Abstract
Background
No clinically useful target molecule has been identified for triple-negative (TN) breast cancer, i.e., estrogen receptor (ER)-negative, progesterone receptor (PgR)-negative, human epidermal growth factor receptor-2 (HER2)-negative phenotype, and its prognosis is poor. Triple-negative cancer consists of two subtypes: basal-like and non-basal-like. The aim of this study is to clarify the clinical and biological characteristics of these two subtypes of TN cancer.
Methods
We examined, by immunohistochemistry, expression of biological markers cytokeratin (CK) 5/6 and epidermal growth factor receptor (EGFR) in triple-negative breast cancer. Basal-like subtype was defined as CK5/6-positive and/or EGFR-positive, and non-basal-like subtype was defined as no expression of these two markers. We studied the correlation between basal-like subtype and several factors related to tumor progression, along with the prognostic value of basal-like subtype and other biological markers in triple-negative cancer.
Results
In the 48 cases of operable triple-negative breast cancer, basal-like subtype was detected in 22 (45.8%) and non-basal-like subtype in 26 (54.2%). Basal-like subtype was significantly correlated with nodal status (P = 0.0475) and nuclear grade (P = 0.0475). Basal-like subtype was also significantly associated with Ki67 labeling index (P = 0.0118), c-kit expression (P = 0.0335), and aurora A expression (P = 0.0020). No association was detected between basal-like cancer and other biological markers. Patients with basal-like subtype of triple-negative cancer showed shorter disease-free survival (P = 0.0049) and overall survival (P = 0.0283) than patients with non-basal-like subtype. No independent prognostic factors were identified among the prognostic factors obtained from univariate analysis.
Conclusions
These findings suggest that basal markers can be used to classify triple-negative breast cancer into at least two subtypes with differing prognoses. It is necessary to develop a novel treatment strategy to improve the prognosis of patients with basal-like subtype of triple-negative breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological markers such as the estrogen receptor (ER) and human epidermal growth factor receptor-2 (HER2) are widely accepted in terms of their clinical importance in breast cancer [1]. Hormone-receptor-positive (i.e., ER-positive and/or PgR-positive) and HER2-positive (i.e., HER2 protein overexpression or gene amplification) breast cancers account for 75–80% and 15–20% of breast cancer cases, respectively [2, 3]. Some studies have found about half of patients with HER2-positive breast cancer to be hormone-receptor-positive [2]. Hormone-receptor-positive and HER2-positive breast cancers have effective therapeutic molecular targets. Hormone-receptor-positive breast cancer is treated with selective ER modulators, gonadotropin-releasing hormone agonists, and aromatase inhibitors. HER2-positive breast cancer is treated with the humanized monoclonal antibody trastuzumab and tyrosine kinase inhibitor lapatinib. The remaining 10–15% of breast cancer cases [4–11] fall into a so-called triple-negative cancer phenotype, which is characterized by absent expression of hormone receptors and nonoverexpressing HER2. Triple-negative breast cancer has been a particular focus of attention because this phenotype has no confirmed therapeutic molecular target and a poor prognosis [12, 13].
DNA microarray profiling studies on breast cancer have identified distinct subtypes of this cancer that are associated with different clinical outcomes. Breast cancer was classified into at least four subtypes: luminal (ER-positive), HER2-overexpressing, normal-breast-like, and basal-like cancers. The HER2-overexpressing and basal-like subtypes had a poor prognosis compared with the luminal subtype [14, 15]. Basal-like cancer is characterized by constitutive expression of genes usually found in normal basal/myoepithelial cells of the breast, including basal cytokeratins (CK5/6, CK14, and CK17), vimentin, P-cadherin, and p63 [14, 16–18]. In addition, basal-like cancer is similar to triple-negative breast cancer because almost all basal-like cancers do not express ER, PgR, and HER2. Basal-like breast cancer has aggressive characteristics, such as high histological grade, p53 mutation, epidermal growth factor receptor (EGFR) overexpression, c-MYC amplification, loss of function of BRCA1, and cytogenetic abnormalities [12, 19].
