Abstract
Background
Variants of DNA repair genes are extensively reported to cause genetic instability and increase the risk of breast cancer. In combination with NBS1, MRE11 and RAD50 constitute an MRN (MRE11–RAD50–NBS1) complex that repairs DNA damage. However, certain genetic alterations in MRE11 and RAD50 produce abnormal protein that affects the repairing process and may result in malignancy. We aimed to investigate the association of MRE11 and RAD50 polymorphisms with breast risk in the female population of Punjab, Pakistan.
Methods
We collected blood samples of 100 breast cancer patients and 100 tumor-free females selected as controls. Extracted DNA was genotyped by tetra ARMS-PCR followed by gel electrophoresis. Results were analyzed by SPSS and SNPstats to analyze the association of different clinical factors and SNPs (single nucleotide polymorphisms) with the risk of breast cancer.
Results
We found that the increased risk of breast cancer is associated with MRE11 variant rs684507 (odds ratio-OR 3.71, 95% confidence interval-CI 1.68–8.18, p value < 0.0001), whereas, RAD50 variant rs28903089 appeared to have protective effect (OR 0.55, CI 0.29–1.02, p value = 0.003). Additionally, clinical factors such as positive family history, life style, and marital status also play significant roles in breast cancer development.
Conclusion
In the present study, strong risk of breast cancer was associated with MRE11 gene. However, RAD50 showed protective effect. Additionally, clinical factors are also pivotal in risk assessment. We anticipate that targeting specific genetic variations confined to ethnic groups would be more effective in future therapeutic approaches for prevention and treatment of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is one of the predominant malignancies worldwide, which is characterized by uncontrolled proliferation of breast cells [1,2,3]. According to an estimate, the breast cancer prevalence is increasing by the rate of 2% each year [4]; however in Pakistan, the incidence rate is 1/9 that is higher than other Asian countries [5]. Although, breast cancer is very common in females but it can occur in male population as well [6, 7]. Several explanatory factors such as age, gender and family history are involved in cancer development [5, 8]. It is well established that the women older than 40 years are more susceptible to breast cancer. Moreover, positive family history also plays a vital role in disease prevalence. Other responsible factors may include genetic alteration, hormonal imbalance, exposure to chemicals, UV radiations, life style, and certain bacteria and viruses [9, 10]. Genetic association in breast cancer has been reported in several studies and suggests that the penetrant genes increase the familial risk of early onset cancer [11]. Mutation in certain genes that participate in cell proliferation, DNA double-strand break repair (DSBR) pathway and activation of cell cycle checkpoint may lead to cancer development [12,13,14].
The MRN complex, comprising of proteins encoded by the genes MRE11, RAD50, and NBS1, plays a vital role in DNA DSBR pathway by recruiting Ataxia telangiectasia mutated (ATM) to damaged sites that activates DNA repair network. Several studies have suggested that the carriers of ATM mutations and polymorphic variants are at higher risk of breast cancer. Principally, the ATM protein is presumed to be a chief activator of DNA DSBR network, and becomes activated only after recruitment to DSB (double-strand break) sites [15].
Among the MRN complex genes, MRE11 encodes specific proteins that are involved in maintenance of telomere length, DSBR pathway and homologous recombination [15]. Another gene of MRN complex “RAD50” is required to keep DNA in close proximity by binding its ends. This gene is responsible to produce a protein that is essential in DNA DSBR pathway [16].
The structure–function studies of the MRN complex suggested that MRE11 subunit is necessary for the aphidicolin (APH)-induced activation of Chk1 [17]. Thomson et al. [18] reported that the MRN complex, particularly the nuclease activity of MRE11, plays a significant role in the activation of Chk1 in response to stalled replication forks. When cells cope with DNA lesions, Chk1 acts as the workhorse of checkpoint response by delaying the cell cycle and controlling the replication apparatus. RAD50 is a member of the ATP-binding cassette family of ATPases and contains coiled-coil domains characteristic of structural maintenance of chromosomes (SMC) proteins. RAD50 possesses both ATPase and adenylate kinase activities. Overall, RAD50 is important for the conformational dynamics of the MRN complex in response to various DNA structures. Lee and Dunphy [17] reported that MRE11–RAD50 (MR) complex without NBS1 was able to support phosphorylation of Chk1 as efficiently as the complete MRN complex. Hence, NBS1 appears to be dispensable for the APH-induced activation of Chk1 under these conditions.
