Abstract
Autophagy is a catabolic process through which organelles and cellular components are sequestered into autophagosomes and degraded via fusion with lysosomes. Autophagy plays a role in many physiological processes, including stress responses, energy homeostasis, elimination of cellular organelles, and tissue remodeling. In addition, autophagy capacity changes in various disease states. A series of studies have shown that autophagy is strictly controlled to maintain homeostatic balance of energy metabolism and cellular organelle and protein turnover. These studies have also shown that this process is post-transcriptionally controlled by small noncoding microRNAs that regulate gene expression through complementary base pairing with mRNAs. Conversely, autophagy regulates the expression of microRNAs. Therefore, dysregulation of the link between autophagy and microRNA expression exacerbates the pathogenesis of various diseases. In this review, we summarize the roles of autophagy and microRNA dysregulation in the course of liver diseases, with the aim of understanding how microRNAs modify key autophagic effector molecules, and we discuss how this dysregulation affects both physiological and pathological conditions. This article may advance our understanding of the cellular and molecular bases of liver disease progression and promote the development of strategies for pharmacological intervention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Autophagy is an evolutionarily conserved, dynamic cellular process that involves lysosomal degradation and recycling of intracellular organelles and proteins to maintain cellular homeostasis under various stresses such as starvation, protein aggregation, viral infection, oxidative stress, and endoplasmic reticulum (ER) stress (Levine and Klionsky 2004). Fundamental to this event is the formation of the autophagosome, a double-membrane cytosolic vesicle, a step required for the sequestration of cytoplasmic cargo and delivery to the lysosomes. As the liver is one of the most dynamic metabolic organs in mammals, it is vulnerable to stress related-autophagy activation. Autophagy functions to remove damaged cellular components, such as mitochondria and long-lived proteins, through lysosome-dependent machinery. Indeed, autophagy is involved in various aspects of liver pathophysiology (Ding 2010; Yin et al. 2008; Codogno and Meijer 2013). Genetic ablation of autophagy in mice led to the accumulation of damaged organelles and aggregated proteins, steatosis, and liver injury. Growing evidence from an increasing number of autophagy studies shows that autophagy is also involved in many liver diseases, including viral hepatitis, alcoholic/nonalcoholic fatty liver, fibrosis, and hepatocellular carcinoma (HCC) (Table 1).
Autophagic pathways
Autophagy, which literally means “self-eating,” was originally introduced in the 1950s and was later systematically studied by de Duve and Wattiaux in the liver of animal models (Terlecky and Dice 1993). Although autophagic degradation has been recognized since the discovery of lysosomes, in-depth studies of autophagic pathways are more recent. In particular, the liver was used as a model of the morphological aspects and hormonal regulation of autophagy. The main drive for the discovery of autophagic molecules originated from a yeast study that led to the identification of autophagy-related genes (Atg) (Klionsky et al. 2003). The novel physiological functions of autophagy were identified by using genetic models of the Atg genes, which connected dysfunction of this lysosomal pathway to various human diseases, such as cancer, neurodegeneration, myopathies, and metabolic disorders (Mizushima et al. 2008; Cuervo 2004).
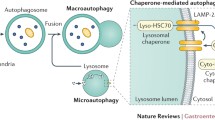
In eukaryotic cells, three distinct types of autophagy have been identified, macroautophagy (herein described as autophagy), microautophagy, and chaperone-mediated autophagy (CMA) (Klionsky 2005). In autophagy, an autophagosome is formed from a portion of engulfed cytosol, and it fuses with a lysosome for enzymatic degradation of sequestered cellular constituents (Mehrpour et al. 2010). The autophagy process is very complicated, and is regulated by the coordinated actions of the Atgs (Mizushima and Levine 2010; Pyo et al. 2012). First, upon induction, an isolated portion of membrane, called a phagophore, makes a small vesicular sac that elongates and subsequently encloses a portion of the cytoplasm, resulting in the formation of an autophagosome. The source of the double membrane might originate from the ER, mitochondria, or plasma membrane. (Hamasaki and Yoshimori 2010). Subsequently, the outer membrane of the autophagosome fuses with a lysosome, also called as an autolysosome, leading to degradation of the enclosed materials along with the inner autophagosomal membrane. Less attention has been paid to microautophagy, which includes the direct engulfment of cytoplasmic cargo at a boundary membrane by autophagic tubes. It is mediated by invagination and vesicle scission into the lumen. The third form of autophagy, CMA, is the direct lysosomal import of unfolded soluble proteins containing a specific pentapeptide sequence. Autophagy and microautophagy degrade proteins and large structures in selective and non-selective manners, whereas CMA selectively degrades a subset of proteins with a KFERQ motif. HSP70 recognizes and binds to this motif (Terlecky and Dice 1993; Agarraberes et al. 1997). In this review, we focused on macroautophagy, the major catabolic mechanism that eukaryotic cells use to degrade long-lived proteins and organelles.
Autophagic complexes
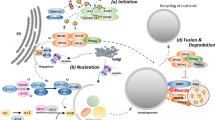
A number of multiprotein complexes mediate the formation and elongation of the double membrane in autophagosomes. This process involves three steps, initiation via the inhibitory mammalian target of rapamycin (mTOR) pathway, nucleation, which involves the beclin1-class III phosphatidylinositol 3-kinase (PI3K) complex, and elongation, which involves the microtubule-associated protein light chain 3 (LC3) (Klionsky et al. 2003; Levine and Kroemer 2008; He and Levine 2010) (Fig. 1). In response to nutrients, particularly amino acids or nutrient-induced insulin, class I PI3K activates Akt and mTOR. This signaling pathway blocks autophagy by engaging mTOR as a protein that inhibits Atg1 from recruiting its partner proteins, such as Atg13 and Atg17 (Neufeld 2010). The Atg1-Atg13-Atg17 complex then recruits other molecules for autophagosome formation (Cheong et al. 2005; Suzuki et al. 2007). Rapamycin, an mTOR inhibitor, is the most commonly used agent to induce autophagy; however, findings from studies using this agent cannot always account for its effect on autophagy because inhibition of mTOR may affect other cellular pathways and components (Sarbassov et al. 2005).
Molecular pathways of autophagy. This process involves three steps including initiation, nucleation, and elongation. In response to various stimuli, class I PI3K inactivates Akt and mTOR upon initiation. This signaling pathway activates autophagy by engaging mTOR as a protein that promotes Atg1 from recruiting its partner proteins such as Atg13 and Atg17. The Atg1–Atg13–Atg17 complex then recruits other molecules for autophagosome formation. It may activate the beclin1/VPS34 complex, which proceeds to make Atg5-Atg12 and LC3/Atg8 complex to the phagophore. Atg5-Atg12 targets the autophagosomes first but then recycles to the cytosol when LC3/Atg8-PE is translocated to the membranes with the isolation membranes closed. Mature autophagosomes are finally fused with the lysosomes called autophagolysosomes
Another pathway that regulates autophagy relies on beclin1, which forms a complex with the class III PI3K Vps34. Activation of the Atg1-Atg13-Atg17 complex induces beclin1-Vps34 complex formation on the lipid membrane (Backer 2008; Liang et al. 1999). This PI3K is distinct from the insulin-activated, class I PI3K that activates mTOR. Vps34 is the target of the autophagy inhibitor 3-methyladenine (Seglen and Gordon 1982). To inhibit autophagy, beclin1 binds to bcl2 and bcl-XL, which is regulated by c-Jun N-terminal kinase 1-mediated phosphorylation of bcl2 (Wei et al. 2008).
The third pathway that mediates autophagosome formation and elongation involves two ubiquitin-like conjugation processes. In the first step, Atg7 and Atg10 mediate the conjugation of Atg12 to Atg5 (Mizushima et al. 1998), which interacts with Atg16 (Ohsumi and Mizushima 2004). The Atg12-Atg5 complex then interacts with the membrane structure, and then dissociates from the autophagosome. The second conjugation reaction involves Atg8 or LC3. Atg4 cleaves the C-terminus of LC3 to generate LC3I, which has a glycine residue at the C-terminus. Then, Atg7 and Atg3 are involved in the conjugation of LC3I to phosphatidylethanolamine in the lipid membrane to generate LC3II (Ichimura et al. 2000). LC3II connects the outer and inner membranes of the autophagosome, where the lipidated protein facilitates membrane elongation and closure. Then, LC3II is shattered after the autophagosome fuses with a lysosome. Finally, the resultant small molecules are transported back to the cytoplasm to maintain homeostasis.
The expression of aberrant microRNAs during the progression of liver disease. Factors such as HBV and HCV infection, metabolic disorder, and alcohol consumption promote the development of chronic liver disease that may often advance to fibrosis/cirrhosis and HCC. The figure shows microRNAs that are dysregulated and involved in different stages of liver diseases. The list includes upregulated microRNAs shown in red, and downregulated microRNAs in blue. Autophagy-related microRNAs were indicated as stars
A link between autophagy and microRNA dysregulation in liver diseases. Interactive relationship between autophagy and microRNAs is involved in liver pathophysiology. However, a clear understanding of the control of autophagy by microRNAs and the biology of microRNAs by autophagy yet remains to be established
Autophagy in liver biology
Autophagy was initially characterized in liver tissue. As the liver is the most sensitive organ to changes in nutrient supply, autophagy is an important step in hepatic adaptation, rapidly responding to cellular stresses. For example, in rodent models, autophagy caused by nutrient starvation elicits degradation of one-third of the total liver proteins within 24 h (Cuervo et al. 1995). When autophagy was inhibited in hepatocytes, liver mass increased due to inhibition of protein degradation (Komatsu et al. 2005). Autophagy occurs at low basal levels in virtually all cells to maintain homeostatic functions. It is swiftly activated when cells require nutrients and energy, such as during starvation, growth factor withdrawal, and under high bioenergetic demands (Mizushima et al. 2008; Mehrpour et al. 2010). The basic metabolites (e.g., amino acids, sugars, and fatty acids) derived from autophagic degradation are transported back to the cytoplasm for use in energy production or to be recycled. When the period of starvation is extended, it attenuates autophagy and generates proto-lysosomal tubules and vesicles, allowing the cell to restore a full complement of lysosomes (Yu et al. 2010).
Autophagy in liver diseases
There is increasing evidence that autophagy also plays a role in the pathogenesis of diverse liver diseases, such as nonalcoholic and alcoholic steatohepatitis, fibrosis, metabolic disorders, and HCC.
Autophagy and nonalcoholic fatty liver disease (NAFLD)
NAFLD refers to the range of liver damage resulting from steatosis to nonalcoholic steatohepatitis (NASH) and cirrhosis, and it is characterized by lipid accumulation within hepatocytes (Marra et al. 2008). Recent studies have suggested a role for autophagy during the development of NAFLD. Autophagy has been shown to regulate intracellular lipid storage through degradation of lipid droplets and the release of fatty acids (FAs) into the cytoplasm in rapid response to starvation (Rautou et al. 2010; Singh et al. 2009; Wang et al. 2010). Thus, disruption of autophagy might be essential to the pathogenesis of NASH (Yang et al. 2010a). Lipids are derived from circulating FAs released by insulin resistance (IR)-induced dysregulation of peripheral lipolysis. FAs are taken up into hepatocytes mainly by membrane-bound transporters (Donnelly et al. 2005). De novo lipogenesis contributes to hepatic steatosis (Postic and Girard 2008). Hepatocellular accumulation of lipotoxic intermediates, such as diacylglycerol and ceramides, causes hepatic IR (Samuel et al. 2010). Autophagy regulates intracellular lipid storage through a process called lipophagy. In response to a moderate increase in lipid availability or during nutrient deprivation, hepatic autophagy degrades lipid droplets. Lipid droplets are translocated by autolipophagosomal vesicles and transported to lysosomes to be degraded into FAs. In contrast, sustained lipid availability induced by a long-lasting high fat diet inhibits hepatic autophagic turnover (Singh et al. 2009). The autophagic marker LC3 reproducibly stains as delineated puncta in normal livers, and LC3 immunostaining decreased with progressing steatosis (Kashima et al. 2013). The expression of cathepsin B, D, and L, which is activated at the low pH in lysosomes, is suppressed in NAFLD patients (Fukuo et al. 2013). The autophagy inhibitors, 3-MA and rapamycin, increase triglyceride (TG) levels in hepatocytes (Singh et al. 2009). Treatment with the lysosomal degradation inhibitor chloroquine (CQ), which increases the lysosomal pH, increases hepatic TG levels upon exposure to free FAs. Inhibition of autophagy by Atg5 knockdown also increases TG content in FA-treated hepatocytes (Mei et al. 2011). In addition, liver-specific Atg7-knockout mice have elevated hepatic TG levels (Singh et al. 2009). Moreover, autophagy is related to the metabolism of lipid droplets in hepatic stellate cells, a cell-type responsible for liver fibrosis (Hernandez-Gea et al. 2012). In addition, other pharmacological compounds, such as carbamazepine and caffeine, reduced steatosis, improving insulin sensitivity in high fat diet-induced non-alcoholic fatty liver conditions via an autophagy-mediated pathway (Lin et al. 2013; Sinha et al. 2014).
