Abstract
Death receptor 4 (DR4) gene is a candidate tumor supressor gene that has a role in apoptotic pathway. It was reported in literature that polymorphisms in DR4 gene lead to susceptibility to many cancers. In accordance with this information, we aimed to investigate the association between G422A, C626G, A683C and A1322G polymorphisms in DR4 gene and lung cancer. We selected 60 patients with lung cancer (LC) and 30 healthy, sex and age matched volunteers randomly. Four polymorhisms, G422A, C626G, A683C and A1322G, in DR4 gene were analyzed with Polymerase Change Reaction (PCR) – Restriction Fragment Lenght Polymorphism (RFLP) and Amplification Refractory Mutation System (ARMS) techniques in both groups. Our results showed that there are no statistically significances between the patients and controls in terms of the G422A, C626G, A683C and A1322G polymorphisms in DR4 gene (p > 0,05). Our findings showed no role of DR4 gene polymorhisms in susceptibility to LC and provide a plausible explanation for DR4 genetic heterogeneity in LC susceptibility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis or porgrammed cell-death is an important physiological process of the organism. Apoptosis occurs in two ways: Cytochrome C-mediated pathway and via death receptors (DR) (Fas, TNFR1, DR3, DR4, DR5, CAR1). Death receptors are transmembrane proteins that contain cytoplasmic death-domain. This death domain characterizes them as apoptotic “death-receptors” and is pivotal for the transduction of the apoptotic signal [1].
Death Receptor 4, DR4, located on chromosome 8p21, was the first DR for APO2L/TRAIL to be identified [2]. The DR4 type I membrane protein contains 486 amino acids, composed of two extracellular cysteine-rich pseudorepeats. Two receptor loops, the 50- and 90-s loops, mediate most of the ligand receptor interactions. One loop consists of mostly a hydrophobic motif that is conserved in ligand receptor complexes throughout the TNF superfamily, and the other loop is unique for each individual complex and appears to be used to control receptor selectivity and cross-reactivity [2]. The principle elements of the DR4 ligand-binding domain are encoded by exons 3 and 4 [3].
The DR4 gene is highly polymorphic, but the most extensively studied polymorphism is the C to G substitution at position 626 (C626G) in the ectodomain region of DR4. In addition, another three DR4 polymorphisms included G422A, A683C, and A1322G are also extensively studied [1–5]. DR4 mutations have been described in different human cancers, such as lung, head and neck cancer, non-Hodgkin’s lymphoma, osteosarcoma and breast cancer [1, 3, 6, 7]. Furthermore, many researchers have studied the role of the DR4 gene polymorphisms in the aetiology of cancers of various organs, including lung, bladder, ovary, colon, stomach, breast, head and neck, prostate, and lymph tissue [3–5, 8–12]. Some cancer types were in association with the DR4 gene polymorphisms, but some were not. In the present study, we aimed to evaluate the association between common genetic variants in DR4 gene and risk of LC in Turkey.
Materials and Methods
Study Population
The study involved 60 Turkish patients with LC referred from Department of Chest Diseases, School of Medicine, Çukurova University, Adana-Turkey. Also it was granted ethical approval by the local health committee and permission from all patients were taken. The patient group consisted of 55 males and 5 females, and their ages ranged from 33 to 77 years with a mean age of 57,97 ± 9,45 years. Tobacco consumption of these patients ranged from 10 to 80 packs × year and the average tobacco consumption was 40,18 ± 25,91 packs x year. It was also recorded that six patients had never used tobacco. All patients didn’t receive chemotherapy or radiotherapy before the present analysis. The control group involved 30 healthy people (one female and 29 male). Their ages ranged from 37 to 71 years with a mean age of 53,37 ± 10,39 years. All of them smoke and the average tobacco consumption was 36,13 ± 19,22 packs x year. There is no statistically difference for tobacco consumptions between the two groups (p > 0,05). For eliminating the cigarette factor, we selected the control group from heavy cigarette users. We also recorded their familial history of cancer. In 21 patients (35 %), familial history of cancers such as lung, breast, stomach, colon, hepatocelluler carcinomas and lymphoma are present. We attended to choice control group without familial history.