It has been well established that the single nucleotide polymorphisms (SNPs) are most frequent genetic variations which may result in harmful effects on an individual [19,20,21]. Therefore, certain SNPs in MRE11 and RAD50 genes are strongly associated with cancer development and could be useful in diagnosis and treatment [22]. In contrast, several studies reported no association of NBS1 with the risk of several malignancies including breast cancer [23,24,25]. Therefore, present study was designed to evaluate the role of MRE11 and RAD50 genes in breast cancer development among the female population of Punjab, Pakistan. For this purpose, we conducted a case–control study to estimate the possible association of rs684507 (MRE11) and rs28903089 (RAD50) with breast cancer incidence.
Materials and methods
Study population
A total of 200 subjects with same age and sex group, including breast cancer patients and healthy individuals were selected. Briefly, 100 patients from different selected hospitals were recruited for blood samples and required information. Inclusion criteria for breast cancer subjects include (1) women with well-diagnosed breast cancer and (2) the parents of selected patients should be of Pakistani origin. The patients with any infectious disease were excluded from the study. The background of patients recruited in present study is given in Table 1. Sampling was carried out during the period of July 2015 to February 2016. The control subjects were taken from general population with normal health status. All the subjects were genetically unrelated and their blood samples were drawn with their consent. An ethical approval was obtained from Ethical Board of Institution to conduct study on human population.
Blood sampling
A 2–3 mL of blood was drawn from median cubital vein of the patient under aseptic conditions and stored in ethylenediaminetetraacetic (EDTA) vials at − 20 °C until proceeded for DNA isolation.
Genotyping
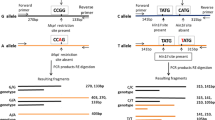
Isolation of genomic DNA was done using modified chloroform; isoamylalcohol protocol followed by gel electrophoresis. Samples were diluted and quantified to obtain the required DNA concentration of 10 ng/µL. Frequently reported SNPs (i.e., rs684507; MRE11 and rs28903089; RAD50) associated with breast cancer were selected for further study. Genotyping was carried out by Tetra-ARMS PCR following the protocol described earlier [26] with minor modifications in the quantity of chemicals that are given in Table 2. The primer sequences used in the study are given below.
MRE11 (rs684507):
Forward inner primer 5′-AGAACCGTATGTGACCCTTTCTGACT
Reverse inner primer 5′-ATGGGAAATTAATGTATGCTAAATAACTG
Forward outer primer 5′-CGTGTTTGTTTATTTACACTCGCTTTAA
Reverse outer primer 5′-GGCAAATTTAGAAGTCTCATTTTCATCT
RAD50 (rs28903089):
Forward inner primer 5′-TCGTGATCAGATTACAAGTAAGGAGGA
Reverse inner primer 5′-TGACAATTTCCTTTGAAGATGTTAACTAGG
Forward outer primer 5′-AGTGACAGCATAATATCCCACTGTATGAA
Reverse outer primer 5′-ACGTTAAATAGCTTGATTTAGCCAGTCC
For MRE11 SNP, PCR program was optimized as follows:
An initial denaturation at 94 °C for 4 min, followed by 35 cycles—denaturation at 94 °C for 1 min, annealing at 59 °C for 1.5 min, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min.
For RAD0 SNP, PCR program was optimized at an initial denaturation at 94 °C for 4 min, followed by 35 cycles—denaturation at 94 °C for 1 min, annealing at 60 °C for 1.5 min, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. Subsequently, PCR products were checked on 2% agarose gel.