During the progression of NAFLD, the “first hit”, excess intracellular lipid levels induced by FA exposure, initiates a “second hit”, induction of ROS and proinflammatory cytokines (Day and James 1998; Day 2006). Pathologic stimuli, such as FAs, ROS, and cytokines, also influence autophagy flux. Unsaturated FAs (e.g., oleic acid; OA) promote the formation of TG-enriched lipid droplets and induce autophagy with minimal effects on apoptosis. In contrast, saturated FAs (e.g., palmitic acid; PA) weakly promote lipid droplet formation, suppress autophagy, and significantly induce apoptosis. OA-induced autophagy is independent of mTOR but dependent on ROS and PI3K. PA-induced apoptosis inhibits autophagy by inducing caspase-dependent beclin1 cleavage, which suggests a cross-talk between apoptosis and autophagy. Induction of autophagy through the formation of triglyceride-enriched lipid droplets may protect cells from FA-induced lipotoxicity. Moreover, oxidative stress potentiates the initiation of autophagy via activation of AMPK/ULK1-related mechanisms, but decreases autophagic flux by inhibiting the fusion of autophagosomes to lysosomes (Mei et al. 2011). Disruption of normal autophagic pathways is also linked to increased production of proinflammatory cytokines (Crisan et al. 2011; Harris et al. 2011; Nakahira et al. 2011; Saitoh et al. 2008; Zhou et al. 2011).
Several reports suggest that mitochondrial function is impaired in NASH, and ultrastructural mitochondrial abnormalities have been found in NASH patients (Caldwell et al. 1999). The mitochondrial quality control process involves an autophagic removal of mitochondria. The removal of aged and damaged mitochondria by autophagy protects cells from dysfunction or injurious processes such as the release of pro-apoptotic proteins from mitochondria, ROS generation, and ATP depletion (Lemasters 2005; Mizushima et al. 2008). The half-life of mitochondria was shown to be 10–25 days in non-proliferating tissues such as the liver (Menzies and Gold 1971). However, in a recent study, the half-life was estimated to be ~2 days (Miwa et al. 2008). In this physiological turnover, mitochondria are removed by mitophagy in balance with new mitochondrial biogenesis. Mitophagy also plays a role in cytoplasmic remodeling. Several signal transduction mechanisms regulate the autophagic processes involved in mitochondrial quality control. Since AMPK activation induces autophagy in response to various stresses, its activation seems to be important for the regulation of autophagy. Moreover, AMPK activation leads to inhibition of the mTOR-dependent pathway, which is a central inhibitory pathway of autophagy (He and Klionsky 2009). AMPK-induced autophagy exerts a cytoprotective effect (Herrero-Martin et al. 2009). Previously, we reported the effects of agents that can activate AMPK, including dithiolethiones and phytochemicals (e.g., resveratrol, isoliquiritigenin, and liquiritigenin). Treatment with these agents showed liver-protective effects against external stimuli as well as therapeutic effects against NAFLD (Yang et al. 2010b). We expect that activation of AMPK by these agents plays a role in autophagy-associated mitochondrial biogenesis.
Autophagy and alcoholic liver disease (ALD)
ALD is a major health problem worldwide. Several studies demonstrate acute ethanol consumption elevates autophagy, perhaps as a protective mechanism against ethanol toxicity. Ni et al. 2013 showed elevation of autophagy-related gene expression and autophagic flux in human hepatocytes by ethanol. Consistently, fluorescent microscopy analyses showed higher green fluorescent autophagosomes in the hepatocytes of ethanol-fed mice than in those of control animals (Ding et al. 2010; Thomes et al. 2012). Inhibition of autophagy augmented ethanol-induced toxicity and steatosis (Ding et al. 2011). In a model using VL-17A, a cell line that metabolizes ethanol via alcohol dehydrogenase (ADH) and CYP2E1, ethanol treatment elevated autophagosome formation as compared to untreated control (Donohue et al. 2006). In the study, inhibition of ethanol metabolism by 4-methyl-pyrazole reduced autophagosome formation, suggesting a link between ethanol metabolism and autophagosomal dysfunction. On the other hand, there are several other proofs that acute ethanol treatment repressed autophagy. Ethanol treatment increased LC3 reduction and beclin1 expression, resulting in autophagy inhibition (Nepal and Park 2013; Noh et al. 2011). In another study using mice, LC3 and Atg7 were both decreased by ethanol, whereas mTOR signaling was activated (Yang et al. 2012a). This discrepancy might be related to the use of different models and varied experimental conditions (e.g., different in vitro models, species, dosage of ethanol, and fasting-time in an in vivo model). Further studies are necessary to clarify the effect of acute ethanol on autophagy.
Autophagy is inhibited by chronic alcohol consumption, which may contribute to the development of alcoholic hepatitis or fibrosis (Rautou et al. 2010). In addition, liver enlargement is frequently observed in alcohol-fed animal models. In a GFP-LC3 transgenic mouse model, chronic ethanol administration resulted in hepatomegaly, steatosis, and elevated liver proteins, suggesting slower catabolism (Lieber and DeCarli 1986). Therefore, chronic ethanol consumption may also inhibit autophagosome degradation (Thomes et al. 2013). It has been suggested that the increased hepatic mass is due to the accumulation of proteins and lipids (Baraona et al. 1975). The increased protein levels may promote pathogenesis because some of the proteins accumulated in the cell are non-functional molecules damaged by reactive free radicals (Bardag-Gorce et al. 2005). In chronic ethanol-fed rats, protein synthetic activity in the liver returns to a normal state (Donohue et al. 1987). However, the degradation rate of hepatic proteins in the ethanol-fed animals was 36–40 % lower than that of the control (Donohue et al. 1989), implying that the autophagic process may also be deregulated by chronic ethanol consumption.
CYP2E1 activity and content were elevated in ethanol-fed mice, which is an indicative of enhanced ethanol metabolism, presumably in association with augmented autophagosomal content. In addition, ethanol feeding suppresses proteasome activity in the liver, which should increase the net liver proteins. In HepG2 cells overexpressing CYP2E1, ethanol treatment increased TG accumulation (Wu et al. 2010), which may have resulted from lipophagy inhibition. In addition, ethanol treatment decreased autophagy in CYP2E1-expressing cells to a greater degree than in control cells, suggesting that the induction of CYP2E1 may impair autophagy, and this process may increase lipid accumulation in response to ethanol (Wu et al. 2012a).
FoxO3a regulates the expression of genes involved in multiple cellular functions, including oxidative stress, apoptosis, DNA repair, and cell-cycle transition (Huang and Tindall 2007; Tzivion et al. 2011). FoxO3a also regulates the expression of Atgs in mouse skeletal muscle (Zhao et al. 2007; Masiero et al. 2009) and cardiomyocytes, promoting cardiomyocyte survival following oxidative stress (Sengupta et al. 2011). Ethanol treatment increased nuclear retention of FoxO3a, a member of the forkhead box O (FoxO) family of transcription factors, which could be mediated by decreased Akt activity. In a genetic animal model, expression of the Atg genes was downregulated, whereas ethanol-induced liver injury was exacerbated (Ni et al. 2013), indicating that FoxO3a may play a role in autophagy in an ethanol-treated model.
Autophagy and viral hepatitis:
Autophagy has an antimicrobial function by which intracellular microbial replication is restricted and/or the viability of infected cells is maintained. As autophagy is often triggered by infection, it is a central component of immune responses. In viral infections, virus survival is linked to its ability to undermine cellular defenses and regulate the processes required for their replication (Ait-Goughoulte et al. 2008).
Hepatitis C viruses subvert the autophagic machinery for use in their own replication (Sir et al. 2008). In contrast, many other viruses have evolved to counteract the antiviral activity of autophagy (Mizushima et al. 2008). HCV infection may advance to liver cirrhosis and in some cases, HCC. The HCV genome encodes polyproteins that are processed into structural and nonstructural (NS) proteins (Reed and Rice 2000). HCV replicates in association with intracellular membrane structures called the membranous web (Egger et al. 2002). Cells infected with HCV accumulate lipid droplets, which play a role in the assembly of HCV particles (Miyanari et al. 2007). HCV core protein recruits NS proteins and replication complexes to the membranes associated with lipid droplets. This step is critical for virus production. In several studies, autophagy was described as a process that is required for productive HCV infection. A recent study showed that HCV RNA replication takes place primarily on autophagosomal membranes in HCV subgenomic RNA replicon cells (Talloczy et al. 2006). HCV-infected cells displayed autophagic vacuole formation (Shepard et al. 2005) and accumulation of autophagic marker proteins, such as LC3B, on vacuole membranes (Shepard et al. 2005; Ogawa et al. 2005; Lee et al. 2008). In addition, both the transcription and protein expression of beclin1, which in complex with Vps34 (the class III PI3K) promotes autophagic vesicle formation, were upregulated. HCV NS5A was sufficient to induce autophagy (Shepard et al. 2005; Moradpour et al. 2007). Moreover, when the expression of autophagy regulatory proteins was blunted, HCV production was reduced. Translation and/or delivery of the incoming viral RNA to the translation apparatus were also blunted in Huh-7 cells transfected with beclin1 and Atg4B shRNAs (Dreux et al. 2009). In another study, human Atg7 and beclin1 RNA inhibition decreased the release of HCV particles into the medium without decreasing the intracellular production of HCV-related proteins and mRNA (Tanida et al. 2009). Guévin et al. showed that Atg5, a protein required for the formation of double membrane vesicles (Mizushima et al. 2001), specifically interacts with HCV NS5B. The interaction between NS5B and Atg5 may be required for the initial onset of HCV replication (Guevin et al. 2010). Moreover, HCV-induced autophagy impairs the innate immune response, whereas HCV infection in autophagy-knockdown cells activates the IFN pathway and apoptosis (Shrivastava et al. 2011). In addition, HCV-induced autophagy affects host lipid metabolism. HCV virion assembly, which necessitates the biogenesis of lipid droplets, could be defective when the autophagy-mediated flux of cholesterol from lysosomes to the endoplasmic reticulum is impaired (Westin et al. 2002). Hence, defective autophagy may contribute to the onset of microvesicular steatosis by accumulating damaged organelles (Vescovo et al. 2012).
HBV is a small DNA virus. After infecting hepatocytes, the ends of the HBV DNA molecule are joined to form a covalently closed circular DNA molecule, which directs the transcription of viral mRNAs. The genome contains S, C, P, and X genes. The S gene encodes the surface antigens, the C gene encodes the core protein and a related protein called the precore protein, the P gene encodes the viral DNA polymerase, and the X gene is a multifunctional regulatory protein. The core protein mRNA (pregenomic RNA; pgRNA) is packaged by the core protein to form a viral particle. It is converted to the partially double-stranded viral genome by viral RNA polymerase, which is also packaged in the core particle. The core particle then interacts with the viral envelope proteins to form a mature virion, which is then released from infected cells (Beck and Nassal 2007). HBV infection enhanced the autophagic process, leading to enhanced virus envelopment (Tian et al. 2011). In addition, HBV small surface proteins were partially colocalized and coimmunoprecipitated with LC3 (Li et al. 2011a). Tian et al. generated HBV transgenic mice with a liver-specific Atg5 knockout. In this model, decreased HBV expression and altered nuclear localization of the core protein were observed. In addition, HBV DNA levels in sera were also reduced by >90 %, and the HBV DNA replicative intermediate was undetectable (Tian et al. 2011). In another study, HBV enhanced the autophagic response without increasing autophagic degradation. This induction of the autophagic response stimulates viral DNA replication, although it has only a slight effect on viral RNA transcription and requires HBx, a protein that binds to PI3KC3 to enhance its activity (Sir et al. 2010). Furthermore, HBX increases autophagy, activating death-associated protein kinase (DAPK), which can induce autophagy by phosphorylating beclin1 (Tang et al. 2009; Zhang et al. 2013a). In contrast, when HBV genomic DNA and HBx were overexpressed in hepatic and hepatoma cells, HBV- or HBx-induced autophagosome formation accompanied by unchanged mTOR activity and decreased degradation of LC3 and SQSTM1/p62, the typical autophagic cargo proteins were observed. In addition, HBx impairs lysosomal acidification leading to a drop in lysosomal degradative capacity and accumulation of immature lysosomes (Tang et al. 2012).
