Abstract
Introduction
In pharmaceutical drug development and manufacturing, the amount and complexity of information of different types, ranging from raw experimental data to lab reports to complex mathematical models that needs to be stored, accessed, validated, manipulated, managed, and used for decision making is staggering. The information is often in different formats, used in different computer tools, making smooth interaction between these tools difficult. A common, explicit, and platform-independent vocabulary that is both machine accessible and human usable is needed to streamline the flow of information and knowledge generation.
Methods
The Purdue Ontology for Pharmaceutical Engineering (POPE) was developed to address this informatics challenge. POPE models information and knowledge and includes models of phases, material properties, molecular structures, experiments, reactions, and unit operations.
Conclusion
In Part 1, we describe the conceptual framework of POPE and in Part 2 its applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In pharmaceutical drug development and manufacturing, the amount and complexity of information of different types, ranging from raw experimental data to lab reports to complex mathematical models, that need to be stored, accessed, validated, manipulated, managed, and used for decision making is staggering. A tremendous amount of information is generated in the form of raw data from analytical instruments, images, spectra, lab notes, various calculations from simulation tools, chemometric models, etc. This information is often in different formats, such as plain text files, Word documents, Excel worksheets, JPEG files, MPEG movies, mathematical models, and so on. A typical FDA filing for a new drug approval requires many hundreds of thousands of pages of documentation of such data and information.
But it is not raw data that we are after. What we desire are in-depth knowledge and mechanistic, first-principles based, understanding of the underlying phenomena that can be modeled to aid us in rational decision making. However, knowledge extraction and model development from this data deluge are major challenges.
Decision making in pharmaceutical product development and manufacturing involves the integration of process modeling tools, effective use of laboratory-generated information, use of knowledge from the scientific literature, as well as development of technical specifications and an information-knowledge base to satisfy regulatory requirements.
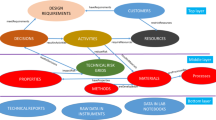
Current and past automation attempts to address various aspects of information management and decision-making (as shown in Fig. 1), such as expert systems [1–6] laboratory information management systems (LIMS) [7, 8], electronic lab notebooks [9], content management systems (CMS) [10], etc. They all have tried to address different slices of the overall problem—data, information, and knowledge management issues were addressed separately leading to stand-alone systems with limited capabilities and integration challenges. Data warehouses often become data graveyards, retrieving LIMS data for development and reporting activities is difficult, and Statistical Process Control (SPC) manufacturing data for trending, control, and decision making can be so challenging that it is drastically under used. Furthermore, little work has been done on supporting mathematical models development which is central to QbD and continual improvement.
To make this work, we need a systematic, integrated, informatics framework based on formal and explicit models of information [11]. In addition, we also need tools that would support rapid extraction of mechanistic, first principles, knowledge from raw data gathered from PAT-like techniques. The information models need to be accessed easily by humans and software tools, and should provide a common understanding for information sharing. Only with such a framework can intelligent model-based decision support systems be developed to assist in real-time decision-making for formulation design, scale-up, control, optimization, and operations.
In this paper, an ontology-based informatics infrastructure (shown in Fig. 2) which supports different activities by streamlining information gathering, data integration, model development and decision making is presented. The foundation of such an infrastructure is explicitly and formally modeled information, called an ontology.
Figure 2 shows the informatics infrastructure that integrates domain knowledge, stored in document repositories or relational databases, through a hardware integration layer (e.g., through a Local Area Network) and a ‘semantic’ or structured information layer, which models the information in a common format and provides a glossary. Multiple tools can then access this structured information and may be collected under a common presentation layer.
The rest of the paper is organized as follows. Ontologies, used as a common information model to describe the domain, are discussed in “Introduction to Ontologies” section. The ontology developed for the domain, the Purdue Ontology for Pharmaceutical Engineering (POPE) and its components are described in “The Purdue Ontology for Pharmaceutical Engineering” section. Applications that make use of POPE are briefly introduced in “Conclusions” section (with a detailed discussion in Part II of this paper).
