plant, Reaumuria soongorica. Plant Physiol Biochem 73:161–167. https://doi.org/10.1016/j.plaphy.2013.09.016
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Ma D, Sun D, Wang C, Li Y, Guo T (2014) Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol Biochem 80:60–66. https://doi.org/10.1016/j.plaphy.2014.03.024
Ma X, Xia H, Liu Y, Wei H, Zheng X, Song C, Chen L, Liu H, Luo L (2016) Transcriptomic and metabolomic studies disclose key metabolism pathways contributing to well-maintained photosynthesis under the drought and the consequent drought-tolerance in rice. Front Plant Sci 7:1886. https://doi.org/10.3389/fpls.2016.01886
Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S (2014) Abscisic acid and abiotic stress tolerance-different tiers of regulation. J Plant Physiol 171:486–496. https://doi.org/10.1016/j.jplph.2013.12.007
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. https://doi.org/10.1038/nmeth.1226
Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77:367–379. https://doi.org/10.1111/tpj.12388
Pizzio GA, Rodriguez L, Antoni R, Gonzalez-Guzman M, Yunta C (2013) The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol 163:441–455. https://doi.org/10.1104/pp.113.224162
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401. https://doi.org/10.1016/j.tplants.2010.04.006
Ranjan A, Pandey N, Lakhwani D, Dubey NK, Pathre UV, Sawant SV (2012) Comparative transcriptomic analysis of roots of contrasting Gossypium herbaceum genotypes revealing adaptation to drought. BMC Genom 13:680. https://doi.org/10.1186/1471-2164-13-680
Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161:1783–1794. https://doi.org/10.1104/pp.112.210773
Saibo NJ, Lourenco T, Oliveira MM (2009) Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot 103:609–623. https://doi.org/10.1093/aob/mcn227
Shakirova F, Allagulova C, Maslennikova D, Fedorova K, Yuldashev R, Lubyanova A (2016) Involvement of dehydrins in 24-epibrassinolide-induced protection of wheat plants against drought stress. Plant Physiol Biochem 108:539–548. https://doi.org/10.1016/j.plaphy.2016.07.013
Soares C, Sousa AD, Pinto A, Azenha M, Teixeira J, Azevedo RA (2016) Effect of 24-epibrassinolide on ROS content, antioxidant system, lipid peroxidation and Ni uptake in Solanum nigrum L. under Ni stress. Environ Exp Bot 122:115–125. https://doi.org/10.1016/j.envexpbot.2015.09.010
Talbi S, Romero-Puertas MC, Hernández A, Terrón L, Ferchichi A, Sandalio LM (2015) Drought tolerance in a Saharian plant Oudneya africana: role of antioxidant defences. Environ Exp Bot 111:114–126. https://doi.org/10.1016/j.envexpbot.2014.11.004
Xu Y, Gao S, Yang Y, Huang M, Cheng L, Qian W (2013) Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genom 14:662–677. https://doi.org/10.1186/1471-2164-14-662
Yamada M, Morishita H, Urano K, Shiozaki N, Yamaguchi-Shinozaki K, Shinozaki K (2005) Effects of free proline accumulation in petunias under drought stress. J Exp Bot 56:1975–1981. https://doi.org/10.1093/jxb/eri195
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281:5310–5318. https://doi.org/10.1074/jbc.M509820200
Yuan GF, Jia CG, Li Z, Sun B, Zhang LP, Liu N (2010) Effect of brassinosteroids on drought resistance and abscisic acid concentration in tomato under water stress. Sci Hortic 126:103–108. https://doi.org/10.1016/j.scienta.2010.06.014
Yuan Y, Qi L, Yang J, Wu C, Liu Y, Huang L (2015) A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tissue Org Cult 120:961–972. https://doi.org/10.1007/s11240-014-0650-x
Zhang CQ, Luo JF, Su YF (2002) The research of drought tolerance on 6 species of Rhododendron. Guihaia 22:174–176 (in Chinese)
Zhang MC, Zhai ZX, Tian XL, Duan L, Li Z (2008) Brassinolide alleviated the adverse effect of water deficits on photosynthesis and the antioxidant of soybean (Glycine max L.). Plant Growth Regul 56:257–264. https://doi.org/10.1007/s10725-008-9305-4
Zhang M, Pan DR, Zhou YF (2012) Analysis of the flavonoids in the leaves of Rhododendron pulchrum Sweet by HPLC-MS. J Anhui Agri Sci 3(6):21–24
Zhang ZF, Li YY, Xiao BZ (2016) Comparative transcriptome analysis highlights the crucial roles of photosynthetic system in drought stress adaptation in upland rice. Sci Rep 6:19349. https://doi.org/10.1038/srep19349
Acknowledgements
Funding was provided by National Natural Science Foundation of China (Grant Nos. 31760229, 31670342), Program of Science and Technology Talents Training in Yunnan Province (Grant No. 2016HA005), Science and Technology Plan of Yunnan Province (Grant No. 2015BB013), and Applied Basic Research Project of Yunnan Province (Grant No. 2016FB058).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, YF., Wang, JH., Zhang, L. et al. Physiological and transcriptomic analysis highlight key metabolic pathways in relation to drought tolerance in Rhododendron delavayi. Physiol Mol Biol Plants 25, 991–1008 (2019). https://doi.org/10.1007/s12298-019-00685-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-019-00685-1