Autophagy and liver fibrosis
Sustained chronic liver injury facilitates fibrosis, cirrhosis, and organ failure. During chronic liver injury, quiescent hepatic stellate cells (HSCs) transdifferentiate into myofibroblast-like cells, which have an increased proliferation rate, decreased number of lipid droplets, and high production of extracellular matrix. Therefore, HSCs have been implicated in intra-hepatic angiogenesis (Friedman 2008). When HSCs were activated, autophagy was increased with a release of lipid droplets, as shown in fibrosis animal models and human liver tissue. Loss of autophagic function in cultured mouse stellate cells, induced by treatment with 3-MA or siAtg7, and in Atg7F/F-GFAP-Cre mice following injury exhibited reduced fibrogenesis and matrix accumulation, indicating that autophagy might provide a critical source of energy substrate in the form of TGs stored within cytoplasmic droplets. OA treatment can partially overcome this effect (Hernandez-Gea et al. 2012). In another study, autophagic flux was significantly increased during HSC activation. When a lysosomal inhibitor bafilomycin A1 was introduced into HSCs, significantly decreased cell proliferation and activation marker expression were observed. In addition, PDGF-BB treatment of quiescent HSCs induced an increase in lipid droplets and LC3B colocalization. Bafilomycin A1 inhibited this phenomenon (Thoen et al. 2011). In addition, various fibrosis mediators are autophagy activators. Transforming growth factor-β1 (TGFβ1), a key regulator of the epithelial-mesenchymal transition, cell proliferation, and extracellular matrix formation (Friedman 1993; Qi et al. 1999; Bataller and Brenner 2005; Zhao et al. 2008), promotes autophagy (Suzuki et al. 2010; Kiyono et al. 2009). Gα12 and Gα13 control cell transformation, cell growth, migration, metastasis, the formation of actin-stress fibers, gene induction, and cell death (Kawanabe et al. 2002; Kang et al. 2003; Fujii et al. 2005; Kelly et al. 2007). Previously, we showed that Gα12 and Gα13 play a role in the induction of TGFβ1 expression through Rho/Rac-dependent AP-1 activation (Lee et al. 2009). Moreover, ligands of the G-protein-coupled receptors that interact with Gα12 members, such as angiotensin-II, thrombin, and sphingosine-1-phosphate, contribute to autophagy activation (Wang et al. 2013; Yadav et al. 2010; Hu et al. 2011; Chang et al. 2009).
Autophagy and HCC
HCC is the third leading cause of cancer death worldwide (Ahn and Flamm 2004). However, the overall survival of patients with HCC has not improved significantly over the past decades, and the mechanisms underlying HCC development and progression should be clarified further (Bruix et al. 2004). Interestingly, the autophagic pathway has been implicated in different, and sometimes contradictory, processes capable of inducing both cancer cell survival and death. As mentioned above, autophagy can be divided into two categories. Constitutive autophagy, which is necessary for intracellular recycling and metabolic regulation, and stress-related autophagy, which is a requisite for the elimination of damaged components. Constitutive autophagy is suppressed in HCC, and in prostate, breast, and ovarian cancers (Bao et al. 2013). In several other studies, autophagy suppression aggravated HCC progression. Liver-specific Atg7-deficient mice and beclin1 knockout mice (beclin1 ±) develop HCC (Qu et al. 2003; Takamura et al. 2011). Based on these observations, autophagy activation has been suggested as an anti-cancer therapy target. Apogossypolone induces both beclin1- and ROS-mediated autophagy and results in HCC cell death (Cheng et al. 2013). Tetrandrine induces autophagy, but not apoptosis, in HCC cells in vitro and in vivo. This autophagy-inducing activity is partially dependent on ROS accumulation and the repression of Atg7 (Gong et al. 2012). 5-FU induced the conversion/turnover of LC3 in SK-HEP-1 cells, whereas depletion of Bmi-1 enhanced 5-FU-induced autophagy. Bmi-1 depletion enhanced the chemosensitivity of HCC cells by inducing apoptosis and autophagy, which may be associated with the PI3K/AKT and bcl-2/beclin1 pathways (Wu et al. 2012b). D9-tetrahydrocannabinol and JWH-015 (a CB2-selective agonist) reduced the viability of HepG2 and Huh-7 cells, an effect that was dependent on the stimulation of autophagy through tribbles homolog 3 (TRB3) upregulation and subsequent inhibition of the Akt/mTOR axis and AMPK stimulation. In vivo studies showed that D9-THC and JWH-015 had an inhibitory effect on HCC xenograft growth (Vara et al. 2011). Interferon-gamma (IFN-γ) is an antiviral and antiproliferative cytokine that modulates immune responses. IFN-γ induced autophagy through autophagosome formation and conversion/turnover of LC3 protein, mediated by beclin1 or Atg5 (Li et al. 2012). Sorafenib (Nexavar), a multiple kinase inhibitor that is approved for the treatment of advanced HCC, activated autophagy in HCC cell lines in a dose- and time-dependent manner. Inhibition of Mcl-1 by sorafenib disrupted the beclin1-Mcl-1 complex and resulted in beclin1 activation (Tai et al. 2013). In contrast, when autophagy is upregulated, it contributes to HCC aggressiveness. Autophagy was shown to be an important mediator of cancer cell invasion through EMT induction regulated by the activation of TGF-β/Smad3-dependent signaling (Li et al. 2013). In humans, LC3 was sharply increased in HCC tissues compared with its levels in noncancerous tissues. Moreover, autophagy contributes to cancer cell survival. Cancer cell lines, which are resistant to TRAIL-mediated apoptosis, display enhanced autophagy upon stimulation through either TRAIL or Fas (Han et al. 2008). Consistently, beclin1 overexpression reduces TRAIL-mediated cell death (Cho et al. 2009). Autophagy inhibited TRAIL-mediated tumor cell death due to proteolytic degradation of caspase 8 (Hou et al. 2010).
Autophagy appears to be activated during metastasis and exploited by metastatic cancer cells for adaptation and survival under unfavorable stresses conditions. For example, autophagy is an adaptive metabolic response to hypoxic conditions (Toshima et al. 2013). When Huh7 cells were treated with an autophagy inhibitor under hypoxia, lower cell viability was observed along with low levels of intracellular ATP due to impaired mitochondrial β-oxidation. Moreover, autophagy may be activated as an alternative energy source to overcome metabolic stress, which is often encountered by metastatic tumor cells, especially those metastasized to organs that provide tumor cells with a poor supply of nutrients (Mathew et al. 2007). LC3 expression was correlated with tumor size, and may serve as a strong prognostic factor of HCC (Lee et al. 2013). LC3 staining of metastasized and primary tumors from HCC patients showed that LC3 expression was significantly higher in metastasized HCC than in primary HCC. Further experiments using immunohistochemical and TEM methods also support that autophagy is enhanced in metastatic colonies compared to that in primary tumors. However, autophagy did not significantly alter the invasion, migration, and detachment of HCC cells in this model (Peng et al. 2013).
The biology of microRNAs
Post-transcriptional and translational regulation play roles in diverse biological functions, including organogenesis and development. MicroRNAs target and regulate essentially all biological processes in all cell types, and influence the complex programs of gene expression for virtually all cellular processes (Kiss 2002). Small RNA molecules can regulate the expression of genes through RNA interference. There are three classes of small noncoding RNA in mammalian cells, namely microRNAs, siRNAs, and PIWI-interacting RNAs (piRNAs) (Grosshans and Filipowicz 2008; Brennecke et al. 2007; Aravin et al. 2007; Chapman and Carrington 2007; Bartel 2004). MicroRNAs, which are 18–25 nucleotides in length, are the most abundant class of small non-coding RNA molecules. They regulate the expression of target genes by inducing translational arrest or mRNA cleavage via interaction with the 3′-UTRs of target mRNAs (Lee et al. 1993; Wightman et al. 1993; He and Hannon 2004). It is well known that mature microRNAs are excised from pri-miRNA transcripts consisting of hairpin-like microRNA precursors. These primary forms of microRNAs are processed by the Drosha/DGCR8 complex, and the resultant 60–70-nucleotide precursor microRNA hairpins (pre-miRNAs) are then exported into the cytoplasm (Kim et al. 2009; Jones-Rhoades et al. 2006).
MicroRNAs in liver biology
Many different microRNAs are expressed in a cell-type specific manner (e.g., hepatocytes, sinusoidal endothelial cells, kupffer cells, and HSCs), and several are involved in different aspects of liver physiology (Wang et al. 2012). Each microRNA targets and affects complex gene expression patterns for virtually all cellular processes. Moreover, dysregulation of microRNAs is closely associated with a variety of diseases. The contribution of microRNAs to the pathogenesis of liver diseases has been studied extensively. One such study showed the effect of Dicer knockdown on the neonatal functions of the liver, and dysregulation of cellular differentiation was observed in this model. The proliferation of liver stem cells is also affected by microRNAs. There was a significant increase in the proliferation of CK19-positive bile ductular cells adjacent to α-fetoprotein and portal tracts (Hand et al. 2009). Insulin-like growth factor 2 was dramatically elevated, promoting hepatocarcinogenesis in livers with oval cell proliferation (Fu et al. 1988). In addition, notable pathological changes were also detected in the liver of Dicer-knockdown mice as the mice grew. Inflammation also occurred in the livers. Therefore, the liver/body weight ratio was higher due to increased hepatocyte proliferation, which exceeded hepatocyte apoptosis. Despite the knockdown of microRNAs, most metabolic liver functions were unchanged, and serum albumin, bilirubin, and cholesterol levels were normal in the animals. Therefore, the remaining microRNAs appear to be sufficient for the basic liver functions to continue. Overall, microRNAs play a key role in maintaining normal liver function, serving as a fine tuner of gene expression.
Hepatic microRNA expression profiling
Many microRNAs are members of families with closely related sequences that may have some common targets. The binding target of a microRNA is determined by its complementarity to the seed (nucleotides 2–8), and an additional 13–16 nucleotides (Pasquinelli et al. 2000; English et al. 1996). Some microRNAs are only detectable in certain tissues, including miR-122 in the liver, miR-1 in muscle, and miR-21 in the heart (Sun et al. 2010). The ten most frequently detected microRNAs in the human liver are listed in Table 2 (Landgraf et al. 2007).
miR-122 is highly enriched in the liver, and is regulated during embryonic development (Reinhart et al. 2000; English et al. 1996). MicroRNAs that are found in a major portion of the liver, such as miR-122, miR-126, miR-16, miR-22, or the let-7 family, do not target human albumin or the α-fetoprotein genes. miR-122 expression seems to occur at an early stage of liver development (Chang et al. 2004), implying that it may control the differentiation of hepatoblasts into hepatocytes. An earlier study showed that miR-122 is conserved in 12 different species (English et al. 1996). Treatment of a mouse model with antagomir-122 upregulated 363 genes and downregulated 305 genes. Cholesterol biogenesis was the most significantly repressed gene functional category. These changes resulted in reduced plasma cholesterol levels in mice and chimpanzees (Esau et al. 2006; Lanford et al. 2010; Krutzfeldt et al. 2005). miR-122 also affects various genes involved in lipid metabolism. miR-122 expression was decreased in the livers of human and mouse NASH models (Cheung et al. 2008; Wang et al. 2009) and in mice with ALD (Bala et al. 2012). Intriguingly, lower levels of serum TGs, serum cholesterol, LDL and HDL were seen in miR-122-knockout mice than in wild-type mice. miR-122-knockout mice showed steatohepatitis and HCC (Tsai et al. 2012; Hsu et al. 2012), supporting the notion that miR-122 plays a central role in maintaining lipid homeostasis in the liver.
miR-126 is the second most abundant microRNA in the liver. miR-126 plays a role in cancer biology and is involved in angiogenenic signaling, cell proliferation, and invasiveness. VCAM-1, p85β, CRKL, IRS-1, VEGF, and TOM-1 are its validated targets (Harris et al. 2008; Guo et al. 2008; Crawford et al. 2008; Zhang et al. 2008; Liu et al. 2009; Oglesby et al. 2010).
The Let-7 family (Let-7a, b, c, d, f, and g) are the third highestly expressed microRNAs in the liver, and have been linked to development and cell differentiation. The Let-7 family also functions as suppressors of tumor and collagen accumulation in HSCs. Its known targets include Ras, HMGA2, Dicer, caspase 3, integrin β3, and TRIM71(Johnson et al. 2005; Meng et al. 2007b; Lin et al. 2007; Shell et al. 2007; Tsang and Kwok 2008; Tokumaru et al. 2008; Nie et al. 2008; Muller and Bosserhoff 2008; Oh et al. 2010; Noh et al. 2009; Takane et al. 2010).
miR-16 is also abundant in the liver and other organs. It has pro-apoptotic activity, inducing hepatocyte and HSC apoptosis and TNF production. Other functions include cell invasion and proliferation. In addition, miR-16 may act as a tumor suppressor, targeting TPPP3, Bcl-2, VEGF, Cyclin D1, D3 and E1, PDCD4, VEGFR2, FGFR1, Cdk6, WT1, Actr1a, Mcl1, UDP-glucose pyrophosphorylase2, Card8, and c-jun (Kiriakidou et al. 2004; Cimmino et al. 2005; Ye et al. 2008; Chen et al. 2008; Calin et al. 2008; Liu et al. 2008; Chamorro-Jorganes et al. 2011).
When miR-22 is highly expressed in the liver, it regulates inflammation and hypoxic signaling in cancer cells, and its validated targets are BMP-7, PPARα, ESR1, PTEN, and MAX (Iliopoulos et al. 2008; Bar and Dikstein 2010; Ting et al. 2010; Xiong et al. 2010a).
Dysregulation of microRNAs in liver diseases
In a number of studies, alterations in microRNAs have been shown to correlate with various liver diseases, and the expression profiles and signatures of these microRNAs are distinct among liver diseases with different etiologies (Fig. 2).