Although the basal-like subtype is identified by a microarray-based gene expression profile, Nielsen et al. [16] immunohistochemically characterized this subtype by analyzing a panel of 21 basal-like tumors that had been previously classified using gene expression profiles. The investigators found that four antibodies (ER, EGFR, HER2, and CK5/6) can be used to identify basal-like cancer immunohistochemically [16]. The basal-like subtype was defined as lacking both ER and HER2 expression, but it was positive for the expression of CK5/6 and EGFR. The panel using these four antibodies showed a specificity of 100% and a sensitivity of 76% for the identification of basal-like cancer. However, Rouzier et al. [20] demonstrated that ER expression and HER2 expression were seen in 5% and 14%, respectively, of basal-like cancers that had been diagnosed by gene expression profiling. Therefore, triple-negative cancer is not necessarily basal-like cancer. Normal-breast-like cancer, as identified by gene expression profiling, also shows absent ER and HER2 expression. Several studies showed that 16–47% of triple-negative breast cancers did not express basal markers [7, 8, 21–24]. Taken together, these data indicate that basal-like cancer is not identical to triple-negative cancer, according to the results of microarray-based gene expression profiling, which is considered to be the gold standard for the identification of basal-like cancer. These reports also indicate that triple-negative cancer is a heterogeneous disease.
Therefore, we sought to clarify the clinicopathological significance of the basal-like and non-basal-like subtypes of triple-negative cancer. We examined the expression of basal markers, CK5/6 and EGFR, using immunohistochemical methods in cases of triple-negative cancer, the correlation between basal-like cancer and clinicopathological factors, and the prognostic implications of basal-like cancer.
Patients, materials, and methods
Patients and tissue samples
Tissue samples were obtained from 218 patients with invasive breast cancer who were diagnosed between 1993 and 2004 at Kumamoto University Hospital in Kumamoto, Japan. A total of 218 specimens of primary invasive carcinoma were obtained from resected tumor. All specimens were fixed in neutral 10% buffered formaldehyde, embedded in paraffin, cut into 4-μm slices, and supplied for this study. None of these cancer patients received treatment prior to surgery. The patients underwent standard and partial mastectomies with fully resected axillary dissections. Patients ≤70 years old received anthracycline-containing chemotherapy if the tumor was node positive. Endocrine therapy was given for 5 years to patients with ER-positive tumors. Median follow-up was 5.5 years (range, 0.3–14.8 years), during which there were 53 relapses and 29 deaths. The HR(+)/HER2(−), HER2(+) and triple-negative breast cancer phenotypes were identified in 147 (67.4%), 23 (10.6%), and 48 (22.0%) of the 218 cases of invasive breast cancer, respectively. Other background data for the 218 cancer patients are shown in Table 1.
Immunohistochemical techniques
The expression of ER, PgR, HER2, CK5/6, EGFR, and other biological markers was determined immunohistochemically in paraffin-embedded tissue specimens. Table 2 summarizes all the antibodies, dilutions, incubation times, and cutoff values used for this analysis. Histological sections (4 μm in thickness) were transferred to silane-coated slides and were air-dried overnight at 37°C. The sections were deparaffinized in xylene and rehydrated through a graded series of decreasing ethanol concentrations. Endogenous peroxidase activity was blocked for 10 min in methanol containing 0.3% hydrogen peroxide, and the slides were then rinsed with Tris-buffered saline (TBS) solution.
Expression of the biological markers, except hypoxia-inducible factor-1α (HIF-1α), was determined by the Histofine Simple Stain MAX-PO® method (Nichirei Biosciences, Tokyo, Japan). This method has been previously described elsewhere [25]. The slides were incubated with primary antibodies and washed in TBS three times. Following this step, the slides were incubated with Histofine Simple Stain MAX-PO®, the labeled polymer (prepared by combining amino acid polymers with peroxidase), and either goat anti-mouse immunoglobulin (Ig) or goat anti-rabbit Ig (which is reduced to Fab’ after incubation for 30 min at room temperature).
Immunohistochemical staining for HIF-1α was performed with the Catalyzed Signal Amplification System (Dako Japan, Tokyo, Japan), which utilizes a streptavidin–biotin–horseradish peroxidase complex. This method has also been previously described elsewhere [25]. After antigen retrieval, the slides were treated in accordance with the manufacturer’s instructions. The slides were incubated with the monoclonal mouse anti-HIF-1α antibody 67 (Novus Biologicals, Littleton, CO, USA) for 30 min at room temperature.
Peroxidase activity was detected by placing 3,3′-diaminobenzidine (Dako Japan, Tokyo, Japan) on each section for 5 min at room temperature. The slides were counterstained with hematoxylin and dehydrated in alcohol and xylene before the slides were mounted. Negative controls were performed using nonimmune serum or phosphate-buffered saline (PBS) in place of the primary antibodies.