Introduction to Ontologies
To describe information explicitly, the syntax as well as semantics for the information must be defined. The explicit description of domain concepts and relationships between these concepts is known as an ontology [12]. One of the definitions of ontology, given by Neches and colleagues [1] is: “An ontology defines the basic terms and relations comprising the vocabulary of a topic area as well as the rules of combining terms and relations to define extensions to the vocabulary.” For the pharmaceutical domain, the ‘basic terms’ could be a ‘material’ and a ‘material property’ and their relations could be ‘<material > has < material property>’. An example of a simple ontology is shown in Fig. 3 below.
Recent developments in the field of ontology have created new software capabilities that facilitate the implementation of the proposed informatics infrastructure. The shared understanding is the basis for a formal encoding of the important entities, attributes, processes, and their inter-relationships in the domain of interest. Ontologies can be used to describe the semantics of the information sources and make the contents explicit, thereby enabling integration of existing information repositories, either by standardizing terminology among the different users of the repositories, or by providing the semantic foundations for translators. Compared to a database schema which targets physical data independence, and an XML schema which targets document structure, an ontology targets agreed upon and explicit semantics of information. As a result, while the functionalities of this infrastructure can be implemented in a traditional client-server framework, the main benefits of this ontology-driven architecture are its openness and semantic richness.
As shown in Fig. 3, the powder flow rate (a material property) of the active pharmaceutical ingredient (API; a material) has an average value of 1 g/s within the range of (0.8, 1.2). The source of the reported value was the experiment ‘API Flow Measurement’ at a given context (78% relative humidity). The collection of the different concepts, e.g., material, material property, etc. and their relation, e.g., has value, comprise an ontology. An ontology defines a common vocabulary for researchers who need to share information in a domain. Ontologies may be thought of as the result of representation evolution proceeding through first order logic, semantic nets, and frames. Ontologies capture the class hierarchy and relationships; they also retain the relationships between the instances of those classes.
Developing an ontology involves defining classes (concepts) in the ontology, arranging the classes in a hierarchy, defining slots (relations), and describing allowed values for these slots and filling in the values for slots for instances. In ontology development, the major steps are determining the scope of the ontology, review (if any) of existing ontologies for possible reuse/integration, enumeration of the important concepts in the domain, definition of the hierarchy of concepts (top-down or bottom-up), and definition of the internal structure of the concepts (slots). The last step is creating individual instances of classes in the hierarchy by creating an individual instance of a given class and filling in the slot values. Classes and slots are inter-related and considered to be the most important steps in building the ontology. The inheritance property of classes allows for significant savings in effort. The developed ontologies were evaluated for consistency, completeness, conciseness, expandability, and robustness to changes [2]. The Web Ontology Language [3] was selected for the modeling of ontologies because of its web accessibility and reasoning tools. For further details on the ontology development process, the reader is referred to Venkatasubramanian et al. [4].
The Purdue Ontology for Pharmaceutical Engineering
The Purdue Ontology for Pharmaceutical Engineering is the first comprehensive attempt in developing an ontology to support decision making in pharmaceutical products development and manufacturing. The ontology is centered on the concepts of materials, experiments and properties and builds on our previous work [4]. Through this ontology, several functions that are difficult to perform like complicated semantic searches, association storage, and reasoning are made possible.
The Purdue Ontology for Pharmaceutical Engineering includes several components as shown in Fig. 4. The expert knowledge is modeled in the form of guidelines in the ontological infrastructure. A guideline models procedural knowledge, which consists of decision logic, information look-up, evaluation of decision variables and provision of recommendations. These components are captured in the POPE ontology [4]. The POPE ontology also describes mathematical knowledge, which consists of the mathematical equations as well as the underlying assumptions on the phenomenon. This separates the declarative and procedural components of mathematical models creation, manipulation and solution [5]. The declarative part consisted of two main ontologies, one which represents the details of a model (model definition) such as the model equations and state variables, and the other which represents the details of its use in modeling a specific processing step (model use).
The information ontologies, as shown in Fig. 4, consist of several categories, which are described below.