MicroRNAs and viral hepatitis
HCV requires miR-122 for its replication. The HCV genomic RNA consists of a 5′-UTR, a single long polyprotein open reading frame (ORF), and a 3′-UTR. The sequence of the 5′-UTR is highly conserved and contains an internal ribosome entry site (IRES) that allows cap-independent translation of the viral ORF (Tsukiyama-Kohara et al. 1992; Reigadas et al. 2001) and regulatory elements that control positive-strand replication (Reigadas et al. 2001). A perfect match to the seed sequence of miR-122 was predicted in close proximity to the 5′ end of the viral genome (Jopling et al. 2005). Direct binding of miR-122 to the HCV RNA induced RNA stabilization and translational enhancement (Conrad et al. 2013). Hence, miR-122 increases HCV replication (Roberts et al. 2011), as supported by the results that treatment of animals with antagomirs and locked nucleic acids (LNAs) reduced HCV replication (Krutzfeldt et al. 2005; Lanford et al. 2010). Therefore, miR-122 may be an antiviral therapy target for HCV patients.
microRNAs and ALD/NAFLD
Alcohol intake stimulates ROS generation in hepatocytes, causing dysregulation of miR-34a, miR-217, and miR-122. Treatment of kupffer cells with lipopolysaccharide influenced miR-125b, miR-146a, and miR-155 through TNF-α production (Baltimore et al. 2008). In addition, dysregulation of miR-1224, miR-182, miR-183, miR-199a, miR-214, miR-320, miR-486, and miR-705 was observed in the livers of chronic alcohol-fed mice (Dolganiuc et al. 2009; McDaniel et al. 2014). Elevated miR-34a levels and decreased miR-122 levels were detected in patients and diet-induced obese mice with NAFLD (Cermelli et al. 2011). These microRNAs could be used as markers for the diagnosis of or as therapeutic targets for ALD and NAFLD.
microRNAs and fibrosis
Dysregulation of a broad variety of microRNAs has been shown in fibrosis models. miR-199a, miR-200, and miR-34a were shown to be profibrotic microRNAs (Murakami et al. 2011; Li et al. 2011b), whereas miR-15b/16, miR-150, miR-19b, miR-194, and miR-29 were the antifibrotic ones (Guo et al. 2009a, b; Venugopal et al. 2010; Roderburg et al. 2011; Kwiecinski et al. 2012; Mannaerts et al. 2013). Dysregulation of miR-15b/16, miR-146a, miR-221, and miR-27 promotes the proliferation of HSCs (Guo et al. 2009a, b; Mannaerts et al. 2013; He et al. 2012; Ogawa et al. 2012). miR-122 dysregulation also activates HSCs (Tsai et al. 2012).
microRNAs and HCC
MicroRNAs can function as either oncogenes or tumor suppressors. MicroRNA dysregulation has been observed in association with sustained proliferation, resistance to cell death, immortality, angiogenesis, invasion, and metastasis in cancer (Hanahan and Weinberg 2011). miR-26a, miR-195, miR-34a, and miR-221 regulate the cell cycle (Xu et al. 2009; Kota et al. 2009; Cheng et al. 2010; Gramantieri et al. 2009; Sharma et al. 2011). miR-125b, miR-224, miR-29, miR-101, and miR-122 were reported as apoptosis regulators in HCC (Zhao et al. 2012; Gong et al. 2013; Zhang et al. 2013b; Xiong et al. 2010b; Su et al. 2009; Lin et al. 2008). Moreover, miR-122 can inhibit angiogenesis and intrahepatic metastasis (Tsai et al. 2009). The PI3K/AKT/mTOR pathway, a major survival pathway, is also controlled by miR-216a, miR-21, miR-148a, miR-221/222, miR-519d, and miR-29a (Wu et al. 2012c; Meng et al. 2007a; Yuan et al. 2012; Fornari et al. 2012; Kong et al. 2011). In addition, dysregulation of miR-29b and miR-125b increased angiogenesis and metastasis (Fang et al. 2011; Alpini et al. 2011).
MicroRNAs and autophagy
The contributions of microRNA-mediated post-transcriptional and translational control of autophagy that have recently emerged suggest the need for further studies about the link between microRNAs and autophagy. Many of the previous studies have been done in the field of cancer. Here, we summarized the current knowledge on the regulation of autophagy by noncoding RNAs, focusing on the pathophysiological roles of microRNAs, particularly, how microRNAs modify key autophagic effector proteins in liver diseases (Table 3).
In tumor cells, several microRNAs have been shown to control autophagy by targeting Atgs (Fig. 3). miR-375 plays an inhibitory role in autophagy and is down-regulated in HCC under hypoxic stress. Therefore, transfection with miR-375 mimic impairs HCC viability by attenuating the protective role of autophagy, which depends on the inhibition of Atg7, an essential autophagy-related gene (Chang et al. 2012). In HCC patients treated with cisplatin, miR-199a-5p was found to be significantly decreased, which may explain the drug resistance as mediated with Atg7-dependent autophagy (Xu et al. 2012). In a study done by Xu et al., miR-101 expression was downregulated in most HCC tissues and cell lines, confirming EZH2 as its direct target gene. In the study, the authors showed that miR-101 inhibited autophagy and with either doxorubicin or fluorouracil, it induced tumor cell death, although they did not suggest autophagy-related genes as targets of miR-101 (Xu et al. 2014). In another study using breast cancer cells, miR-101 potently inhibited autophagy by regulating STMN1, RAB5A, and Atg4D (Frankel et al. 2011). STMN1 encodes Stathmin/Oncoprotein18, a cytosolic phosphoprotein that regulates microtubule dynamics (Marklund et al. 1996; Rana et al. 2008), and is identified as an autophagy regulator. RAB5A, a small GTPase that regulates early endocytosis, was previously suggested to act at an early stage of autophagosome formation (Ravikumar et al. 2008). Finally, Atg4D is a member of the Atg4 family of cysteine proteases that regulate autophagosome biogenesis through LC3 processing, allowing its conjugation to phosphatidylethanolamine on autophagosomal membranes (Marino et al. 2003; Scherz-Shouval et al. 2007).
In turn, autophagy may regulate microRNA biology (Fig. 3). miR-224 was preferentially recruited and degraded during autophagic progression. Autophagy was inversely correlated with miR-224 expression in specimens from patients with HBV-associated HCC. In the study, autophagy was shown to mediate miR-224 degradation and liver tumor suppression (Lan et al. 2014). Nevertheless, the roles of autophagy in the regulation of microRNAs, particularly cell-type specific regulation, in the liver remain largely unknown.
Conclusion and implications
Based on our current understanding of autophagy, we know that autophagy plays a role in liver pathophysiology, and that both basal and stress-induced autophagy are important. Autophagy is involved in the removal of misfolded proteins, organelle turnover, energy homeostasis, lipid homeostasis, and immunity against intracellular and extracellular aggressors. Autophagy might be increased in liver diseases such as liver fibrosis and viral hepatitis. Increased levels of intracellular proteins and damaged organelles due to decreased autophagy may lead to alcoholic/nonalcoholic steatosis and cancer. However, we still do not completely understand how autophagy acts against numerous adaptive physiological conditions in most diseases. Therefore, further research is needed to determine the effects of autophagy on liver pathobiology and the underlying regulatory mechanisms in more disease-specific contexts.
Post-transcriptional regulation plays roles during stress. Numerous studies have provided important information on the roles of microRNAs both under normal cellular functions and in diseases. The study of microRNAs as post-transcriptional regulators in liver disease is a rapidly growing field. The major microRNA expressed in liver, miR-122, controls lipid and glucose metabolism, and its dysregulation is involved in most liver diseases, including viral hepatitis, steatosis, fibrosis, and HCC. Some microRNAs are globally expressed, whereas others are expressed in an organ- or cell-specific manner. It is expected that globally expressed microRNAs may have a variety of functions that act to modulate many target genes.
Certain microRNAs such as miR-375, miR-199-5p, and miR-101 regulate autophagy-related genes and decrease autophagy levels in HCC models, which demonstrates the effects of microRNAs on autophagy. In addition, autophagy affects microRNAs, as shown by the example of miR-224, a microRNA that is selectively recruited and degraded through the autophagosome-lysosome pathway. We have clearly learned that autophagy and microRNAs have reciprocal interactions in the regulation of liver pathophysiology. Nonetheless, we need to understand the control of autophagy by microRNAs and the biology of microRNA regulation by autophagy in greater depth. Identification of autophagy and microRNA dysregulation profiles that are specific for a particular cell type during liver disease, in conjunction with the identification of the associated cellular functions, will benefit our understanding of the pathologic processes of liver diseases. It is also be realistic to expect that autophagy- and microRNA-based therapies will become useful tools for liver diseases.
References
Agarraberes, F.A., S.R. Terlecky, and J.F. Dice. 1997. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. Journal of Cell Biology 137: 825–834.
Ahn, J., and S.L. Flamm. 2004. Hepatocellular carcinoma. Disease-A-Month 50: 556–573.
Ait-Goughoulte, M., T. Kanda, K. Meyer, J.S. Ryerse, R.B. Ray, and R. Ray. 2008. Hepatitis C virus genotype 1a growth and induction of autophagy. Journal of Virology 82: 2241–2249.
Akao, Y., Y. Nakagawa, and T. Naoe. 2007. MicroRNA-143 and -145 in colon cancer. DNA and Cell Biology 26: 311–320.
Alpini, G., S.S. Glaser, J.P. Zhang, H. Francis, Y. Han, J. Gong, A. Stokes, T. Francis, N. Hughart, L. Hubble, S.M. Zhuang, and F. Meng. 2011. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. Journal of Hepatology 55: 1339–1345.
Aravin, A.A., G.J. Hannon, and J. Brennecke. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764.
Backer, J.M. 2008. The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochemical Journal 410: 1–17.
Bala, S., J. Petrasek, S. Mundkur, D. Catalano, I. Levin, J. Ward, H. Alao, K. Kodys, and G. Szabo. 2012. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 56: 1946–1957.
Baltimore, D., M.P. Boldin, R.M. O’connell, D.S. Rao, and K.D. Taganov. 2008. MicroRNAs: New regulators of immune cell development and function. Nature Immunology 9: 839–845.
Bao, L., P.K. Chandra, K. Moroz, X. Zhang, S.N. Thung, T. Wu, and S. Dash. 2013. Impaired autophagy response in human hepatocellular carcinoma. Experimental and Molecular Pathology 96: 149–154.
Bar, N., and R. Dikstein. 2010. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS ONE 5: e10859.
Baraona, E., M.A. Leo, S.A. Borowsky, and C.S. Lieber. 1975. Alcoholic hepatomegaly: Accumulation of protein in the liver. Science 190: 794–795.
Bardag-Gorce, F., J. Li, B.A. French, and S.W. French. 2005. The effect of ethanol-induced CYP2E1 on proteasome activity: The role of 4-hydroxynonenal. Experimental and Molecular Pathology 78: 109–115.
Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297.
Bataller, R., and D.A. Brenner. 2005. Liver fibrosis. The Journal of Clinical Investigation 115: 209–218.
Beck, J., and M. Nassal. 2007. Hepatitis B virus replication. World Journal of Gastroenterology 13: 48–64.
Brennecke, J., A.A. Aravin, A. Stark, M. Dus, M. Kellis, R. Sachidanandam, and G.J. Hannon. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103.
Bruix, J., L. Boix, M. Sala, and J.M. Llovet. 2004. Focus on hepatocellular carcinoma. Cancer Cell 5: 215–219.
Caldwell, S.H., R.H. Swerdlow, E.M. Khan, J.C. Iezzoni, E.E. Hespenheide, J.K. Parks, and W.D. Parker Jr. 1999. Mitochondrial abnormalities in non-alcoholic steatohepatitis. Journal of Hepatology 31: 430–434.
Calin, G.A., A. Cimmino, M. Fabbri, M. Ferracin, S.E. Wojcik, M. Shimizu, C. Taccioli, N. Zanesi, R. Garzon, R.I. Aqeilan, H. Alder, S. Volinia, L. Rassenti, X. Liu, C.G. Liu, T.J. Kipps, M. Negrini, and C.M. Croce. 2008. MiR-15a and miR-16-1 cluster functions in human leukemia. Proceedings of the National Academy of Sciences of the United States of America 105: 5166–5171.
Cermelli, S., A. Ruggieri, J.A. Marrero, G.N. Ioannou, and L. Beretta. 2011. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE 6: e23937.
Chamorro-Jorganes, A., E. Araldi, L.O. Penalva, D. Sandhu, C. Fernandez-Hernando, and Y. Suarez. 2011. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arteriosclerosis, Thrombosis, and Vascular Biology 31: 2595–2606.
Chang, C.L., M.C. Ho, P.H. Lee, C.Y. Hsu, W.P. Huang, and H. Lee. 2009. S1P(5) is required for sphingosine 1-phosphate-induced autophagy in human prostate cancer PC-3 cells. Cell Physiology: American Journal of Physiology 297: C451–C458.
Chang, J., E. Nicolas, D. Marks, C. Sander, A. Lerro, M.A. Buendia, C. Xu, W.S. Mason, T. Moloshok, R. Bort, K.S. Zaret, and J.M. Taylor. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biology 1: 106–113.
Chang, Y., W. Yan, X. He, L. Zhang, C. Li, H. Huang, G. Nace, D.A. Geller, J. Lin, and A. Tsung. 2012. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 143(e8): 177–187.