Death Receptor 4 (DR4) Genotyping
A 3-ml sample of blood was taken from all patients and controls for genotyping DR4 gene polymorphisms including G422A, C626G, A683C and A1322G. Genomic DNA was isolated from 0.2 ml of whole blood using QIAMP-DNA isolation kit (Qiagen). Primers for G422A, C626G, A683C regions selected from literatures [1, 3] and for A1322G, they were dizayned using gene runner programme. The primers used and the product sizes were shown in Table 1.
For detecting DR4 gene polymorphisms, PCR-RFLP technique was used for G422A, C626G, A683C polymorphisms and PCR-ARMS for A1322G. Because of the method for A1322G, two different reverse primers were dizayned: one stops with cytosine and the other with thymine.
PCR-RFLP Methods for G422A, C626G and A683C Polymorphisms
PCR conditions for G422A, C626G, and A683C polymorphisms were 94 °C for 5 min, followed by 94 °C for 30s, 62 °C for 30s (for A683C, annealing temperature is 58 °C), and 72 °C for 30s (35 cycles). The PCR mixture (25 μl) included 1XPCR Buffer, 1.25 mM MgCl2, 0.2 mM dNTPs, 5pmol primers, 50 ng DNA, and 2U Taq Polimerase (Fermantas). The PCR products were electrophoresed in 10 % polyacrilamid gels in 1XTBE and visualized with ethidiumbromide. The product sizes were 230 bp for G422A, 220 bp for C626G, and 201 bp for A683C. The amplified DNA fragments include G422A and A683C were incubated with 5U of the restriction enzymes FokI and TaqI, respectively at 65 °C for an overnight, and for detect C626G polymorphism, the product were incubated with 5U of the restriction enzyme DraIII at 37 °C for an overnight. Restriction digests were evaluated using 10 % polyacrilamid gels in 1XTBE, and visualised by ethidium bromide staining. To determine the size of the banding patterns, pUC18/HaeIII marker were loaded together with the digested samples and then compared with it.
PCR-ARMS Method for A1322C Polymorphism
PCR conditions for A1322C polymorphism were 95 °C for 10 min, followed by 95 °C for 1 min, 56 °C for 1 min, and 72 °C for 1 min (34 cycles). The PCR mixture (50 μl) included 1XPCR Buffer, 2 mM MgCl2, 0.2 mM dNTPs, 5pmol primers, 50 ng DNA, and 2U Taq Polimerase (Fermantas). Two different tubes were prepared for this purpose. Forward primer was added to the two tubes, but the reverse primers (ended with C and T) added separately. Also for this study, internal control [CYP19 (Cytochrome P450, subfamily XIX) gene; the second exon included the 39th codon; forward primer: 5′-atctgtactgtacagcacc-3′, reverse primer: 5′-ctccaagtcctcatttgct-3′] was used. Two primers were dizayned and added to the tubes. The PCR products were 254 bp for A1322C and 427 bp for internal control. The PCR products were electrophoresed in 10 % polyacrilamid gels in 1XTBE and visualized with ethidiumbromide. Then, compared with pUC18/HaeIII marker loaded together with PCR products.
Results
Our study included 60 confirmed Turkish LC patients, of whom 91,6 % were men and 8,4 % were women, and 30 healthy individuals, of whom 96,6 % were men and 3,4 % were women. There were no differences between study groups according to age and gender. DR4 gene polymorphisms, G422A, C626G, A683C and A1322G, were analyzed and compared between the study groups.
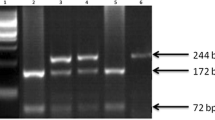
In the G422A polymorphism, the G to A transversion results in an amino acid change of a histidine for an arginine and creates a new and unique FokI restriction site and therefore yields a 160 bp and a 70 bp fragment upon incubation with FokI (Fig. 1a). The homozygous G/G variant in the G422A locus was detected in 15 (25 %) patients and in 10 (33,3 %) control subjects in our study. The heterozygous G/A and homozygous A/A variants were found in 34 (56,7 %) and 11 (18,3 %) patients and, 12 (40 %) and 8 (26,7 %) control subjects, respectively (Table 2). In direct comparision, the GG, GA and AA alleles were almost equally distributed in patients and controls (p = 0,324; Fig. 1a).