Material Ontology
There had been some work done to describe materials in an explicit manner. Stephanopoulos, Henning, and Leone [6] presented the Model. LA framework in which a material is defined to have a composition of components and phases. The Standard for the Exchange of Product (STEP) Data (ISO 10303) [7] included a representation of engineering product data modeled experimental, material, and chemical reaction data. Nielsen, Abildskov, Harper, Papaeconomou, and Gani [8] presented a structure in which compounds in a database were classified into categories including polar compounds, non-associating compounds, electrolytes and steroids. In the ontology defined by Batres, Aoyama, and Naka [9], a material is defined to have components with compositions defined as ‘component_in_mixture’ properties. FIX (physico-chemical ontology for biology) [10] included a classification of molecular matter by phases. Mixtures were divided into homogeneous and heterogeneous mixtures. Yang and Marquardt [13] presented OntoCAPE, which included descriptions of phases, chemical components and reactions. The Purdue Ontology for Material Entities (POME) builds on previous work as shown in Fig. 5. The material has two manifestations: one which is intrinsic and does not depend on conditions external to the material like temperature and pressure, called the substance; the other, dependent on the external conditions called the phase system. The intrinsic presence is described through the constitutional aspect. As uniqueness is required, the material can have, at most, one substance associated with it. For instance, the substance of water would be H2O. Substance includes atomic species like He, ionic species like H+, and polymeric species through the AtomContainer construct, which is described in the Purdue Ontology for Molecular Structure (POMS). The phase systems for H2O would be (liquid) water, ice, and steam and would include polymorphs for solids as they have different crystal structures and thus different material properties. A description of phase includes mention of the aggregation state, which is a mention of whether or not the given phase is a solid, liquid, or gas. In drug products (composed of multiple compounds), materials frequently have roles to play (what the material contributes to the drug product). These roles could include being an API, assisting flow (flow aid) among others [14].
Composition of a phase system is described at two levels: the composition of phases (phase composition) and the composition of compounds (substance composition) within each phase. Substance composition includes tuples of substance and concentration (which includes mass and mole fractions). Phase composition includes tuples of single-phase phase systems and a concentration description. Impurities were captured under this scheme as new substances. Blend uniformity was considered in the ontology for material properties. In addition, the substance has properties which include molecular mass and critical temperature, which are modeled further in the Purdue Ontology for Material Properties (POMP).
Molecular Structure Ontology
Ontologies have previously been developed for molecular structure. Fernandez-Lopez, Gomez-Perez, Pazos-Sierra and Pazos-Sierra [15] developed the Chemicals Ontology for the description of the periodic table by classifying the elements with descriptions of their physical properties. The EcoCyc Ontology [16] contains an ontology of compounds based on function (metabolite or not) and structure (alcohols, amines, aldehydes, acids, aromatics, and their derivatives). Murray-Rust, Rzepa, and Wright [17] presented the Chemical Markup Language (CML), which represented molecules in terms of a set of atoms and their spatial position. A possibility of parsing other molecular description formats was discussed. The FIX ontology [10] provided a description of compounds as atoms, molecules, ions, or radicals with further description of subatomic particles. Co-Ordination of Metals [18] represents the ontology for bioinorganic and other small molecule centers in complex proteins. Feldman, Dumontier, Ling, Haider, and Hogue [19] used a list of functional groups to classify compounds into a chemical ontology. Hsu, Krishnamurthy, Rao, Zhao, Jagannathan, Caruthers, and Venkatasubramanian [20] described molecules as atom containers, which consist of atoms and electrons. The atoms were further described by their position as ‘Atom_in_Ring’, ‘Carbon’, ‘Hydrogen’, etc. [21] presented ontologies for organic compound, reactions and reagents, the latter classified through their action. CML was used for the class descriptions. Villanueva-Rosales, and Dumontier [22] presented an ontology for functional groups which described molecules as having atom constituents connected through bonds. Chemical Entities of Biological Interest (ChEBI) included ontologies for describing molecular structure hierarchically, going from molecular structure to constituent atoms and subatomic particles to improve access to the ChEBI database [23].
In the pharmaceutical domain, Solomon, Wroe, Rogers, and Rector [24] developed a formal classification ontology for drug substances to support the drug knowledge database. Schuffenhauer, Zimmermann, Stoop, van der Vyver, Lecchini, and Jacoby [25] defined an ontology for pharmaceutical ligands to allow annotation-based searching of the database.