Chapman, E.J., and J.C. Carrington. 2007. Specialization and evolution of endogenous small RNA pathways. Nature Reviews Genetics 8: 884–896.
Chen, R.W., L.T. Bemis, C.M. Amato, H. Myint, H. Tran, D.K. Birks, S.G. Eckhardt, and W.A. Robinson. 2008. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood 112: 822–829.
Chen, X., X. Guo, H. Zhang, Y. Xiang, J. Chen, Y. Yin, X. Cai, K. Wang, G. Wang, Y. Ba, L. Zhu, J. Wang, R. Yang, Y. Zhang, Z. Ren, K. Zen, J. Zhang, and C.Y. Zhang. 2009. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28: 1385–1392.
Cheng, J., L. Zhou, Q.F. Xie, H.Y. Xie, X.Y. Wei, F. Gao, C.Y. Xing, X. Xu, L.J. Li, and S.S. Zheng. 2010. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics 10: 1557–1572.
Cheng, P., Z. Ni, X. Dai, B. Wang, W. Ding, A. Rae Smith, L. Xu, D. Wu, F. He, and J. Lian. 2013. The novel BH-3 mimetic apogossypolone induces Beclin-1- and ROS-mediated autophagy in human hepatocellular carcinoma [corrected] cells. Cell Death and Disease 4(2): e489.
Cheong, H., T. Yorimitsu, F. Reggiori, J.E. Legakis, C.W. Wang, and D.J. Klionsky. 2005. Atg17 regulates the magnitude of the autophagic response. Molecular Biology of the Cell 16: 3438–3453.
Cheung, O., P. Puri, C. Eicken, M.J. Contos, F. Mirshahi, J.W. Maher, J.M. Kellum, H. Min, V.A. Luketic, and A.J. Sanyal. 2008. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 48: 1810–1820.
Cho, D.H., Y.K. Jo, J.J. Hwang, Y.M. Lee, S.A. Roh, and J.C. Kim. 2009. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Letters 274: 95–100.
Cimmino, A., G.A. Calin, M. Fabbri, M.V. Iorio, M. Ferracin, M. Shimizu, S.E. Wojcik, R.I. Aqeilan, S. Zupo, M. Dono, L. Rassenti, H. Alder, S. Volinia, C.G. Liu, T.J. Kipps, M. Negrini, and C.M. Croce. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America 102: 13944–13949.
Clape, C., V. Fritz, C. Henriquet, F. Apparailly, P.L. Fernandez, F. Iborra, C. Avances, M. Villalba, S. Culine, and L. Fajas. 2009. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS ONE 4: e7542.
Codogno, P., and A.J. Meijer. 2013. Autophagy in the liver. Journal of Hepatology 59: 389–391.
Conrad, K.D., F. Giering, C. Erfurth, A. Neumann, C. Fehr, G. Meister, and M. Niepmann. 2013. MicroRNA-122 dependent binding of Ago2 protein to hepatitis C virus RNA is associated with enhanced RNA stability and translation stimulation. PLoS ONE 8: e56272.
Crawford, M., E. Brawner, K. Batte, L. Yu, M.G. Hunter, G.A. Otterson, G. Nuovo, C.B. Marsh, and S.P. Nana-Sinkam. 2008. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochemical and Biophysical Research Communications 373: 607–612.
Crisan, T.O., T.S. Plantinga, F.L. Van De Veerdonk, M.F. Farcas, M. Stoffels, B.J. Kullberg, J.W. Van Der Meer, L.A. Joosten, and M.G. Netea. 2011. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PLoS ONE 6: e18666.
Cuervo, A.M. 2004. Autophagy: In sickness and in health. Trends in Cell Biology 14: 70–77.
Cuervo, A.M., E. Knecht, S.R. Terlecky, and J.F. Dice. 1995. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. American Journal of Physiology 269: C1200–C1208.
Day, C.P. 2006. From fat to inflammation. Gastroenterology 130: 207–210.
Day, C.P., and O.F. James. 1998. Steatohepatitis: A tale of two “hits”? Gastroenterology 114: 842–845.
Ding, W.X. 2010. Role of autophagy in liver physiology and pathophysiology. World Journal of Biological Chemistry 1: 3–12.
Ding, W.X., M. Li, X. Chen, H.M. Ni, C.W. Lin, W. Gao, B. Lu, D.B. Stolz, D.L. Clemens, and X.M. Yin. 2010. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 139: 1740–1752.
Ding, W.X., S. Manley, and H.M. Ni. 2011. The emerging role of autophagy in alcoholic liver disease. Experimental Biology and Medicine 236: 546–556.
Dolganiuc, A., J. Petrasek, K. Kodys, D. Catalano, P. Mandrekar, A. Velayudham, and G. Szabo. 2009. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcoholism, Clinical and Experimental Research 33: 1704–1710.
Donnelly, K.L., C.I. Smith, S.J. Schwarzenberg, J. Jessurun, M.D. Boldt, and E.J. Parks. 2005. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. Journal of Clinical Investigation 115: 1343–1351.
Donohue Jr, T.M., M.F. Sorrell, and D.J. Tuma. 1987. Hepatic protein synthetic activity in vivo after ethanol administration. Alcoholism, Clinical and Experimental Research 11: 80–86.
Donohue Jr, T.M., R.K. Zetterman, and D.J. Tuma. 1989. Effect of chronic ethanol administration on protein catabolism in rat liver. Alcoholism, Clinical and Experimental Research 13: 49–57.
Donohue, T.M., N.A. Osna, and D.L. Clemens. 2006. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. International Journal of Biochemistry & Cell Biology 38: 92–101.
Dreux, M., P. Gastaminza, S.F. Wieland, and F.V. Chisari. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America 106: 14046–14051.
Egger, D., B. Wolk, R. Gosert, L. Bianchi, H.E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. Journal of Virology 76: 5974–5984.
English, J.J., E. Mueller, and D.C. Baulcombe. 1996. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8: 179–188.
Esau, C., S. Davis, S.F. Murray, X.X. Yu, S.K. Pandey, M. Pear, L. Watts, S.L. Booten, M. Graham, R. Mckay, A. Subramaniam, S. Propp, B.A. Lollo, S. Freier, C.F. Bennett, S. Bhanot, and B.P. Monia. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabolism 3: 87–98.
Esau, C., X. Kang, E. Peralta, E. Hanson, E.G. Marcusson, L.V. Ravichandran, Y. Sun, S. Koo, R.J. Perera, R. Jain, N.M. Dean, S.M. Freier, C.F. Bennett, B. Lollo, and R. Griffey. 2004. MicroRNA-143 regulates adipocyte differentiation. Journal of Biological Chemistry 279: 52361–52365.
Fabani, M.M., and M.J. Gait. 2008. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 14: 336–346.
Fang, J.H., H.C. Zhou, C. Zeng, J. Yang, Y. Liu, X. Huang, J.P. Zhang, X.Y. Guan, and S.M. Zhuang. 2011. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology 54: 1729–1740.
Ferretti, E., E. De Smaele, E. Miele, P. Laneve, A. Po, M. Pelloni, A. Paganelli, L. Di Marcotullio, E. Caffarelli, I. Screpanti, I. Bozzoni, and A. Gulino. 2008. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO Journal 27: 2616–2627.
Fornari, F., M. Milazzo, P. Chieco, M. Negrini, E. Marasco, G. Capranico, V. Mantovani, J. Marinello, S. Sabbioni, E. Callegari, M. Cescon, M. Ravaioli, C.M. Croce, L. Bolondi, and L. Gramantieri. 2012. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. The Journal of Pathology 227: 275–285.
Frankel, L.B., J. Wen, M. Lees, M. Hoyer-Hansen, T. Farkas, A. Krogh, M. Jaattela, and A.H. Lund. 2011. microRNA-101 is a potent inhibitor of autophagy. EMBO Journal 30: 4628–4641.
Friedman, S.L. 1993. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. New England Journal of Medicine 328: 1828–1835.
Friedman, S.L. 2008. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiological Reviews 88: 125–172.
Fu, X.X., C.Y. Su, Y. Lee, R. Hintz, L. Biempica, R. Snyder, and C.E. Rogler. 1988. Insulinlike growth factor II expression and oval cell proliferation associated with hepatocarcinogenesis in woodchuck hepatitis virus carriers. Journal of Virology 62: 3422–3430.
Fujii, T., N. Onohara, Y. Maruyama, S. Tanabe, H. Kobayashi, M. Fukutomi, Y. Nagamatsu, N. Nishihara, R. Inoue, H. Sumimoto, F. Shibasaki, T. Nagao, M. Nishida, and H. Kurose. 2005. Galpha12/13-mediated production of reactive oxygen species is critical for angiotensin receptor-induced NFAT activation in cardiac fibroblasts. Journal of Biological Chemistry 280: 23041–23047.
Fukuo, Y., Yamashina, S., Sonoue, H., Arakawa, A., Nakadera, E., Aoyama, T., Uchiyama, A., Kon, K., Ikejima, K., and Watanabe, S. (2013) Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatology Research.. doi:10.1111/hepr.12282.
Gong, J., J.P. Zhang, B. Li, C. Zeng, K. You, M.X. Chen, Y. Yuan, and S.M. Zhuang. 2013. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 32: 3071–3079.
Gong, K., C. Chen, Y. Zhan, Y. Chen, Z. Huang, and W. Li. 2012. Autophagy-related gene 7 (ATG7) and reactive oxygen species/extracellular signal-regulated kinase regulate tetrandrine-induced autophagy in human hepatocellular carcinoma. Journal of Biological Chemistry 287: 35576–35588.
Gramantieri, L., M. Ferracin, F. Fornari, A. Veronese, S. Sabbioni, C.G. Liu, G.A. Calin, C. Giovannini, E. Ferrazzi, G.L. Grazi, C.M. Croce, L. Bolondi, and M. Negrini. 2007. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Research 67: 6092–6099.
Gramantieri, L., F. Fornari, M. Ferracin, A. Veronese, S. Sabbioni, G.A. Calin, G.L. Grazi, C.M. Croce, L. Bolondi, and M. Negrini. 2009. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clinical Cancer Research 15: 5073–5081.
Grosshans, H., and W. Filipowicz. 2008. Molecular biology: The expanding world of small RNAs. Nature 451: 414–416.
Guevin, C., D. Manna, C. Belanger, K.V. Konan, P. Mak, and P. Labonte. 2010. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 405: 1–7.
Guo, C., J.F. Sah, L. Beard, J.K. Willson, S.D. Markowitz, and K. Guda. 2008. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes, Chromosomes & Cancer 47: 939–946.
Guo, C.J., Q. Pan, B. Jiang, G.Y. Chen, and D.G. Li. 2009a. Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis 14: 1331–1340.
Guo, C.J., Q. Pan, D.G. Li, H. Sun, and B.W. Liu. 2009b. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. Journal of Hepatology 50: 766–778.
Guo, X., Y. Wu, and R.S. Hartley. 2009c. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biology 6: 575–583.
Hamasaki, M., and T. Yoshimori. 2010. Where do they come from? Insights into autophagosome formation. FEBS Letter 584: 1296–1301.
Han, J., W. Hou, L.A. Goldstein, C. Lu, D.B. Stolz, X.M. Yin, and H. Rabinowich. 2008. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. Journal of Biological Chemistry 283: 19665–19677.
Hanahan, D., and R.A. Weinberg. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674.
Hand, N.J., Z.R. Master, J. Le Lay, and J.R. Friedman. 2009. Hepatic function is preserved in the absence of mature microRNAs. Hepatology 49: 618–626.
Harris, J., M. Hartman, C. Roche, S.G. Zeng, A. O’shea, F.A. Sharp, E.M. Lambe, E.M. Creagh, D.T. Golenbock, J. Tschopp, H. Kornfeld, K.A. Fitzgerald, and E.C. Lavelle. 2011. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. Journal of Biological Chemistry 286: 9587–9597.
Harris, T.A., M. Yamakuchi, M. Ferlito, J.T. Mendell, and C.J. Lowenstein. 2008. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proceedings of the National Academy of Sciences of the United States of America 105: 1516–1521.
He, C., and D.J. Klionsky. 2009. Regulation mechanisms and signaling pathways of autophagy. Annual Review of Genetics 43: 67–93.
He, C., and B. Levine. 2010. The Beclin 1 interactome. Current Opinion in Cell Biology 22: 140–149.
He, L., and G.J. Hannon. 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nature Reviews Genetics 5: 522–531.
He, Y., C. Huang, X. Sun, X.R. Long, X.W. Lv, and J. Li. 2012. MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cellular Signalling 24: 1923–1930.
Hernandez-Gea, V., Z. Ghiassi-Nejad, R. Rozenfeld, R. Gordon, M.I. Fiel, Z. Yue, M.J. Czaja, and S.L. Friedman. 2012. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 142: 938–946.
Herrero-Martin, G., M. Hoyer-Hansen, C. Garcia-Garcia, C. Fumarola, T. Farkas, A. Lopez-Rivas, and M. Jaattela. 2009. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO Journal 28: 677–685.
Hou, W., J. Han, C. Lu, L.A. Goldstein, and H. Rabinowich. 2010. Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy 6: 891–900.