Evaluation of immunohistochemical staining
The expression of ER or PgR was designated as positive when at least 10% of the tumor nuclei showed positive staining. The expression of HER2 was classified according to the HercepTest® assay’s scoring system, which includes four categories, namely 0, 1+, 2+ and 3+, based on the intensity and proportion of membrane staining in tumor cells. Positivity was defined as a HER2 score of 3+ for immunostaining or a ≥2-fold increase in HER2 gene amplification, as determined by fluorescence in situ hybridization (FISH). The expression of Ki67 was measured as the percentage of stained nuclei in 1,000 tumor cell nuclei. The measurements were performed in five randomly selected fields, with the Ki67 labeling index determined as the average of these values. Microvessel density (MVD) was evaluated based on the number of endothelial cell deposits per mm2 in the areas considered to be most active for neovascularization (hot spots). The counts were performed in five fields, with the average of the highest three counts (of the five) used as the value for MVD. The expression of CK5/6, CK18/8, EGFR, vascular endothelial growth factor (VEGF), p53, COX-2 or c-kit was designated as positive when at least 10% of the tumor cells showed positive staining (cytoplasmic staining for CK5/6, CK8/18, EGFR, VEGF, COX-2, and c-kit; nuclear staining for p53). The expression of HIF-1α was designated as positive when at least 5% of the tumor cells showed nuclear positive staining according to Bos et al. [26]. Expression of aurora A was designated as positive when at least 1% of the tumor cells showed cytoplasmic positive staining according to Royce et al. [27]. Two independent observers evaluated the immunohistochemical data without knowledge of the clinical outcomes of the patients.
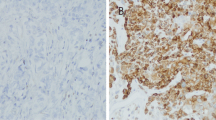
ER-positive and/or PgR-positive expression combined with HER2-negative expression was defined as HR(+)/HER2(−). HER2-positive expression, irrespective of the hormone receptor status, was defined as HER2(+). The combination of ER-negative, PgR-negative, and HER2-negative expression was defined as triple-negative breast cancer. CK5/6-positive and/or EGFR-positive expression in cases of triple-negative breast cancer was defined as the basal-like subtype. On the other hand, CK5/6-negative and EGFR-negative expression in cases of triple-negative breast cancer was defined as the non-basal-like subtype.
Histology
Histological examinations were performed on slides with paraffin-embedded samples stained by hematoxylin–eosin, according to the criteria of the Japanese Breast Cancer Society, which are based on International Union against Cancer (UICC) tumor–node–metastasis (TNM) classification criteria.
Statistical analysis
For statistical analysis, the chi-squared test and unpaired t test were used for analysis of two unpaired samples. Disease-free survival and overall survival rates and time periods after surgical resection were calculated by the Kaplan–Meier method, and differences in survival curves were assessed by the log-rank test. The Cox proportional hazards model was used for multivariate analysis. All analyses were performed with the JMP® IN version 5.1 software package (SAS® Institute Japan, Tokyo, Japan). A P value of less than 0.05 was regarded as statistically significant. All statistical tests were two-sided.
Results
Basal-like and non-basal-like breast cancer
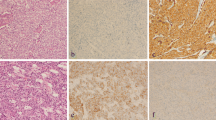
Triple-negative breast cancer was classified into two subtypes (basal-like and non-basal-like breast cancer) according to the expression of CK5/6 and EGFR. Immunohistochemical staining of CK5/6 and/or EGFR was identified in the cytoplasmic membrane of tumor cells (Fig. 1). The expression of CK5/6 or EGFR was found in 15 (31.3%) and 16 (33.3%) of the 48 cases of triple-negative cancer, respectively. Basal-like or non-basal-like cancer was detected in 22 (45.8%) and 26 (54.2%) cases of triple-negative cancer, respectively (Table 3).
Correlations between basal-like subtype and clinicopathological factors
Correlations between the basal-like versus non-basal-like subtype and clinicopathological factors are shown in Table 4. The basal-like subtype was positively correlated with nodal metastasis (P = 0.0475) and high nuclear grade (P = 0.0475). These results suggest that the basal-like subtype has more aggressive phenotype compared with the non-basal-like subtype in triple-negative breast cancer. Basal-like cancer was marginally associated with histological type (P = 0.0519). For example, non-basal-like cancer had higher rates of solid-tubular carcinoma than basal-like cancer. Three cases of other histological types of carcinoma in basal-like cancer were two cases of spindle cell carcinoma and one case of adenoid cystic carcinoma. No association with basal subtype was found in terms of age, menopausal status, tumor size or clinical stage.