POMS (Purdue Ontology for Molecular Structures) builds on the above for the pharmaceutical domain by making use of common molecular fragments. These fragments, shown in Fig. 6, represent the set of atoms which participate in the chemical reaction and are derived from the set of most common drug degradation reactions [26, 27]. Molecular structures are represented in POMS as shown in Fig. 7. For instance, the molecular structure of cycloserine may be described as a collection of molecular fragments (amine, carbonyl, ether) as shown in Fig. 8.
Each fragment is part of a ‘fragment-entity’ which might participate in a reaction and is connected to (or identified as) a backbone group. This ontology can be coupled with the Reaction Ontology (PORE) to represent chemical systems and with POME to describe a material during product development.
Reaction Ontology
Some work had been done previously to describe chemical reactions. Gasteiger, Pförtner, Sitzmann, Höllering, Sacher, Kostka, and Karg [28] developed the Elaboration of Reactions for Organic Synthesis (EROS) system to model chemical reactions where a reactant could be made to react with every other reactant or with a select set, as defined through a reaction mode. Murray-Rust, Rzepa, and Wright [17] used XHTML tables to represent reactions as pictures of arrows, with information on reaction conditions, attached to the arrows. Angele, Moench, Oppermann, Staab, and Wenke [29] developed an ontology in which a reaction was described with respect to its participants (instances of a molecule class) and exist as part of a mixture. Borodina, Sadym, Filimonov, Blinova, Dmitriev, and Poroikov [30] suggested a representation for biomolecular transformation as a tuple of (X, reaction), with optional description of the enzyme. Hsu, Krishnamurthy, Rao, Zhao, Jagannathan, Caruthers, and Venkatasubramanian [22] modeled a reaction to have reactants, products and catalysts. Reactants and products were considered to be atom containers, which in turn consist of atoms and electrons. Sankar and Aghila [23] represented a chemical reaction as a set including a substrate, attacking reagent, transition state, and products in CML The authors used chemical relations such as “is isomeric with” and “reacts to form” to capture additional information.
PORE (Purdue Ontology for Reaction Engineering) was developed, based on previous work, to represent reactions as interactions between functional groups/phase systems as shown in Fig. 9. A change in the substance identity, e.g., polymerization is a chemical reaction while change in phase system, e.g., boiling is considered in physical reactions. Each reaction would have a physical context, which describes the pertinent descriptors of the reaction, e.g., at what temperature it occurs, at what pressure, pH, etc. Several restrictions such as the requirement of at least one reactant and one product for a reaction were put in place. Properties like the enthalpy of reaction would be computed elsewhere (POPE-Mm) and are currently outside the scope of the discussion.
Property Ontology
Previous work on explicit modeling of material properties includes Model-LA [8], STEP [9], CAPEC [10], and OntoCAPE [15]. POMP (Purdue Ontology for Material Properties) extends the properties in OntoCAPE to include inter-property relations and solid material properties as shown in Figs. 10, 11, and 12. The property structure includes generic properties like heat, mass, and momentum transfer properties (e.g., heat capacity, diffusivity, and density, respectively) as well as a separate description for solid properties. Solid properties were described at three levels; substance properties (pertaining to the molecular level, e.g., molecular structure), particle properties (pertaining to single crystals or amorphous particles, e.g., unit cell dimensions) and powder (bulk) properties (e.g., particle size distribution). Each property value would be correlated to a set of environmental conditions during measurement (e.g., temperature, pressure) and a source (experiment, mathematical model, or literature).
A property would have a value, reported for a given set of other material properties and physical parameters. An example would be the bulk density of a powder, which is dependent on particle size distribution (a material property as shown in Fig. 11) and the relative humidity of the air (physical parameter). These relations capture the dependencies in a qualitative manner; a mathematical relationship would be captured by the ‘mathematical’ model as a source of the property value. The list of properties used to develop the ontology is shown in Fig. 12 and spans properties of particular concern to pharmaceutical processing, like the Bonding Index and generic properties like specific heat capacities.