Hsu, S.H., B. Wang, J. Kota, J. Yu, S. Costinean, H. Kutay, L. Yu, S. Bai, K. La Perle, R.R. Chivukula, H. Mao, M. Wei, K.R. Clark, J.R. Mendell, M.A. Caligiuri, S.T. Jacob, J.T. Mendell, and K. Ghoshal. 2012. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. The Journal of Clinical Investigation 122: 2871–2883.
Hu, S., G. Xi, H. Jin, Y. He, R.F. Keep, and Y. Hua. 2011. Thrombin-induced autophagy: A potential role in intracerebral hemorrhage. Brain Research 1424: 60–66.
Huang, H., and D.J. Tindall. 2007. Dynamic FoxO transcription factors. Journal of Cell Science 120: 2479–2487.
Huang, J.C., T. Babak, T.W. Corson, G. Chua, S. Khan, B.L. Gallie, T.R. Hughes, B.J. Blencowe, B.J. Frey, and Q.D. Morris. 2007. Using expression profiling data to identify human microRNA targets. Nature Methods 4: 1045–1049.
Huang, L., J. Luo, Q. Cai, Q. Pan, H. Zeng, Z. Guo, W. Dong, J. Huang, and T. Lin. 2011. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. International Journal of Cancer 128: 1758–1769.
Ichimura, Y., T. Kirisako, T. Takao, Y. Satomi, Y. Shimonishi, N. Ishihara, N. Mizushima, I. Tanida, E. Kominami, M. Ohsumi, T. Noda, and Y. Ohsumi. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492.
Iliopoulos, D., K.N. Malizos, P. Oikonomou, and A. Tsezou. 2008. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE 3: e3740.
Johnson, C.D., A. Esquela-Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. Wang, J. Shelton, J. Shingara, L. Chin, D. Brown, and F.J. Slack. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Research 67: 7713–7722.
Johnson, S.M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K.L. Reinert, D. Brown, and F.J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647.
Jones-Rhoades, M.W., D.P. Bartel, and B. Bartel. 2006. MicroRNAS and their regulatory roles in plants. Annual Review of Plant Biology 57: 19–53.
Jopling, C.L., M. Yi, A.M. Lancaster, S.M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–1581.
Kang, K.W., S.Y. Choi, M.K. Cho, C.H. Lee, and S.G. Kim. 2003. Thrombin induces nitric-oxide synthase via Galpha12/13-coupled protein kinase C-dependent I-kappaBalpha phosphorylation and JNK-mediated I-kappaBalpha degradation. Journal of Biological Chemistry 278: 17368–17378.
Kashima, J., Shintani-Ishida, K., Nakajima, M., Maeda, H., Unuma, K., Uchiyama, Y., and Yoshida, K. I. (2013) Immunohistochemical study of the autophagy marker microtubule-associated protein 1 light chain 3 in normal and steatotic human livers. Hepatology Research. doi:10.1111/hepr.12183.
Kawanabe, Y., Y. Okamoto, K. Nozaki, N. Hashimoto, S. Miwa, and T. Masaki. 2002. Molecular mechanism for endothelin-1-induced stress-fiber formation: Analysis of G proteins using a mutant endothelin(A) receptor. Molecular Pharmacology 61: 277–284.
Kelly, P., P.J. Casey, and T.E. Meigs. 2007. Biologic functions of the G12 subfamily of heterotrimeric g proteins: growth, migration, and metastasis. Biochemistry 46: 6677–6687.
Kim, V.N., J. Han, and M.C. Siomi. 2009. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology 10: 126–139.
Kiriakidou, M., P.T. Nelson, A. Kouranov, P. Fitziev, C. Bouyioukos, Z. Mourelatos, and A. Hatzigeorgiou. 2004. A combined computational-experimental approach predicts human microRNA targets. Genes & Development 18: 1165–1178.
Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148.
Kiyono, K., H.I. Suzuki, H. Matsuyama, Y. Morishita, A. Komuro, M.R. Kano, K. Sugimoto, and K. Miyazono. 2009. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Research 69: 8844–8852.
Klionsky, D.J. 2005. Autophagy. Current Biology 15: R282–R283.
Klionsky, D.J., J.M. Cregg, W.A. Dunn Jr, S.D. Emr, Y. Sakai, I.V. Sandoval, A. Sibirny, S. Subramani, M. Thumm, M. Veenhuis, and Y. Ohsumi. 2003. A unified nomenclature for yeast autophagy-related genes. Developmental Cell 5: 539–545.
Klusmann, J.H., Z. Li, K. Bohmer, A. Maroz, M.L. Koch, S. Emmrich, F.J. Godinho, S.H. Orkin, and D. Reinhardt. 2010. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes & Development 24: 478–490.
Komatsu, M., S. Waguri, T. Ueno, J. Iwata, S. Murata, I. Tanida, J. Ezaki, N. Mizushima, Y. Ohsumi, Y. Uchiyama, E. Kominami, K. Tanaka, and T. Chiba. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. Journal of Cell Biology 169: 425–434.
Kong, G., J. Zhang, S. Zhang, C. Shan, L. Ye, and X. Zhang. 2011. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS ONE 6: e19518.
Koscianska, E., V. Baev, K. Skreka, K. Oikonomaki, V. Rusinov, M. Tabler, and K. Kalantidis. 2007. Prediction and preliminary validation of oncogene regulation by miRNAs. BMC Molecular Biology 8: 79.
Kota, J., R.R. Chivukula, K.A. O’donnell, E.A. Wentzel, C.L. Montgomery, H.W. Hwang, T.C. Chang, P. Vivekanandan, M. Torbenson, K.R. Clark, J.R. Mendell, and J.T. Mendell. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017.
Krutzfeldt, J., N. Rajewsky, R. Braich, K.G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689.
Kwiecinski, M., N. Elfimova, A. Noetel, U. Tox, H.M. Steffen, U. Hacker, R. Nischt, H.P. Dienes, and M. Odenthal. 2012. Expression of platelet-derived growth factor-C and insulin-like growth factor I in hepatic stellate cells is inhibited by miR-29. Laboratory Investigation 92: 978–987.
Lan, S.H., S.Y. Wu, R. Zuchini, X.Z. Lin, I.J. Su, T.F. Tsai, Y.J. Lin, C.T. Wu, and H.S. Liu. 2014. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 59: 505–517.
Landgraf, P., M. Rusu, R. Sheridan, A. Sewer, N. Iovino, A. Aravin, S. Pfeffer, A. Rice, A.O. Kamphorst, M. Landthaler, C. Lin, N.D. Socci, L. Hermida, V. Fulci, S. Chiaretti, R. Foa, J. Schliwka, U. Fuchs, A. Novosel, R.U. Muller, B. Schermer, U. Bissels, J. Inman, Q. Phan, M. Chien, D.B. Weir, R. Choksi, G. De Vita, D. Frezzetti, H.I. Trompeter, V. Hornung, G. Teng, G. Hartmann, M. Palkovits, R. Di Lauro, P. Wernet, G. Macino, C.E. Rogler, J.W. Nagle, J. Ju, F.N. Papavasiliou, T. Benzing, P. Lichter, W. Tam, M.J. Brownstein, A. Bosio, A. Borkhardt, J.J. Russo, C. Sander, M. Zavolan, and T. Tuschl. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414.
Lanford, R.E., E.S. Hildebrandt-Eriksen, A. Petri, R. Persson, M. Lindow, M.E. Munk, S. Kauppinen, and H. Orum. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201.
Le, M.T., N. Shyh-Chang, S.L. Khaw, L. Chin, C. Teh, J. Tay, E. O’day, V. Korzh, H. Yang, A. Lal, J. Lieberman, H.F. Lodish, and B. Lim. 2011. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genetics 7: e1002242.
Lee, R.C., R.L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854.
Lee, S.J., J.W. Yang, I.J. Cho, W.D. Kim, M.K. Cho, C.H. Lee, and S.G. Kim. 2009. The gep oncogenes, Galpha(12) and Galpha(13), upregulate the transforming growth factor-beta1 gene. Oncogene 28: 1230–1240.
Lee, Y.J., Y.J. Ha, Y.N. Kang, K.J. Kang, J.S. Hwang, W.J. Chung, K.B. Cho, K.S. Park, E.S. Kim, H.Y. Seo, M.K. Kim, K.G. Park, and B.K. Jang. 2013. The autophagy-related marker LC3 can predict prognosis in human hepatocellular carcinoma. PLoS ONE 8: e81540.
Lee, Y.R., H.Y. Lei, M.T. Liu, J.R. Wang, S.H. Chen, Y.F. Jiang-Shieh, Y.S. Lin, T.M. Yeh, C.C. Liu, and H.S. Liu. 2008. Autophagic machinery activated by dengue virus enhances virus replication. Virology 374: 240–248.
Lee, Y.S., and A. Dutta. 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & Development 21: 1025–1030.
Lemasters, J.J. 2005. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 8: 3–5.
Levine, B., and D.J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental Cell 6: 463–477.
Levine, B., and G. Kroemer. 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42.
Li, J., Y. Liu, Z. Wang, K. Liu, Y. Wang, J. Liu, H. Ding, and Z. Yuan. 2011a. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. Journal of Virology 85: 6319–6333.
Li, J., B. Yang, Q. Zhou, Y. Wu, D. Shang, Y. Guo, Z. Song, Q. Zheng, and J. Xiong. 2013. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis 34: 1343–1351.
Li, P., Q. Du, Z. Cao, Z. Guo, J. Evankovich, W. Yan, Y. Chang, L. Shao, D.B. Stolz, A. Tsung, and D.A. Geller. 2012. Interferon-gamma induces autophagy with growth inhibition and cell death in human hepatocellular carcinoma (HCC) cells through interferon-regulatory factor-1 (IRF-1). Cancer Letters 314: 213–222.
Li, W.Q., C. Chen, M.D. Xu, J. Guo, Y.M. Li, Q.M. Xia, H.M. Liu, J. He, H.Y. Yu, and L. Zhu. 2011b. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS Journal 278: 1522–1532.
Liang, X.H., S. Jackson, M. Seaman, K. Brown, B. Kempkes, H. Hibshoosh, and B. Levine. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676.
Lieber, C.S., and L.M. Decarli. 1986. The feeding of ethanol in liquid diets. Alcoholism, Clinical and Experimental Research 10: 550–553.
Lin, C.J., H.Y. Gong, H.C. Tseng, W.L. Wang, and J.L. Wu. 2008. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochemical and Biophysical Research Communications 375: 315–320.
Lin, C.W., H. Zhang, M. Li, X. Xiong, X. Chen, X.C. Dong, and X.M. Yin. 2013. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. Journal of Hepatology 58: 993–999.
Lin, Y.C., L.C. Hsieh, M.W. Kuo, J. Yu, H.H. Kuo, W.L. Lo, R.J. Lin, A.L. Yu, and W.H. Li. 2007. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Molecular Biology and Evolution 24: 2525–2534.
Liu, B., X.C. Peng, X.L. Zheng, J. Wang, and Y.W. Qin. 2009. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 66: 169–175.
Liu, Q., H. Fu, F. Sun, H. Zhang, Y. Tie, J. Zhu, R. Xing, Z. Sun, and X. Zheng. 2008. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Research 36: 5391–5404.
Mannaerts, I., N. Eysackers, O.O. Onyema, K. Van Beneden, S. Valente, A. Mai, M. Odenthal, and L.A. Van Grunsven. 2013. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS ONE 8: e55786.
Marino, G., J.A. Uria, X.S. Puente, V. Quesada, J. Bordallo, and C. Lopez-Otin. 2003. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. Journal of Biological Chemistry 278: 3671–3678.
Marklund, U., N. Larsson, H.M. Gradin, G. Brattsand, and M. Gullberg. 1996. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO Journal 15: 5290–5298.
Marra, F., A. Gastaldelli, G. Svegliati Baroni, G. Tell, and C. Tiribelli. 2008. Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends in Molecular Medicine 14: 72–81.
Masiero, E., L. Agatea, C. Mammucari, B. Blaauw, E. Loro, M. Komatsu, D. Metzger, C. Reggiani, S. Schiaffino, and M. Sandri. 2009. Autophagy is required to maintain muscle mass. Cell Metabolism 10: 507–515.
Mathew, R., V. Karantza-Wadsworth, and E. White. 2007. Role of autophagy in cancer. Nature Reviews Cancer 7: 961–967.
Mcdaniel, K., L. Herrera, T. Zhou, H. Francis, Y. Han, P. Levine, E. Lin, S. Glaser, G. Alpini, and F. Meng. 2014. The functional role of microRNAs in alcoholic liver injury. Journal of Cellular and Molecular Medicine 18: 197–207.
Mehrpour, M., A. Esclatine, I. Beau, and P. Codogno. 2010. Autophagy in health and disease. 1. Regulation and significance of autophagy: An overview. Cell Physiology: American Journal of Physiology 298: C776–C785.
Mei, S., H.M. Ni, S. Manley, A. Bockus, K.M. Kassel, J.P. Luyendyk, B.L. Copple, and W.X. Ding. 2011. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. Journal of Pharmacology and Experimental Therapeutics 339: 487–498.