Correlations between basal-like subtype and biological markers
Correlations between basal-like cancer and the biological markers are summarized in Table 5. The basal-like subtype was positively correlated with expression of c-kit (P = 0.0335) and aurora A (P = 0.0020). In addition, the basal-like subtype was positively associated with a high Ki67 labeling index (P = 0.0118) and marginally associated with the infiltration of CD68-positive stromal cells in tumor tissue (P = 0.0551). However, the basal-like subtype was not correlated with CK8/18, p53, MVD, VEGF-A, HIF-1α or COX-2.
Survival analysis
In univariate survival analysis, patients with basal-like cancer had significantly shorter disease-free survival (P = 0.0049) and overall survival (P = 0.0283) than patients with non-basal-like cancer (Figs. 2, 3). In multivariate analysis of disease-free survival by Cox regression analysis, no independent prognostic factor was identified among the prognostic factors that had been obtained from univariate analysis (Table 6).
Discussion
We demonstrated that triple-negative breast cancer can be classified into at least two subtypes of cancer according to the expression of basal markers CK5/6 and EGFR. In addition, we showed the clinicopathological significance of these subtypes, including their prognostic impact on survival in patients with triple-negative breast cancer.
In our study, the basal-like tumor subtype was detected in 46% of triple-negative cancers. This proportion is less than in previously published data, which included basal-like cancer rates ranging from 53% to 84% in cases of triple-negative breast cancer [7, 8, 21–24]. The definition of triple-negative cancer in this study was ER expression in <10% of tumor nuclei, PgR expression in <10% of tumor nuclei, and nonoverexpression of HER2. In addition, the cutoff value for CK5/6 and EGFR expression was ≥10% of the cytoplasmic membrane staining positive for these two markers. Our study’s cutoff values for these markers differed from previously reported definitions. For example, Nielsen et al. [16] reported that ER positivity was identified by any staining of tumor nuclei, HER2 positivity was defined as strong membranous staining, and CK5/6 and EGFR positivity was defined as any cytoplasmic and/or nuclei staining in tumor specimens. Our definition of triple-negative cancer was based on published guidelines for the primary treatment of breast cancer, such as those from the St. Gallen International Consensus Conference of 2007 [28], and this definition is widely accepted in the adjuvant setting.
Basal-like cancer correlated with lymph node metastasis and high nuclear grade in this study. Sasa et al. [24] showed that basal-like cancer was associated with tumor size and nuclear grade but not nodal status. Cheang et al. [23] also demonstrated that the basal-like subtype was significantly associated with high grade (87% of cases were grade 3) and young age (18.8% were <40 years old) when the basal-like subtype was compared with cancer that was negative for all five markers (ER, PgR, HER2, CK5/6, and EGFR). The investigators showed that the five-marker classification of the basal-like subtype identified a subset of particularly high-risk patients [23].
We also showed that the basal-like subtype was associated with a high Ki67 labeling index, c-kit expression, and aurora A expression. The high Ki67 labeling index is related to the mitotic index and high levels of cell proliferation [29]. The proto-oncogene c-KIT encodes for a surface membrane tyrosine kinase receptor, and its ligand is the stem cell factor. In gastrointestinal stromal tumors, activating mutations in the c-KIT gene play a crucial role in tumor progression [30]. The c-kit protein is highly expressed in normal epithelium, and its expression is decreased in invasive cancer [31, 32]. Nielsen et al. [16] reported that expression of the c-kit protein was found in approximately 30% of basal-like cancers. The role of c-kit does not fully elucidate breast cancer initiation and progression. Aurora A is one of the serine/threonine protein kinases and is involved in multiple mitotic events: regulation of the spindle assembly checkpoint pathway, function of centrosomes and the cytoskeleton, and cytokinesis [33]. In experimental rodent models, aberrant expression of aurora A was found to be oncogenic [34, 35]. Thus, overexpression of aurora A may lead to cancer. Aurora A was not identified a prognostic factor for DFS, although aurora A was significantly associated with basal-like subtype in this study. Royce et al. [27] demonstrated that overexpression of aurora A was associated with nuclear grade but not prognosis in 112 invasive breast cancer cases. On the other hand, Nadler et al. [36] recently showed that aurora A was an independent prognostic factor in 638 breast cancer patients during 15-year follow-up. These results suggest that basal-like tumors have an aggressive phenotype compared with non-basal-like tumors in cases of triple-negative cancer.
In univariate analysis, three factors: nodal status (P = 0.0023), Ki67 labeling index (P = 0.020), and basal markers (P = 0.0064), were identified as prognostic factors affecting disease-free survival. However, in multivariate analysis, no independent prognostic factor was identified. One of the reasons is that only a small number of patients were examined in this study, making it difficult to reach statistical significance. In a previous large study, basal markers (CK5/6 and/or EGFR) were identified as independent prognostic factors [7, 23]. Taken together, the available data suggest that triple-negative breast cancer can be classified into at least two subgroups based on basal markers, which are predictive of different prognosis.