Statistical analyses
Chi-square test of association was done to analyze the association of different clinical factors with the risk of breast cancer in patients in comparison with controls using SPSS (Version 22). Exact test for Hardy–Weinberg equilibrium was applied for both genetic variants. Allelic and genotypic frequencies were calculated using SNPStats.
Results
We found that breast cancer is significantly associated with positive family history, married women and house wives in patients compared to controls (Table 3). In the present study, two SNPs were genotyped by tetra-ARMS PCR to reveal their association with breast cancer. The MRE11 SNP showed significant association with cancer development (Exact test for Hardy–Weinberg equilibrium, p value < 0.0001; Table 4). Detailed analysis showed that the individual alleles are not significantly associated with disease development (OR 1.27, CI 0.69–2.36, p value = 0.43) as the proportion of variant C is almost same in both groups (i.e., patients and controls). However, the genotype frequency showed significant association with breast cancer (OR 3.71, CI 1.68–8.18, p value < 0.00001; Table 5) and can cause more than three times higher risk for disease development. In detail, MRE11 SNP showed overdominant model (logistic regression model, p value < 0.0001) and confirmed that the heterozygous TC was more responsible for disease compared to homozygotes (TT, CC; see Table 6).
The SNP rs28903089 of RAD50 showed a slight association with the disease (Exact test for Hardy–Weinberg equilibrium, p value 0.07; Table 4). Allelic frequencies showed significant association of variant with breast cancer (OR 2.3976, CI 1.32–4.32, p = 0.0037). The genotype frequencies also showed significant association with disease (OR 0.55, CI 0.29–1.02, p = 0.0036) acting as a protective allele.
As the result of regression analysis, single copy of variant C was mainly responsible (logistic regression, p value < 0.005) to decrease the risk of disease whereas heterozygous (AC) and homozygous genotypes had the same effect (Table 7).
Discussion
Breast cancer is the most frequently diagnosed malignancy and second leading cause of cancer death among women worldwide. Beside alterations in certain genes, several factors such as family history, marital status and lifestyle play significant roles in cancer development [27]. In the present study, we found that the risk of breast cancer is associated with patients that have positive family history, married and house wives compared to controls (Table 3). Due to the importance of MRN complex genes in maintenance of the genomic integrity and DNA damage repair to prevent malignancy, we analyzed the significance of two MRN complex polymorphisms for the risk of breast cancer. Both of the SNPs showed significant association with malignancy of breast tissues. The polymorphisms of RAD50 and MRE11 have been widely associated with cancer as they are among the major DNA damage repair genes [28].
We observed that the MRE11 SNP showed significant association with elevated risk of breast cancer and the heterozygous TC was mainly responsible for disease (OR 3.71, CI 1.68–8.18, p value < 0.00001). In contrast, the SNP rs28903089 of RAD50 showed a slight association with the disease and single copy of variant C is giving protective effect against cancer development (OR 0.55, CI 0.29–1.02, p = 0.0036). It has been well established that variations in DNA damage repair genes are highly associated with increased risk of breast cancer [29]. Basically, alteration in genes of DNA DSBR pathway affects DNA double-strand repair and maintenance of telomere length [3, 30,31,32]. Dysregulation of these pathways may lead to cancer development. Several studies have reported the involvement of RAD50 and MRE11 SNPs in breast cancer, nevertheless variation may occur among different populations [33]. Kuschel et al. [34] and Heikkinen et al. [33] reported a strong association of MRE11 and RAD50 SNPs with breast cancer in specific populations.
In contrast, our results showed no significant association of RAD50 SNP rs28903089 with increased risk of breast cancer. It might be due to the fact that the RAD50 SNP rs28903089 is rare in breast cancer and have low penetrance. Furthermore, its association could not be detected because of limited sample size. Similar results have been reported elsewhere [35, 36]. These data indicate that RAD50 mutations are rare in familial breast cancer and either carry no, or a very small, increased risk of cancer. Altogether, these results suggest RAD50 can only be making a very minor contribution to familial breast cancer predisposition. MRE11 and RAD50 polymorphisms are well studied in different populations but their association patterns are controversial [37]. Some studies report these SNPs playing protective role while other associate them with increased risk of disease [6, 38,39,40].