Meng, F., R. Henson, H. Wehbe-Janek, K. Ghoshal, S.T. Jacob, and T. Patel. 2007a. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–658.
Meng, F., R. Henson, H. Wehbe-Janek, H. Smith, Y. Ueno, and T. Patel. 2007b. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. Journal of Biological Chemistry 282: 8256–8264.
Menzies, R.A., and P.H. Gold. 1971. The turnover of mitochondria in a variety of tissues of young adult and aged rats. Journal of Biological Chemistry 246: 2425–2429.
Miwa, S., C. Lawless, and T. Von Zglinicki. 2008. Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: Application of a simple dynamic model. Aging Cell 7: 920–923.
Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nature Cell Biology 9: 1089–1097.
Mizushima, N., and B. Levine. 2010. Autophagy in mammalian development and differentiation. Nature Cell Biology 12: 823–830.
Mizushima, N., B. Levine, A.M. Cuervo, and D.J. Klionsky. 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075.
Mizushima, N., T. Noda, T. Yoshimori, Y. Tanaka, T. Ishii, M.D. George, D.J. Klionsky, M. Ohsumi, and Y. Ohsumi. 1998. A protein conjugation system essential for autophagy. Nature 395: 395–398.
Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. Journal of Cell Biology 152: 657–668.
Moradpour, D., F. Penin, and C.M. Rice. 2007. Replication of hepatitis C virus. Nature Reviews Microbiology 5: 453–463.
Muller, D.W., and A.K. Bosserhoff. 2008. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27: 6698–6706.
Murakami, Y., H. Toyoda, M. Tanaka, M. Kuroda, Y. Harada, F. Matsuda, A. Tajima, N. Kosaka, T. Ochiya, and K. Shimotohno. 2011. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 6: e16081.
Nakahira, K., J.A. Haspel, V.A. Rathinam, S.J. Lee, T. Dolinay, H.C. Lam, J.A. Englert, M. Rabinovitch, M. Cernadas, H.P. Kim, K.A. Fitzgerald, S.W. Ryter, and A.M. Choi. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature Immunology 12: 222–230.
Nepal, S., and P.H. Park. 2013. Activation of autophagy by globular adiponectin attenuates ethanol-induced apoptosis in HepG2 cells: Involvement of AMPK/FoxO3A axis. Biochimica et Biophysica Acta 1833: 2111–2125.
Neufeld, T.P. 2010. TOR-dependent control of autophagy: Biting the hand that feeds. Current Opinion in Cell Biology 22: 157–168.
Ng, E.K., W.P. Tsang, S.S. Ng, H.C. Jin, J. Yu, J.J. Li, C. Rocken, M.P. Ebert, T.T. Kwok, and J.J. Sung. 2009. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. British Journal of Cancer 101: 699–706.
Ni, H.M., K. Du, M. You, and W.X. Ding. 2013. Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. American Journal of Pathology 183: 1815–1825.
Nie, K., M. Gomez, P. Landgraf, J.F. Garcia, Y. Liu, L.H. Tan, A. Chadburn, T. Tuschl, D.M. Knowles, and W. Tam. 2008. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in Hodgkin lymphomas. American Journal of Pathology 173: 242–252.
Noh, B.K., J.K. Lee, H.J. Jun, J.H. Lee, Y. Jia, M.H. Hoang, J.W. Kim, K.H. Park, and S.J. Lee. 2011. Restoration of autophagy by puerarin in ethanol-treated hepatocytes via the activation of AMP-activated protein kinase. Biochemical and Biophysical Research Communications 414: 361–366.
Noh, S.J., S.H. Miller, Y.T. Lee, S.H. Goh, F.M. Marincola, D.F. Stroncek, C. Reed, E. Wang, and J.L. Miller. 2009. Let-7 microRNAs are developmentally regulated in circulating human erythroid cells. Journal of Translational Medicine 7: 98.
Ogawa, M., T. Yoshimori, T. Suzuki, H. Sagara, N. Mizushima, and C. Sasakawa. 2005. Escape of intracellular Shigella from autophagy. Science 307: 727–731.
Ogawa, T., M. Enomoto, H. Fujii, Y. Sekiya, K. Yoshizato, K. Ikeda, and N. Kawada. 2012. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut 61: 1600–1609.
Oglesby, I.K., I.M. Bray, S.H. Chotirmall, R.L. Stallings, S.J. O’neill, N.G. Mcelvaney, and C.M. Greene. 2010. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. The Journal of Immunology 184: 1702–1709.
Oh, J.S., J.J. Kim, J.Y. Byun, and I.A. Kim. 2010. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. International Journal of Radiation Oncology Biology Physics 76: 5–8.
Ohsumi, Y., and N. Mizushima. 2004. Two ubiquitin-like conjugation systems essential for autophagy. Seminars in Cell & Developmental Biology 15: 231–236.
Pan, W., P. Xin, and G.A. Clawson. 2010. MicroRNAs align with accessible sites in target mRNAs. Journal of Cellular Biochemistry 109: 509–518.
Pasquinelli, A.E., B.J. Reinhart, F. Slack, M.Q. Martindale, M.I. Kuroda, B. Maller, D.C. Hayward, E.E. Ball, B. Degnan, P. Muller, J. Spring, A. Srinivasan, M. Fishman, J. Finnerty, J. Corbo, M. Levine, P. Leahy, E. Davidson, and G. Ruvkun. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89.
Peng, Y., J. Laser, G. Shi, K. Mittal, J. Melamed, P. Lee, and J.J. Wei. 2008. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Molecular Cancer Research 6: 663–673.
Peng, Y.F., Y.H. Shi, Y.H. Shen, Z.B. Ding, A.W. Ke, J. Zhou, S.J. Qiu, and J. Fan. 2013. Promoting colonization in metastatic HCC cells by modulation of autophagy. PLoS ONE 8: e74407.
Pogue, A.I., J.G. Cui, Y.Y. Li, Y. Zhao, F. Culicchia, and W.J. Lukiw. 2010. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neuroscience Letters 476: 18–22.
Postic, C., and J. Girard. 2008. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: Lessons from genetically engineered mice. The Journal of Clinical Investigation 118: 829–838.
Pyo, J.O., J. Nah, and Y.K. Jung. 2012. Molecules and their functions in autophagy. Experimental & Molecular Medicine 44: 73–80.
Qi, Z., N. Atsuchi, A. Ooshima, A. Takeshita, and H. Ueno. 1999. Blockade of type beta transforming growth factor signaling prevents liver fibrosis and dysfunction in the rat. Proceedings of the National Academy of Sciences of the United States of America 96: 2345–2349.
Qu, X., J. Yu, G. Bhagat, N. Furuya, H. Hibshoosh, A. Troxel, J. Rosen, E.L. Eskelinen, N. Mizushima, Y. Ohsumi, G. Cattoretti, and B. Levine. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of Clinical Investigation 112: 1809–1820.
Rana, S., P.B. Maples, N. Senzer, and J. Nemunaitis. 2008. Stathmin 1: A novel therapeutic target for anticancer activity. Expert Review of Anticancer Therapy 8: 1461–1470.
Rautou, P.E., A. Mansouri, D. Lebrec, F. Durand, D. Valla, and R. Moreau. 2010. Autophagy in liver diseases. Journal of Hepatology 53: 1123–1134.
Ravikumar, B., S. Imarisio, S. Sarkar, C.J. O’kane, and D.C. Rubinsztein. 2008. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. Journal of Cell Science 121: 1649–1660.
Reed, K.E., and C.M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Current Topics in Microbiology and Immunology 242: 55–84.
Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. European Journal of Biochemistry 268: 5857–5867.
Reinhart, B.J., F.J. Slack, M. Basson, A.E. Pasquinelli, J.C. Bettinger, A.E. Rougvie, H.R. Horvitz, and G. Ruvkun. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906.
Roberts, A.P., A.P. Lewis, and C.L. Jopling. 2011. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Research 39: 7716–7729.
Roderburg, C., G.W. Urban, K. Bettermann, M. Vucur, H. Zimmermann, S. Schmidt, J. Janssen, C. Koppe, P. Knolle, M. Castoldi, F. Tacke, C. Trautwein, and T. Luedde. 2011. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53: 209–218.
Saitoh, T., N. Fujita, M.H. Jang, S. Uematsu, B.G. Yang, T. Satoh, H. Omori, T. Noda, N. Yamamoto, M. Komatsu, K. Tanaka, T. Kawai, T. Tsujimura, O. Takeuchi, T. Yoshimori, and S. Akira. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456: 264–268.
Samuel, V.T., K.F. Petersen, and G.I. Shulman. 2010. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet 375: 2267–2277.
Sarbassov, D.D., S.M. Ali, and D.M. Sabatini. 2005. Growing roles for the mTOR pathway. Current Opinion in Cell Biology 17: 596–603.
Scherz-Shouval, R., E. Shvets, E. Fass, H. Shorer, L. Gil, and Z. Elazar. 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO Journal 26: 1749–1760.
Schultz, J., P. Lorenz, G. Gross, S. Ibrahim, and M. Kunz. 2008. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Research 18: 549–557.
Scott, G.K., A. Goga, D. Bhaumik, C.E. Berger, C.S. Sullivan, and C.C. Benz. 2007. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. Journal of Biological Chemistry 282: 1479–1486.
Seglen, P.O., and P.B. Gordon. 1982. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America 79: 1889–1892.
Sengupta, A., J.D. Molkentin, J.H. Paik, R.A. Depinho, and K.E. Yutzey. 2011. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. Journal of Biological Chemistry 286: 7468–7478.
Sharma, A.D., N. Narain, E.M. Handel, M. Iken, N. Singhal, T. Cathomen, M.P. Manns, H.R. Scholer, M. Ott, and T. Cantz. 2011. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology 53: 1651–1661.
Shell, S., S.M. Park, A.R. Radjabi, R. Schickel, E.O. Kistner, D.A. Jewell, C. Feig, E. Lengyel, and M.E. Peter. 2007. Let-7 expression defines two differentiation stages of cancer. Proceedings of the National Academy of Sciences of the United States of America 104: 11400–11405.
Shepard, C.W., L. Finelli, and M.J. Alter. 2005. Global epidemiology of hepatitis C virus infection. The Lancet Infectious Diseases 5: 558–567.
Shi, L., J. Zhang, T. Pan, J. Zhou, W. Gong, N. Liu, Z. Fu, and Y. You. 2010. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Research 1312: 120–126.
Shi, X.B., L. Xue, J. Yang, A.H. Ma, J. Zhao, M. Xu, C.G. Tepper, C.P. Evans, H.J. Kung, and R.W. Devere White. 2007. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proceedings of the National Academy of Sciences of the United States of America 104: 19983–19988.
Shrivastava, S., A. Raychoudhuri, R. Steele, R. Ray, and R.B. Ray. 2011. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53: 406–414.
Singh, R., S. Kaushik, Y. Wang, Y. Xiang, I. Novak, M. Komatsu, K. Tanaka, A.M. Cuervo, and M.J. Czaja. 2009. Autophagy regulates lipid metabolism. Nature 458: 1131–1135.
Sinha, R.A., B.L. Farah, B.K. Singh, M.M. Siddique, Y. Li, Y. Wu, O.R. Ilkayeva, J. Gooding, J. Ching, J. Zhou, L. Martinez, S. Xie, B.H. Bay, S.A. Summers, C.B. Newgard, and P.M. Yen. 2014. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology 59: 1366–1380.
Sir, D., W.L. Chen, J. Choi, T. Wakita, T.S. Yen, and J.H. Ou. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48: 1054–1061.
Sir, D., Y. Tian, W.L. Chen, D.K. Ann, T.S. Yen, and J.H. Ou. 2010. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proceedings of the National Academy of Sciences of the United States of America 107: 4383–4388.
Su, H., J.R. Yang, T. Xu, J. Huang, L. Xu, Y. Yuan, and S.M. Zhuang. 2009. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Research 69: 1135–1142.
Sun, W., Y.S. Julie Li, H.D. Huang, J.Y. Shyy, and S. Chien. 2010. microRNA: a master regulator of cellular processes for bioengineering systems. Annual Review of Biomedical Engineering 12: 1–27.
Suzuki, H.I., K. Kiyono, and K. Miyazono. 2010. Regulation of autophagy by transforming growth factor-beta (TGF-beta) signaling. Autophagy 6: 645–647.
Suzuki, K., Y. Kubota, T. Sekito, and Y. Ohsumi. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes to Cells 12: 209–218.
Tai, W.T., C.W. Shiau, H.L. Chen, C.Y. Liu, C.S. Lin, A.L. Cheng, P.J. Chen, and K.F. Chen. 2013. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death and Disease 4: e485.
Takamura, A., M. Komatsu, T. Hara, A. Sakamoto, C. Kishi, S. Waguri, Y. Eishi, O. Hino, K. Tanaka, and N. Mizushima. 2011. Autophagy-deficient mice develop multiple liver tumors. Genes & Development 25: 795–800.
Takanabe, R., K. Ono, Y. Abe, T. Takaya, T. Horie, H. Wada, T. Kita, N. Satoh, A. Shimatsu, and K. Hasegawa. 2008. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochemical and Biophysical Research Communications 376: 728–732.