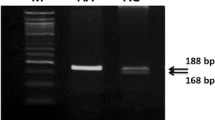
RFLP analysis of exon3, exon 4, and exon5 of DR4 and analysis of exon10 of DR4 with ARMS technique: a Analysis of exon 3 with Fok1 restriction enzyme. The polymorphism at position 422 reveal a new restriction site for Fok1 restriction enzyme. The Fok1 restriction enzyme does not cut wild type samples and shows a single band (230 bp). When the G-- > A transition occurs at 422 position of exon3, two bands (160 bp and 70 bp) are seen after gel-electrophoresis of digested samples, b Analysis of exon4 with DraIII restriction enzyme. The polymorphism at position 626 (C-- > G) eliminates a restriction site for the DraIII enzyme. Wild type (CC) samples are cut into two fragments (164 bp and 56 bp), whereas the C-- > G transition at position 626 bloks the cutting (220 bp), c Analysis of exon5 with TaqI restriction enzyme. The polymorphism at 683 position of exon5 eliminates a restriction site for TaqI restriction enzyme. Wild type samples are cut into two fragments, 110 bp and 91 bp, whereas poymorphic region blocks the cutting and yields only a band 201 bp, d Analysis of exon10 with PCR-ARMS technique using internal control
In the C626G polymorphism, there was a C to G transversion and results in an amino acid change of an arginine to threonine. The C to G transversion eliminates a unique DraIII restriction site. Incubation with DraIII restriction endonuclease, the CC variant shows two bands, 164 bp and 56 bp, and the GG variant shows one unique band with the size of 220 bp (Fig. 1b). In our patient group, C/C, C/G and G/G variants detected in 12 (20 %), 34 (56,7 %), and 14 (23,3 %) cases, respectively. In the controls, the homozygous C/C, heterozygous C/G, and homozygous G/G variants found in 10 (33,3 %), 12 (40 %), and 8 (26,7 %), respectively (Table 2). It was found that there were no significant association between the two groups (p = 0,271).
The A683C polymorphism occurs in the exon 5 of the DR4 gene. The transversion of the A to C eliminates a unique TaqI restriction site. Incubation with TaqI endonuclease, A/A variant shows two bants, 110 bp and 91 bp, whereas C/C variant shows a 201 bp unique bant. In our study population, the A/A variant was seen in 41 (68,3 %) patients and 21 (70 %) controls, and the A/C variant in 19 (31,7 %) patients and 7 (23,3 %) controls. The C/C variant didn’t seen in the patient group, whereas 2 control individuals has it (Table 2, Fig. 1c). When the two groups compared for the A683C polymorphism, no association was observed significantly (p = 0,086).
For the A1322G polymophism, we used the PCR-ARMS technique in 60 patients and 30 controls. Most of the patients and controls [53 (88,3 %) patients and 24 (80 %) controls] showed AA genotype, and the others AG [6 (10 %) patients, 3 (10 %) controls] and GG [1 (1,7 %) patient and 3 (10 %) controls)] genotypes (Table 2, Fig. 1d). When these results evaluated statistically, no association was found (p = 0,230).
Discussion
DR4 (TRAIL-R1), a member of the tumor necrosis factor receptor superfamily, is a cell surface receptor that triggers the apoptotic machinery upon binding to its ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). TRAIL is a potent inducer of apoptosis in malignant but not in normal cells. It binds to the proapoptotic death receptor DR4 and DR5, also to decoy receptors DcR1 and DcR2 for the survival of cells [4].
DR4 was the first identified death receptor for TRAIL. It is a type I membrane protein and contains 486 amino acids. The principle elements of the DR4 ligand-binding domain are encoded by exon 3 and 4 of the gene [3, 13]. DR4 gene has highly polymorphic regions. The most extensively studied polymorphism is C626G, the C to G substitution at position 626 in the ectodomain region of DR4. In addition, another DR4 polymorphisms, studied most frequent, are G422A, A683C and A1322G. The G422A polymorhism occurs in the ectodomain region of DR4, whereas A683C in extracellular cycteine-rich domain and A1322G in the death domain of DR4 [14]. Recently, many studies on DR4 gene polymorphisms were performed on different cancer types, included bladder, lymph tissue, ovary, colon, stomach, breast, lung, head and neck, prostate cancers, and osteosarcoma tumor [1–5, 10–12, 15]. It is therefore possible that point mutations in DR4 might be found in lung cancer.