Experiment Ontology
Previous ontologies for experiments include the Experimental Molecular Biology ontology [31] for the representation of molecular biology experiments. Pouchard, Rana, and Walker [32] presented an experiment ontology which included a description of the start and end times, experiment requestors, instrument types and approved operating conditions. In the STEP data model [9], experimental data was defined to include data entry, data quality, data source and the data, which might be raw or smoothed. Hughes, Mills, de Roure, Frey, Moreau, Schraefel et al. [33] developed a laboratory ontology which captured the relationship between materials and processes, which involved a hierarchy of actions like mix, separate, etc. [33] presented EXPO, a generic ontology in which experiments are defined be either physical or computational, have a goal, belong to an experiment classification hierarchy and include administrative information. In addition, there are descriptive languages based on XML like mzXML [34] for mass spectrometry, the Generalized Analytical Markup Language [33] and the Joint Committee on Atomic and Molecular Physical Data Exchange data exchange format for plots and tabular data [35] While ontologies and data representations have been developed for a wide range of experiments, none of the above applications are directly applicable in the pharmaceutical product development domain, which requires a framework that can adequately describe not only experiments but material properties and chemical reactions in a semantically rich manner. The Purdue Ontology for Description of Experiments (PODE) was developed to address this need (Fig. 13).
The description of experiments includes generic descriptors like the time and place of the experiment as well as the identity of the people who performed the experiment. The equipment and procedure would, however, vary between different experiments. Equipments are described in the Purdue Ontology for Characterization of Equipment (POCE). Two levels of procedures were defined: an overarching Experimental Procedure (which may take the form: operate equipment 1, operate equipment 2, etc.) and the Experimental Equipment Procedure which is specific to the equipment. The former describes the sequence at which the equipments are to be used, while the latter describe how equipment is used. In general, the Experimental Procedure changes with the property measured while the Experiment Equipment Procedure is expected to stay relatively constant. Both procedures were modeled as a collection of actions, which could be observation/measurement actions, processing actions (e.g., mix, separate) or operation actions. These actions may occur in series, in parallel or as part of a ‘cluster’, e.g., heat while mixing. The interrelations between adjacent actions are described by precedence (predecessor, successor) or conditionality (starting, ending, and failure conditions). ‘Process’ actions describe unit operations and would thus be linked to instances of the Purdue Ontology for the Description of Unit Operations (PODUP). The connection between pieces of equipment was captured through equipment adjacency. Each piece of equipment has a setting that is specific to the data collection made.
Unit Operations Ontology
There have been several data models developed for unit operations. In the ISO 10303 formalism [9], a unit operation is defined by the process description and stream data, which includes material information, and port information. Model.LA [16] included descriptions of a generic unit, a port and streams which are associated with ports. In the Multidimensional Design Framework developed by Batres, Lu and Naka [36], each unit has structural and behavioral aspects linked to physical units (structural aspect) or mathematical models (behavioral aspect). The Conceptual Lifecycle Process Model (CLiP) developed by Bayer, Krobb and Marquardt [35], involved the description of a chemical process system that included a unit operation, process ports and process states. The OntoCAPE ontology [15] includes a description of unit operations in a hierarchy and use of ports to describe streams and also distinguishes between the behavioral and structural aspects of the unit operation.
PODUP builds on these models as shown in Fig. 14. Each unit operation is considered to occur in equipment and involves at least one inlet and/or outlet stream. The unit operation (involving mass/heat/momentum transfer) may be expressed as a ‘reaction’ (e.g., evaporation) involving the inlet and outlet streams. The streams are characterized by terminal ports, a phase system that is ‘carried’ (and described using POME) and a flow rate associated with the stream. Here, the ports may be physical (actual opening in the vessel) or virtual (walls through which heat is transferred, e.g., boiling in a vessel).
Equipment Ontology
Equipments are used for both unit operations and experiments and thus are defined separately for consistency. There has been previous work on equipment ontologies. In the CLiP data model [35], an equipment may be a fixture or a plant item which can be an apparatus (e.g., a column, heat exchanger) or machine (pump). Pouchard, Rana and Walker [34] presented an equipment ontology which included a description of availability, required training, location, and equipment settings. Sunagawa, Kozaki, Kitamura, and Mizoguchi [37] described an equipment ontology in which each component is a conduit with a flow of heat, mass or information. Ansaldi, Bragatto, Camossi, Giannini, Monti, and Pittiglio [38] presented an ontology for pressure equipment, where a vessel has subclasses of vessels with and without a stable volume. In the ontology developed by Lohse, Hirani, and Ratchev [39], the equipment could be a system with subcomponents and interface relationships with the ports.