Takane, K., K. Fujishima, Y. Watanabe, A. Sato, N. Saito, M. Tomita, and A. Kanai. 2010. Computational prediction and experimental validation of evolutionarily conserved microRNA target genes in bilaterian animals. BMC Genomics 11: 101.
Talloczy, Z., H.W.T. Virgin, and B. Levine. 2006. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2: 24–29.
Tang, H., L. Da, Y. Mao, Y. Li, D. Li, Z. Xu, F. Li, Y. Wang, P. Tiollais, T. Li, and M. Zhao. 2009. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology 49: 60–71.
Tang, S.W., A. Ducroux, K.T. Jeang, and C. Neuveut. 2012. Impact of cellular autophagy on viruses: Insights from hepatitis B virus and human retroviruses. Journal of Biomedical Science 19: 92.
Tanida, I., M. Fukasawa, T. Ueno, E. Kominami, T. Wakita, and K. Hanada. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5: 937–945.
Terlecky, S.R., and J.F. Dice. 1993. Polypeptide import and degradation by isolated lysosomes. Journal of Biological Chemistry 268: 23490–23495.
Thoen, L.F., E.L. Guimaraes, L. Dolle, I. Mannaerts, M. Najimi, E. Sokal, and L.A. Van Grunsven. 2011. A role for autophagy during hepatic stellate cell activation. Journal of Hepatology 55: 1353–1360.
Thomes, P.G., R.A. Ehlers, C.S. Trambly, D.L. Clemens, H.S. Fox, D.J. Tuma, and T.M. Donohue. 2013. Multilevel regulation of autophagosome content by ethanol oxidation in HepG2 cells. Autophagy 9: 63–73.
Thomes, P.G., C.S. Trambly, G.M. Thiele, M.J. Duryee, H.S. Fox, J. Haorah, and T.M. Donohue Jr. 2012. Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochemical and Biophysical Research Communications 417: 262–267.
Tian, Y., D. Sir, C.F. Kuo, D.K. Ann, and J.H. Ou. 2011. Autophagy required for hepatitis B virus replication in transgenic mice. Journal of Virology 85: 13453–13456.
Ting, Y., D.J. Medina, R.K. Strair, and D.G. Schaar. 2010. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochemical and Biophysical Research Communications 394: 606–611.
Tokumaru, S., M. Suzuki, H. Yamada, M. Nagino, and T. Takahashi. 2008. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29: 2073–2077.
Toshima, T., Shirabe, K., Matsumoto, Y., Yoshiya, S., Ikegami, T., Yoshizumi, T., Soejima, Y., Ikeda, T., and Maehara, Y. (2013). Autophagy enhances hepatocellular carcinoma progression by activation of mitochondrial beta-oxidation. Journal of Gastroenterology. doi:10.1007/s00535-013-0835-9.
Tsai, W.C., P.W. Hsu, T.C. Lai, G.Y. Chau, C.W. Lin, C.M. Chen, C.D. Lin, Y.L. Liao, J.L. Wang, Y.P. Chau, M.T. Hsu, M. Hsiao, H.D. Huang, and A.P. Tsou. 2009. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 49: 1571–1582.
Tsai, W.C., S.D. Hsu, C.S. Hsu, T.C. Lai, S.J. Chen, R. Shen, Y. Huang, H.C. Chen, C.H. Lee, T.F. Tsai, M.T. Hsu, J.C. Wu, H.D. Huang, M.S. Shiao, M. Hsiao, and A.P. Tsou. 2012. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. The Journal of Clinical Investigation 122: 2884–2897.
Tsang, W.P., and T.T. Kwok. 2008. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 13: 1215–1222.
Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. Journal of Virology 66: 1476–1483.
Tzivion, G., M. Dobson, and G. Ramakrishnan. 2011. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochimica et Biophysica Acta 1813: 1938–1945.
Tzur, G., A. Israel, A. Levy, H. Benjamin, E. Meiri, Y. Shufaro, K. Meir, E. Khvalevsky, Y. Spector, N. Rojansky, Z. Bentwich, B.E. Reubinoff, and E. Galun. 2009. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS ONE 4: e7511.
Vara, D., M. Salazar, N. Olea-Herrero, M. Guzman, G. Velasco, and I. Diaz-Laviada. 2011. Anti-tumoral action of cannabinoids on hepatocellular carcinoma: Role of AMPK-dependent activation of autophagy. Cell Death and Differentiation 18: 1099–1111.
Venugopal, S.K., J. Jiang, T.H. Kim, Y. Li, S.S. Wang, N.J. Torok, J. Wu, and M.A. Zern. 2010. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Gastrointestinal and Liver Physiology: American Journal of Physiology 298: G101–G106.
Vescovo, T., A. Romagnoli, A.B. Perdomo, M. Corazzari, F. Ciccosanti, T. Alonzi, R. Nardacci, G. Ippolito, M. Tripodi, C. Garcia-Monzon, O. Lo Iacono, M. Piacentini, and G.M. Fimia. 2012. Autophagy protects cells from HCV-induced defects in lipid metabolism. Gastroenterology 142(e3): 644–665.
Wang, B., S. Majumder, G. Nuovo, H. Kutay, S. Volinia, T. Patel, T.D. Schmittgen, C. Croce, K. Ghoshal, and S.T. Jacob. 2009. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 50: 1152–1161.
Wang, X., Y. Dai, Z. Ding, M. Khaidakov, F. Mercanti, and J.L. Mehta. 2013. Regulation of autophagy and apoptosis in response to angiotensin II in HL-1 cardiomyocytes. Biochemical and Biophysical Research Communications 440: 696–700.
Wang, X.W., N.H. Heegaard, and H. Orum. 2012. MicroRNAs in liver disease. Gastroenterology 142: 1431–1443.
Wang, Y., R. Singh, Y. Xiang, and M.J. Czaja. 2010. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology 52: 266–277.
Wei, Y., S. Pattingre, S. Sinha, M. Bassik, and B. Levine. 2008. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Molecular Cell 30: 678–688.
Westin, J., H. Nordlinder, M. Lagging, G. Norkrans, and R. Wejstal. 2002. Steatosis accelerates fibrosis development over time in hepatitis C virus genotype 3 infected patients. Journal of Hepatology 37: 837–842.
Wightman, B., I. Ha, and G. Ruvkun. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862.
Wu, D., X. Wang, R. Zhou, and A. Cederbaum. 2010. CYP2E1 enhances ethanol-induced lipid accumulation but impairs autophagy in HepG2 E47 cells. Biochemical and Biophysical Research Communications 402: 116–122.
Wu, D., X. Wang, R. Zhou, L. Yang, and A.I. Cederbaum. 2012a. Alcohol steatosis and cytotoxicity: The role of cytochrome P4502E1 and autophagy. Free Radical Biology and Medicine 53: 1346–1357.
Wu, J., D. Hu, and R. Zhang. 2012b. Depletion of Bmi-1 enhances 5-fluorouracil-induced apoptosis and autophagy in hepatocellular carcinoma cells. Oncology Letters 4: 723–726.
Wu, K., J. Ding, C. Chen, W. Sun, B.F. Ning, W. Wen, L. Huang, T. Han, W. Yang, C. Wang, Z. Li, M.C. Wu, G.S. Feng, W.F. Xie, and H.Y. Wang. 2012c. Hepatic transforming growth factor beta gives rise to tumor-initiating cells and promotes liver cancer development. Hepatology 56: 2255–2267.
Wu, L., and J.G. Belasco. 2005. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Molecular and Cellular Biology 25: 9198–9208.
Xiong, J., D. Yu, N. Wei, H. Fu, T. Cai, Y. Huang, C. Wu, X. Zheng, Q. Du, D. Lin, and Z. Liang. 2010a. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS Journal 277: 1684–1694.
Xiong, Y., J.H. Fang, J.P. Yun, J. Yang, Y. Zhang, W.H. Jia, and S.M. Zhuang. 2010b. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 51: 836–845.
Xu, H., J.H. He, Z.D. Xiao, Q.Q. Zhang, Y.Q. Chen, H. Zhou, and L.H. Qu. 2010. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 52: 1431–1442.
Xu, L., S. Beckebaum, S. Iacob, G. Wu, G.M. Kaiser, A. Radtke, C. Liu, I. Kabar, H.H. Schmidt, X. Zhang, M. Lu, and V.R. Cicinnati. 2014. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. Journal of Hepatology 60: 590–598.
Xu, N., J. Zhang, C. Shen, Y. Luo, L. Xia, F. Xue, and Q. Xia. 2012. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochemical and Biophysical Research Communications 423: 826–831.
Xu, T., Y. Zhu, Y. Xiong, Y.Y. Ge, J.P. Yun, and S.M. Zhuang. 2009. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 50: 113–121.
Yadav, A., S. Vallabu, S. Arora, P. Tandon, D. Slahan, S. Teichberg, and P.C. Singhal. 2010. ANG II promotes autophagy in podocytes. Cell Physiology: American Journal of Physiology 299: C488–C496.
Yang, L., P. Li, S. Fu, E.S. Calay, and G.S. Hotamisligil. 2010a. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metabolism 11: 467–478.
Yang, L., D. Wu, X. Wang, and A.I. Cederbaum. 2012a. Cytochrome P4502E1, oxidative stress, JNK, and autophagy in acute alcohol-induced fatty liver. Free Radical Biology and Medicine 53: 1170–1180.
Yang, Y.M., C.Y. Han, Y.J. Kim, and S.G. Kim. 2010b. AMPK-associated signaling to bridge the gap between fuel metabolism and hepatocyte viability. World Journal of Gastroenterology 16: 3731–3742.
Yang, Y.M., S.Y. Seo, T.H. Kim, and S.G. Kim. 2012b. Decrease of microRNA-122 causes hepatic insulin resistance by inducing protein tyrosine phosphatase 1B, which is reversed by licorice flavonoid. Hepatology 56: 2209–2220.
Ye, W., Q. Lv, C.K. Wong, S. Hu, C. Fu, Z. Hua, G. Cai, G. Li, B.B. Yang, and Y. Zhang. 2008. The effect of central loops in miRNA: MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS ONE 3: e1719.
Yin, X.M., W.X. Ding, and W. Gao. 2008. Autophagy in the liver. Hepatology 47: 1773–1785.
Yu, L., C.K. Mcphee, L. Zheng, G.A. Mardones, Y. Rong, J. Peng, N. Mi, Y. Zhao, Z. Liu, F. Wan, D.W. Hailey, V. Oorschot, J. Klumperman, E.H. Baehrecke, and M.J. Lenardo. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946.
Yuan, K., Z. Lian, B. Sun, M.M. Clayton, I.O. Ng, and M.A. Feitelson. 2012. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS ONE 7: e35331.
Zhang, H., X. Cai, Y. Wang, H. Tang, D. Tong, and F. Ji. 2010. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncology Reports 24: 1363–1369.
Zhang, H. T., Chen, G. G., Hu, B. G., Zhang, Z. Y., Yun, J. P., He, M. L., and Lai, P. B. (2013a) Hepatitis B virus x protein induces autophagy via activating death-associated protein kinase. Journal of Viral Hepatitis. doi:10.1111/jvh.12191.
Zhang, J., Y.Y. Du, Y.F. Lin, Y.T. Chen, L. Yang, H.J. Wang, and D. Ma. 2008. The cell growth suppressor, mir-126, targets IRS-1. Biochemical and Biophysical Research Communications 377: 136–140.
Zhang, X., S. Liu, T. Hu, Y. He, and S. Sun. 2009. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50: 490–499.
Zhang, X.H., Y.N. Zhang, H.B. Li, C.Y. Hu, N. Wang, P.P. Cao, B. Liao, X. Lu, Y.H. Cui, and Z. Liu. 2012. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. American Journal of Respiratory and Critical Care Medicine 185: 140–151.
Zhang, Y., S. Takahashi, A. Tasaka, T. Yoshima, H. Ochi, and K. Chayama. 2013b. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. Journal of Gastroenterology and Hepatology 28: 565–575.
Zhao, A., Q. Zeng, X. Xie, J. Zhou, W. Yue, Y. Li, and X. Pei. 2012. MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. Journal of Genetics and Genomics 39: 29–35.
Zhao, J., J.J. Brault, A. Schild, P. Cao, M. Sandri, S. Schiaffino, S.H. Lecker, and A.L. Goldberg. 2007. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metabolism 6: 472–483.
Zhao, S., K. Venkatasubbarao, J.W. Lazor, J. Sperry, C. Jin, L. Cao, and J.W. Freeman. 2008. Inhibition of STAT3 Tyr705 phosphorylation by Smad4 suppresses transforming growth factor beta-mediated invasion and metastasis in pancreatic cancer cells. Cancer Research 68: 4221–4228.
Zhou, R., A.S. Yazdi, P. Menu, and J. Tschopp. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature 469: 221–225.
Acknowledgments
This work was supported by the fund from Ministry of Health & Welfare (No. A111641).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, K.M., Kim, S.G. Autophagy and microRNA dysregulation in liver diseases. Arch. Pharm. Res. 37, 1097–1116 (2014). https://doi.org/10.1007/s12272-014-0439-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-014-0439-9