We identified missense alterations in the DR4 ectodomain, extracellular cycteine-rich domain and death domain. The C626G polymorphism is the most extensively studied polymorphism in the literatures. In our study, the wild type (CC) was seen in 20 % of patients and 33,3 % of controls, whereas the CG and GG allelles seen in 56,7 % patients and 40 % controls, and 23,3 % patients and 26,7 % controls, respectively. The differences in both groups for C626G polymorphisms were not significant (p = 0,271). Fisher et al [3]. showed that two alterations (G422C and C626G) in ectodomain of DR4 occur an increased frequency in LC, HNSCC, and gastric adenocarsinoma samples. In another study with bladder cancer, no differences in genotype or haplotype distribution for C626G polymorphism in DR4 gene were found between patients with bladder cancer and controls in Spain [4]. Furthermore, Hazra et al [2]. showed an increased risk of C to G transition in 626 position of exon 4 of DR4 gene in bladder cancer. In Ovarian cancer, alteration of the DR4 gene including C626G do not lead to clinically relevant ovarian cancer predispositon [5]. Frank et al [10]. reported that the heterozygous carriers of DR4 626C > G showed a significant association with a decreased colorectal cancer risk. A meta-analysis by Chen et al [14]. indicated that the C626G polymorphism in DR4 gene is associated with cancer susceptibility.
The other missense alterations in the DR4 ectodomain is G422A transition. Like C626G, G422A is also the TRAIL binding region on DR4 receptor. Perhaps, this one or two alteration in 422 and 626 positions of exon 4 of DR4 gene may alter the TRAIL binding site on DR4 receptor and result in deficient apoptotic signaling. We are found this transition in 56,7 % heterozygous and 18,3 % homozygous in the patients, 40 % heterozygous and 26,7 % homozygous in the control group. Statistical analysis showed no differences in the two groups (p = 0,324). It was reported by Fisher et al [3]. that the G422A polymorphism was found more frequently in some primary tumors, including LC and head and neck cancer. Moreover, Horak et al [5]. demonstrated no association between G422A polymorphism and ovarian cancer.
The A683C polymorphism is extensively studied, but to a lesser extent than the C626G polymorphism. The A683C polymorphism are located in the extracellular cycteine-rich domain of DR4. An evidence-based meta-analysis by Chen et al [14]. showed an association between an increased cancer risk and A683C polymorphism in different genetic models. Especially the AC and CC variants were associated with increased risk of all types of cancer. In our study, the AC variant was seen in 19 (31,7 %) patients and 7 (23,3 %) controls, whereas the CC variant seen in only 2 controls (p = 0,086). The CC variant may be the protective variant in our study population.
The A1322G polymorphism is located in the death domain of DR4. This domain is responsible from the transmission of the apoptotic signal to the TRADD (TNFR-associated death domain) and induct the apoptosis. Chen et al [14]. reported that the AG and GG variant genotypes in 1322 position of exon 10 of DR4 were associated with marginally and statistically significant increased risk of all types of cancer. In our study, most of our patients and controls (88,3 % and 80 % respectively) were AA in 1322 position. Only 7 patients and 6 controls had AG and GG variants at this position (p = 0,230). In a study with ovarian cancer, the AA variant was seen most frequently like our study, and the did not find statistically significant [5]. Martinez-Ferrandis et al [4]. were also found no association between A1322G polymorphism and breast cancer in Spanish women, whereas the DR4 death domain A1322G polymorphism was significantly more frequent in Mantle Cell Lymphoma and chronic lymphocytic leukemia patients than in a sex and age-adjusted healthy population [15].
To summarize, our investigation on DR4 gen polymorphisms, including G422A, C626G, A683C and A1322G, showed no statistically significances between patients with LC and controls. Most of genes, had a role on apoptotic pathway, must be researched and the study population must be kept great.