The POCE builds on the ideas described above. Equipment are classified into actuating (for control purposes), analytical, flow, processing, storage equipment as well as fixtures used for structural support as shown in Fig. 15. Equipment would have specifications including dimensional (e.g., vessel volume), material of construction and safety specifications as well as settings for experiments and/or unit operations (e.g., solvent flow-rate in HPLC).
Value Ontology
The description of value (either of material properties or environmental conditions), is a central component of POPE. The major types of ‘value’ modeled are single values with units (e.g., atomic number), range of values (e.g., angle of repose), list (e.g., crystal system), table (e.g., fractional coordinates of atoms in a crystal) or pictures. There is some precedent for the development of a numerical value ontology. Gruber and Olsen [40] defined the EngMath ontology where a physical quantity is defined as a constant or a function quantity with physical dimensions. The Verfahrenstechnisches Datenmodell [41] includes a description of value as a tensor value also linked to a scalar variable linked to a reference. The STEP data model [9] defined lower, nominal, and upper values alongside data accuracy and standard deviation. Lam, Li and Xu [41] presented a model for an equation element as a matrix. There have been table ontologies developed by Olajide [39] and Embley, Hurst, Lopresti and Nagy [41].
The Purdue Ontology for Value Description (POVDE) includes descriptions of values, physical context like temperature, pressure, etc. (which are used in POME, PORE, POMP, PODE, PODUP) ontology for physical dimensions and an ontology for documents, including related documents, related concepts and author similar to those developed for clinical documents [42] and for organizational documents [43] as shown in Fig. 16. In POVDE, single values were treated as a tuple of numerical and string fields. Ranges were defined as tuples of Single values and tables ordered sets of cells containing single values or ranges. Pictures are represented through URLs to the respective files. Previously developed ontologies (EngMath [39], UnitDim [44]) were adapted for POVDE. In both approaches, a set of base units (describing the fundamental measures length, mass, time, electric current, temperature, amount of substance, and luminous intensity) were used to build composite units that relate the base units through multiplication and exponential relations.
Conclusions
The POPE was developed to assist pharmaceutical product development by providing an explicit model for information exchange. This is the first comprehensive informatics system developed to address the needs and challenges in the pharmaceutical domain. It lays down the conceptual foundations for ontological informatics in the pharmaceutical domain. POPE includes components for the description of information and knowledge in both guideline and mathematical model forms. The information description component of POPE (POPE-Im) includes descriptions of materials, molecular structure, reactions, properties, experiments, unit operations, and equipment. POPE is expected to provide a common information template for data, information, knowledge, and tool integration as well as information processing for better pharmaceutical product development. It is hoped that future efforts can benefit from the POPE experience.
POPE has been used in four applications involving decision support for product formulation [4], unit operation model integration [4], reaction prediction, and experiment analysis. Formulation is the selection of a manufacturing route, set, and amounts of appropriate excipients to be used in a drug product. The developed decision support system models the guidelines used for selection using the POPE-Km component and accesses the POPE-Im component for populating values of material properties like bulk density. Unit operation model integration involves the modeling of mathematical model knowledge in terms of the components of a mathematical model (variables, parameters, assumptions) and its use in POPE-Mm, which is connected to POPE-Im for material property data. The reaction prediction application deals with the modeling of molecular and reaction information (part of POPE-Im) in a manner that allows for semantic search and similarity comparison. Finally, the experiment analysis application makes use of experiment information modeling (POPE-Im) to compare experiments with respect to procedure, equipment settings, and data quality. The reaction prediction and experiment analysis applications are discussed in further detail in f Part II of this communication.
References
Gomez-Perez A, Fernandez-Lopez M, Corcho O. Ontological engineering: with examples from the areas of knowledge management, e-Commerce and the Semantic Web. London: Springer; 2004.
Gomez-Perez A. Evaluation of ontologies. Intl J Int Sys. 2001;16:391–409.