More comprehensive studies with larger sample size are required to determine broader range of different factors involved in breast cancer. These studies will aim to eradicate the limitations of present study and also to validate its results and findings. Despite of having some limitations, the findings of the present study are remarkable. Both markers showed association with the onset of breast cancer. Due to the findings of this research, the markers which are studied here can open up ways to improve the present treatment strategies against breast cancer in upcoming future.
Conclusion
In conclusion, our results suggest that not every polymorphism is associated with the increased risk of disease as in the case of RAD50 SNP but the SNP of MRE11 is involved in increased risk of breast cancer development as it plays a significant role in DNA damage repair. Both SNPs, more or less, are associated with response subjects in the studied population. Major proportion of patients was heterozygous showing the effectiveness of variants in disease population with dominant and over dominant model.
References
Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15(5):R92.
Tariq A, Majeed I, Khurshid A. Types of cancers prevailing in Pakistan and their management evaluation. Asian Pac J Cancer Prev. 2015;16(9):3605–16.
Majeed W, Aslam B, Javed I, Khaliq T, Muhammad F, Ali A, et al. Breast cancer: major risk factors and recent developments in treatment. Asian Pac J Cancer Prev. 2014;15(8):3353–8.
DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62.
Asif HM, Sultana S, Akhtar N, Rehman JU, Rehman RU. Prevalence, risk factors and disease knowledge of breast cancer in Pakistan. Asian Pac J Cancer Prev. 2014;15(11):4411–6.
Amirali Z, Edgar K, Azim ZA, Sadruddin S. Breast cancer in Pakistani females. Imanagers J Nurs. 2014;4(2):11.
Oboho IK, Tomczyk SM, Al-Asmari AM, Banjar AA, Al-Mugti H, Aloraini MS, et al. 2014 MERS-CoV outbreak in Jeddah—a link to health care facilities. N Engl J Med. 2015;372(9):846–54.
Gokdemir-Yazar O, Yaprak S, Colak M, Yildirim E, Guldal D. Family history attributes and risk factors for breast cancer in Turkey. Asian Pac J Cancer Prev. 2014;15(6):2841–6.
Hilakivi-Clarke L, de Assis S, Warri A. Exposures to synthetic estrogens at different times during the life, and their effect on breast cancer risk. J Mammary Gland Biol Neoplasia. 2013;18(1):25–42.
Mozaffarian D, Fahimi S, Singh GM, Micha R, Khatibzadeh S, Engell RE, et al. Global sodium consumption and death from cardiovascular causes. N Engl J Med. 2014;371(7):624–34.
Cipollini G, Tommasi S, Paradiso A, Aretini P, Bonatti F, Brunetti I, et al. Genetic alterations in hereditary breast cancer. Ann Oncol. 2004;15(suppl_1):i7–13.
Petrini JH, Walsh ME, DiMare C, Chen X-N, Korenberg JR, Weaver DT. Isolation and characterization of the human MRE11 homologue. Genomics. 1995;29(1):80–6.
Venkatesan P, Puvvada N, Dash R, Kumar BP, Sarkar D, Azab B, et al. The potential of celecoxib-loaded hydroxyapatite–chitosan nanocomposite for the treatment of colon cancer. Biomaterials. 2011;32(15):3794–806.
Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–5.
Hsu H-M, Wang H-C, Chen S-T, Hsu G-C, Shen C-Y, Yu J-C. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Prev Biomark. 2007;16(10):2024–32.
Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12(2):90–103.
Lee J, Dunphy WG. The Mre11–Rad50–Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell. 2013;24(9):1343–53.
Thomson AM, Gillespie PJ, Blow JJ. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol JCB. 2010;188(2):209–21. https://doi.org/10.1083/jcb.200911037.
Mizoo T, Taira N, Nishiyama K, Nogami T, Iwamoto T, Motoki T, et al. Effects of lifestyle and single nucleotide polymorphisms on breast cancer risk: a case–control study in Japanese women. BMC Cancer. 2013;13(1):565.