References
Dechant MJ, Fellenberg J, Scheuerpflug CG, Ewerbeck V, Debatin K-M (2004) Mutation Analysis of the apoptotic “death-receptors” and the adaptors TRADD and FADD/MORT-1 in osteosarcoma tumor samples and osteosarcoma cell lines. Int J Cancer 109:661–667
Hazra A, Chamberlain RM, Grossman HB, Zhu Y, Spitz MR, Wu X (2003) Death receptor 4 and bladder cancer risk. Cancer Res 63:1157–1159
Fisher MJ, Virmani AK, Wu L, Aplenc R, Harper JC, Powell SM, Rebbeck TR, Sidransky D, Gazdar AF, El-Deiry WS (2001) Nucleotide substitution in the ectodomain of TRAIL receptor DR4 is associated with lung cancer and head and neck cancer. Clin Cancer Res 7:1688–1697
Martinez-Ferrandis JI, Rodriguez-Lopez R, Milne RL, Gonzalez E, Cebolla E, Chirivella I, Zamora P, Arias JI, Palacios S, Cervantes A, Diez O, Benitez J, Armengod ME (2007) Polymorphisms in TRAIL receptor genes and risk of breast cancer in Spanish women. Cancer Biomark 3(2):89–93
Horak P, Pils D, Roessler M, Tomek S, Elandt K, Zeillinger R, Zielinski C, Krainer M (2005) Common death receptor 4 (DR4) polymorphisms do not predispose to ovarian cancer. Gynecol Oncol 97:514–518
Seitz S, Wassmuth P, Fischer J, Nothnagel A, Jandrig B, Schlag PM, Scherneck S (2002) Mutation analysis and mRNA expression of trail-receptors in human breast cancer. Int J Cancer 102:117–128
Lee SH, Shin MS, Kim HS, Lee HK, Park WS, Kim SY, Lee JH, Han SY, Park JY, Oh RR, Kang CS, Kim KM, Jang JJ, Nam SW, Lee JY, Yoo NJ (2001) Somatic mutations of TRAIL-receptor 1 and TRAIL-receptor 2 genes in non-Hodgkin’s lymphoma. Oncogene 20(3):399–403
Ulybina YM, Kuligina ES, Mitiushkina NV, Rozanov ME, Ivantsov AO, Ponomariova DN, Togo AV, Levchenko EV, Shutkin VA, Brenister SI, Devilee P, Zhivotovsky B, Hirvonen A, Imyanitov EN (2009) Coding polymorphisms in Casp5, Casp8 and DR4 genes may play a role in predisposition to lung cancer. Cancer Lett 278(2):183–191
Wang M, Wang M, Cheng G, Zhang Z, Fu G, Zhang Z (2009) Genetic variants in the death receptor 4 gene contribute to susceptibility to bladder cancer. Mutat Res 661:85–92
Frank B, Shanmugam KS, Beckmann L, Hemminki K, Brenner H, Hoffmeister M, Chang-Claude J, Burwinkel B (2006) Death receptor 4 variants and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 15(10):2002–2005
Lu G, Bing X, Qi S, Chun L (2007) Association of DR4 gene polymorphism in Chinese patients with gastroduodenal diseases. Med J Wuhan Univ 28:93–95
Wolf S, Mertens D, Pscherer A, Schroeter P, Winkler D, Gröne HJ, Hofele C, Hemminki K, Kumar R, Steineck G, Döhner H, Stilgenbauer S, Lichter P (2006) Ala228 variant of trail receptor 1 affecting the ligand binding site is associated with chronic lymphocytic leukemia, mantle cell lumphoma, prostate cancer, head and neck squamous cell carcinoma and bladder cancer. Int J Cancer 118:1831–1835
Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O’Connell M, Kelley RF, Ashkenazi A, de Vos AM (1999) Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell 4:563–571
Chen B, Liu S, Wang X-L, Xu W, Li Y, Zhao W-H, Wu (2009) TRAIL-R1 polymorphisms and cancer susceptibility: an evidence-based meta-analysis. Eur J Cancer 45:2598–2605
Fernandez V, Jares P, Bea S, Salaverria I, Guino E, de Sanjose S, Colomer D, Ott G, Montserrat E, Campo E (2004) Frequent polymorphic changes but not mutations of TRAIL receptors DR4 and DR5 in mantle cell lymphoma and other B-cell lymphoid neoplasms. Haematologica 89:1322–1331
Acknowledgments
This work was funded, in part, by a grant from the University of Çukurova, Adana, Turkey (TF2005D1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Taştemir-Korkmaz, D., Demirhan, O., Kuleci, S. et al. There is no Significant Association Between Death Receptor 4 (DR4) Gene Polymorphisms and Lung Cancer in Turkish Population. Pathol. Oncol. Res. 19, 779–784 (2013). https://doi.org/10.1007/s12253-013-9643-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-013-9643-z