OWL (Web Ontology Language): http://www.w3c.org
Venkatasubramanian V, Zhao C, Joglekar G, Jain A, Hailemariam L, Suresh P, et al. Ontological informatics infrastructure for chemical product design and process development. Comp Chem Engg. 2006;30(10–12):1482–96.
Zhao C, Jain A, Hailemariam L, Suresh P, Akkisetty P, Joglekar G, et al. Towards intelligent decision support for pharmaceutical product development. J Ph Inn. 2006;1(1):23–35.
Stephanopoulos G, Henning G, Leome H. MODEL.LA. A language for process engineering. Part I and II. Comp Chem Engg. 1990;14(8):813–86.
Sousa P, Jardim-Goncëalves R, Pimentao JP, Pamieâ S-Teixeira J, Steiger-Garceao P. Seeking intelligent product development—an integrator environment based on STEP. J Int Man. 1999;10:313–21.
Nielsen TL, Abildskov J, Harper PM, Papaeconomou I, Gani R. The CAPEC database. J Chem Engg Data. 2001;46(5):1041–4.
Batres R, Aoyama A, Naka Y. A life-cycle approach for model reuse and exchange. Comp Chem Engg. 2002;26:487–98.
Degtyarenko K. Chemical Vocabularies and Ontologies for Bioinformatics. Proceedings of the 2003 International Chemical Information Conference. Nîmes, France. 2003;19–22:1–22
ClinicalTrials: http://clinicaltrials.gov
Gruber T. Toward principles for the design of ontologies used for knowledge sharing. Int J Human Comput Stud. 1993;43:907–928.
Yang A, Marquardt W. An ontology-based approach to conceptual process modeling. In: Barbarosa-Póvoa A, Matos H editors. Proceedings of the European symposium on computer-aided process engineering 14; 2004. pp. 1159–64
Fung KY, Ng KM. Product-centered processing: pharmaceutical tablets and capsules. AIChE J. 2003;49(5):1193–215.
Fernandez-Lopez M, Gomez-Perez AJ, Pazos-Sierra A. Building a chemical ontology using methontology and the ontology development environment. IEEE Int Sys. 1999;14(1):37–46.
EcoCyc: http://ecocyc.org/
Murray-Rust P, Rzepa HS, Wright M. Development of chemical markup language (CML) as a system for handling complex chemical content. New J Chem. 2001;25:618–34.
Degtyarenko K, Contrino S. COMe: the ontology of bioinorganic proteins. BioMed Cent Str Bio. 2004;4:1–10.
Feldman HJ, Dumontier M, Ling S, Haider N, Hogue CWV. CO: a chemical ontology for identification of functional groups and semantic comparison of small molecules. FEBS Lett. 2005;579:4685–91.
Hsu SH, Krishnamurthy B, Rao P, Zhao C, Jagannathan S, Caruthers J, et al. A systematic approach for automated reaction network generation. Marquardt W, Pantelides C (Eds) Proceedings of the 16th European Symposium on Computer-aided Process Engineering and 9th International Symposium on Process Systems Engineering. 2006; 973–8.
Sankar P, Aghila GJ. Design and development of chemical ontologies for reaction representation. J Chem Info Mod. 2006;46(6):2355–68.
Villanueva-Rosales N, Dumontier M. Describing chemical functional groups in OWL-DL for the classification of chemical compounds. OWL: experiences and directions Innsbruck, Austria; 2007.
Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nuc Acids Res. 2008;36:D344–50.
Solomon W, Wroe C, Rogers JE, Rector A. A reference terminology for drugs. J Am Med Inf Ass. 1999;152–5.
Schuffenhauer A, Zimmermann J, Stoop R, van der Vyver JJ, Lecchini S, Jacoby E. An ontology for pharmaceutical ligands and its application for in silico screening and library design. J Chem Inf Comp Sci. 2002;42:947–55.
March J. Advanced organic chemistry: reactions, mechanisms and structures. 4th ed. USA: Wiley; 1992.
Baertschi S. Pharmaceutical stress testing: predicting drug degradation. In: Drugs and the Pharmaceutical Sciences Vol. 153 Taylor and Francis Group, Boca Raton FL. 2005.
Gasteiger J, Pförtner M, Sitzmann M, Höllering R, Sacher O, Kostka T, et al. Computer-assisted synthesis and reaction planning in combinatorial chemistry. Persp Drug Disc Des. 2000;20:245–64.