Campa D, Barrdahl M, Gaudet MM, Black A, Chanock SJ, Diver WR, et al. Genetic risk variants associated with in situ breast cancer. Breast Cancer Res. 2015;17(1):82.
Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, et al. The African genome variation project shapes medical genetics in Africa. Nature. 2015;517(7534):327–32.
Kooshyar MM, Nassiri M, Nasiri K. Hereditary genes and SNPs associated with breast cancer. Asian Pac J Cancer Prev. 2013;14(6):3403–409.
Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta. 2005;359(1):150–5.
Carlomagno F, Chang-Claude J, Dunning AM, Ponder BA. Determination of the frequency of the common 657Del5 Nnijmegen breakage syndrome mutation in the German population: no association with risk of breast cancer. Genes Chromosomes Cancer. 1999;25(4):393–5.
Gorski B, Dębniak T, Masojć B, Mierzejewski M, Mędrek K, Cybulski C, et al. Germline 657del5 mutation in the NBS1 gene in breast cancer patients. Int J Cancer. 2003;106(3):379–81.
Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):e88.
Montazeri A, Vahdaninia M, Harirchi I, Harirchi AM, Sajadian A, Khaleghi F, et al. Breast cancer in Iran: need for greater women awareness of warning signs and effective screening methods. Asia Pac Fam Med. 2008;7(1):6.
McTiernan A, Kooperberg C, White E, Wilcox S, Coates R, Adams-Campbell LL, et al. Recreational physical activity and the risk of breast cancer in postmenopausal women: the Women’s Health Initiative Cohort Study. JAMA. 2003;290(10):1331–6.
Kim H, Cho D, Choi D, Jung G, Shin I, Park W, et al. Abstract P1-08-08: heterozygous germline mutations in RAD50 among Korean patients with high-risk breast cancer negative for BRCA1/2 mutation. AACR. 2016. https://doi.org/10.1158/1538-7445
Cancer CGoHFiB. Menarche, menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol. 2012;13(11):1141–51.
Van Ravesteyn NT, Miglioretti DL, Stout NK, Lee SJ, Schechter CB, Buist DS, et al. Tipping the balance of benefits and harms to favor screening mammography starting at age 40 years: a comparative modeling study of risk. Ann Intern Med. 2012;156(9):609–17.
Sineshaw HM, Gaudet M, Ward EM, Flanders WD, Desantis C, Lin CC, et al. Association of race/ethnicity, socioeconomic status, and breast cancer subtypes in the National Cancer Data Base (2010–2011). Breast Cancer Res Treat. 2014;145(3):753–63.
Heikkinen K, Rapakko K, Karppinen S-M, Erkko H, Knuutila S, Lundán T, et al. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27(8):1593–9.
Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11(12):1399–407.
Tommiska J, Seal S, Renwick A, Barfoot R, Baskcomb L, Jayatilake H, et al. Evaluation of RAD50 in familial breast cancer predisposition. Int J Cancer. 2006;118(11):2911–6.
Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22.
Assi HA, Khoury KE, Dbouk H, Khalil LE, Mouhieddine TH, El Saghir NS. Epidemiology and prognosis of breast cancer in young women. J Thorac Dis. 2013;5(Suppl 1):S2.
Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18(7):748–54.
Venkitaraman AR. Tumour suppressor mechanisms in the control of chromosome stability: insights from BRCA2. Mol Cells. 2014;37(2):95.
Amgiasvasanth A, Patil PS. Profile of breast cancer patients attending a tertiary care centre: a cross-sectional study. Int J Community Med Public Health. 2017;3(3):663–7.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
About this article
Cite this article
Khan, R.T., Siddique, A., Shahid, N. et al. Breast cancer risk associated with genes encoding DNA repair MRN complex: a study from Punjab, Pakistan. Breast Cancer 25, 350–355 (2018). https://doi.org/10.1007/s12282-018-0837-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-018-0837-9