In conclusion, our study showed triple-negative breast cancer to be a heterogeneous disease that consists of at least two phenotypes based on the expression of basal markers (CK5/6 and/or EGFR). Basal-like breast cancer is an aggressive phenotype and has an extremely poor prognosis, although all the study patients were treated with standard therapy according to conventional practice guidelines. Based on the poor prognosis associated with the basal-like subtype, a novel treatment strategy for this type of breast cancer is urgently needed.
Abbreviations
- ER:
-
Estrogen receptor
- PgR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor-2
- EGFR:
-
Epidermal growth factor receptor
References
Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–312.
Konecny G, Pauletti G, Pegram M, Untch M, Dandekar S, Aguila Z, et al. Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst. 2003;95:142–53.
Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12.
Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34.
Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–7.
Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32.
Tischkowitz M, Brunet JS, Begin LR, Huntsman DG, Cheang MC, Akslen LA, et al. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134.
Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8.
Harris LN, Broadwater G, Lin NU, Miron A, Schnitt SJ, Cowan D, et al. Molecular subtypes of breast cancer in relation to paclitaxel response and outcomes in women with metastatic disease: results from CALGB 9342. Breast Cancer Res. 2006;8:R66.
Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer. 2007;110:876–84.
Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–44.
Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–18.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74.
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–23.
Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74.
van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–6.
Matos I, Dufloth R, Alvarenga M, Zeferino LC, Schmitt F. p63, cytokeratin 5, and P-cadherin: three molecular markers to distinguish basal phenotype in breast carcinomas. Virchows Arch. 2005;447:688–94.
Kobayashi S. Basal-like subtype of breast cancer: a review of its unique characteristics and their clinical significance. Breast Cancer. 2008;15:153–8.
Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–85.
Bidard FC, Conforti R, Boulet T, Michiels S, Delaloge S. Andre F.egative phenotype accurately identify basal-like tumour? An immunohistochemical analysis based on 143 ‘triple-negative’ breast cancers. Ann Oncol. 2007;18:1285–6.
Tan DS, Marchio C, Jones RL, Savage K, Smith IE, Dowsett M, et al. Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res Treat. 2008;111:27–44.
Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–76.
Sasa M, Bando Y, Takahashi M, Hirose T, Nagao T. Screening for basal marker expression is necessary for decision of therapeutic strategy for triple-negative breast cancer. J Surg Oncol. 2008;97:30–4.
Yamamoto Y, Ibusuki M, Okumura Y, Kawasoe T, Kai K, Iyama K, et al. Hypoxia-inducible factor 1alpha is closely linked to an aggressive phenotype in breast cancer. Breast Cancer Res Treat. 2008;110:465–75.
Bos R, van der Groep P, Greijer AE, Shavarts A, Meijer S, Pinedo HM, et al. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–81.
Royce ME, Xia W, Sahin AA, Katayama H, Johnston DA, Hortobagi G, et al. STK15/Aurora-A expression in primary breast tumors is correlated with nuclear grade but not with prognosis. Cancer. 2003;100:12–9.
Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. J Clin Oncol. 2003;21:3357–65.
Spyratos F, Ferrero-Pous M, Trassard M, Hacene K, Phillips E, Tubiana-Hulin M, et al. Correlation between MIB-1 and other proliferation markers: clinical implications of the MIB-1 cutoff value. Cancer. 2002;94:2151–9.
Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–80.
Natali PG, Nicotra MR, Sures I, Mottolese M, Botti C, Ullrich A. Breast cancer is associated with loss of the c-kit oncogene product. Int J Cancer. 1992;52:713–7.
Chui X, Egami H, Yamashita J, Kurizaki T, Ohmachi H, Yamamoto S, et al. Immunohistochemical expression of the c-kit proto-oncogene product in human malignant and non-malignant breast tissues. Br J Cancer. 1996;73:1233–6.
Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Gandara DR Jr. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14:1639–48.
Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–65.
Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–93.
Nadler Y, Camp RL, Schwartz C, Rimm DL, Kluger HM, Kluger Y. Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin Cancer Res. 2008;14:4455–62.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Yamamoto, Y., Ibusuki, M., Nakano, M. et al. Clinical significance of basal-like subtype in triple-negative breast cancer. Breast Cancer 16, 260–267 (2009). https://doi.org/10.1007/s12282-009-0150-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-009-0150-8