Angele J, Moench E, Oppermann H, Staab S, Wenke D. Ontology-based query and answering in chemistry: ontonova @ project halo. Lect Notes Comp Sci. 2003;2870:913–28.
Borodina Y, Sadym A, Filimonov D, Blinova V, Dmitriev A, Poroikov V. Predicting biotransformation potential from molecular structure. J Chem Info Comp Sci. 2003;43:1636–46.
Noy N, Hafner C. Ontological foundations for experimental science knowledge bases. App Art Int. 2000;14:565–618.
Pouchard LC, Rana OF, Walker DW. An ontology for user support in the materials microcharacterization collaboratory. Proceedings of 5th International Conference on Autonomous Agents. Montreal, Canada. 2001.
Generalized Analytical Markup Language: http://www.gaml.org
Bayer B, Krobb C, Marquardt W. A data model for design data in chemical engineering/information models. Tech Rep. (LPT-2001-15). RWTH Aachen 2001
Cammack R, Fann Y, Lancashire RJ, Maher JP, Mcintyre PS, Morse R. CAMP-DX for electron magnetic resonance (EMR). Pure App Chem. 2006;78(3):613–31.
Batres R, Lu ML, Naka Y. A multidimensional design framework and its implementation in an engineering design environment. J Conc Engg 1999; 7(1)
Lam CP, Li H, Xu D. A model-centric approach for the management of model evolution in chemical process modeling. Comp Chem Engg. 2007;31:1633–62.
Ansaldi S, Bragatto P, Camossi E, Giannini F, Monti M, Pittiglio P. A knowledge-based tool for risk prevention on pressure equipments. Comp Aided Des App. 2006;3(1-4):99–108.
Olajide WO. An aid to convert spreadsheets to higher quality presentations. MS Thesis. Texas A & M University. 2004.
Gruber TR, Olsen GR. An ontology for engineering mathematics. In Proceedings Fourth International Conference on Principles of Knowledge Representation and Reasoning. Doyle J, Torasso P, Sandewall E. (eds) Stanford, CA 1994. pp 258–269.
Embley DW, Hurst M, Lopresti D, Nagy G. Table-processing paradigms: a research survey. Int J Doc Anal. 2006;8(2):66–86.
Frazier P, Rossi-Mori A, Dolin RH, Alschuler L, Huff SM. The creation of an ontology of clinical document names. Studies in Health Technology and Informatics In: Patel V, Rogers R, Haux R, editors. Amsterdam, The Netherlands; 2001. p. 84.
Slota R, Majewska M, Dziewierz M, Krawczyk K, Laclavik M, Balogh Z, et al. Ontology assisted access to document repositories in public sector organizations. Lect Notes Comp Sci. 2004;3019:700–5.
UnitDim: http://www.atoapps.nl/foodinformatics.
Haas LM, Schwarz PM, Koda P, Kotla E, Rice JE, Swope WC. DiscoveryLink, a system for integrated access to life sciences. IBM Sys J. 2001;40(2):489–510.
Neumann EK, Quan D. Biodash: a semantic web dashboard for drug development. Proc Pac Sym Biocomp. 2006;11:176–87.
Pedrioli PGA, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotech. 2004;22, 11:1459–66.
Acknowledgements
The work was done through the financial support of the Engineering Research Center for Structured Organic Particulate Systems (ERC-SOPS), the Indiana 21st Century Fund and Eli Lilly and Company. The authors thank Aktham Aburub, Pavan Akkisetty, Ahmad Almaya, Steven Baertschi, Arun Giridhar, Brian Good, Intan Hamdan, Gus Hartauer, Henry Havel, Shuo-Huan Hsu, Girish Joglekar, Balachandra Krishnamurthy, David Long, Prabir Basu, Kenneth Morris, Gintaras Reklaitis, Pradeep Suresh, and Chunhua Zhao for their input.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hailemariam, L., Venkatasubramanian, V. Purdue Ontology for Pharmaceutical Engineering: Part I. Conceptual Framework. J Pharm Innov 5, 88–99 (2010). https://doi.org/10.1007/s12247-010-9081-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12247-010-9081-3