Abstract
Japanese knotweed s.l. (Fallopia spp.) is a highly invasive clonal plant, best known from roadside and riparian habitats. Its expansion into beaches on Long Island, NY, USA, represents a major habitat shift. I surveyed populations from beaches and wetlands and conducted a common garden experiment to test for variation in drought tolerance and phenotype among populations and habitats. All populations were composed mostly of first- and later-generation hybrids. I found significant variation among populations in growth, lamina size, specific leaf area (SLA), and biomass allocation, in both the field and the common garden. Lamina size, growth, and root-to-shoot responded plastically to drought treatment. Wetland populations tolerated drought as well as beach populations. Differentiation in SLA between habitats suggests that some selection for beach genotypes may have occurred. It appears that both hybridization and phenotypic plasticity are contributing to the expansion of Fallopia spp. into novel habitat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Invasion by nonnative species represents a serious threat to the health of coastal ecosystems, on par with pollution, overharvesting, and rising sea level (Williams and Grosholz 2008). Coastal habitats have undergone dramatic changes as a result of invasions by nonnative plants such as hybrid cordgrass (Spartina spp.), giant reed (Phragmites australis), and European beach grass (Ammophila arenaria; Amsberry et al. 2000; Chambers et al. 1999; Daehler and Strong 1997; Minchinton et al. 2006; Wiedemann and Pickart 1996). In this study, I describe invasive populations of Japanese knotweed sensu lato (Fallopia spp.) from beach and wetland habitats on Long Island, NY, USA. Japanese knotweed is a vigorously growing, clonal, herbaceous perennial, native to eastern Asia, with known negative impacts on native plant and animal species (Maerz et al. 2005, Lecerf et al. 2007, Siemens and Blossey 2007, Topp et al. 2008). It is considered one of the most invasive plants in North America and Europe (Bailey 2003; Huebner et al. 2004; Pyšek et al. 2001). Invasion of coastal habitats by Japanese knotweed is particularly alarming because of its poor ability to prevent erosion in riparian zones (Tickner et al. 2001), which could translate into significant ecological impacts in coastal areas. Roadsides and riparian zones are considered typical habitat for invasive Japanese knotweed. Although Zika and Jacobson (2003) discussed coastal populations in the northwestern USA and Richards et al. (2008) recently described coastal salt marsh populations on Long Island, there are no previously published reports of Japanese knotweed on beaches in the eastern USA. A spread from mesic and poorly drained environments into very-well-drained beach environments represents a major habitat shift for Japanese knotweed. The ability to expand into novel habitat conditions raises general questions about the mechanisms that enable invasive species to succeed in nonnative ranges.
The establishment and spread of Japanese knotweed are classic examples of the paradox of invasive species: despite limited genetic variability due to population bottlenecks during establishment, invasive species not only overcome the deleterious effects of the bottleneck but are often more successful than locally adapted species (Allendorf and Lundquist 2003). Some researchers have suggested that the need to accumulate additive genetic variation for local adaptation is a prerequisite to invasion and is responsible for the commonly observed lag time between the establishment and spread of an invasive species (Ellstrand and Schierenbeck 2000; Lee 2002). Hybridization has been proposed as an important mechanism for generating heritable variation in invasive species (Ellstrand and Schierenbeck 2000; Galatowitsch et al. 1999; Mooney and Cleland 2001), and recent studies have demonstrated that novel combinations of traits or transgressive trait expression in hybrids can allow plants to expand into habitats that are inhospitable to the parent species (Facon et al. 2005; Fitzpatrick and Shaffer 2007; Johnston et al. 2001; Rieseberg et al. 1999, 2007; Schierenbeck and Ellstrand 2009). Alternatively, phenotypic plasticity may provide broad ecological tolerances and explain the ability of exotic species with low genetic variability to successfully invade new habitats (Baker 1965; Parker et al. 2003; Xu et al. 2003). While a number of studies have demonstrated the importance of broad tolerances and plasticity in the success of invasive species (Collyer et al. 2007; Dybdahl and Kane 2005; Funk 2008; Parker et al. 2003; Rapson and Wilson 1992; Richards et al. 2008; Schweitzer and Larson 1999; Xu et al. 2003), others have shown that rapid contemporary evolution may also contribute to the success of invasive plants (Blair and Wolfe 2004; Collyer et al. 2007; Lavergne and Molofsky 2007; Leger and Rice 2003; Maron et al. 2004; Wolfe et al. 2004). In this study, I used a combination of comparative and experimental approaches to evaluate the importance of adaptation and phenotypic plasticity in the invasion success of Japanese knotweed into a novel beach environment. Understanding the relative importance of these factors is essential for invasive species management, both for predicting how a species will respond to biological control and for predicting their potential ecological impact (Sakai et al. 2001; Stockwell et al. 2003; Williams and Grosholz 2008).
Japanese knotweed sensu lato includes the taxa Fallopia japonica (Houtt.) Ronse Decraene (synonyms Polygonum cuspidatum Sieb. and Zucc. and Reynoutria japonica Houttn.), Fallopia sachalinensis (F. Schmidt) Ronse Decraene, their hybrid Fallopia × bohemica (Chrtek and Chrtkova) J. Bailey, and any F2 or backcross offspring (Bailey et al. 2007, 2009). Although hybridization between F. japonica and F. sachalinensis is uncommon in their native range, native populations of the two species do not form two monophyletic groups and some history of interbreeding is likely (Inamura et al. 2000). The invasion history of Japanese knotweed in its introduced range has been thoroughly reviewed by Bailey et al. (2009). Japanese knotweed was introduced through horticulture to both Europe and North America in the mid-nineteenth century and was well established by the early twentieth century (Barney 2006; Bailey et al. 2009). While less is known about the introduction history and distribution of Japanese knotweed in North America, evidence suggests that it was similar to Europe (Barney et al. 2006, 2009; Barney 2006; Seiger and Merchant 1997). Japanese knotweed species are known to hybridize in both Europe and North America (Bailey et al. 2007; Hollingsworth et al. 1999; Mandak et al. 2005; Tiebre et al. 2007; Zika and Jacobson 2003). The high rates of hybridization in Europe may have been driven by the absence of male fertile individuals of F. japonica, but it is still unknown if male fertile F. japonica were introduced to North America (Bailey et al. 2009; Forman and Kesseli 2003). Both European and North American populations produce viable seeds that can establish in the wild (Bailey et al. 2009; Bram and McNair 2004; Forman and Kesseli 2003), but sexual reproduction appears more important in North America, perhaps because ecological conditions are more suitable for seedling establishment (Bailey et al. 2009). Recent molecular and morphological work has confirmed that Japanese knotweed populations in the northeastern USA can occur as mixed-taxon swarms, with a range of phenotypes that includes both parental forms and many intermediate forms (Gammon et al. 2007; Grimsby et al. 2007).
On one hand, the invasion history of Japanese knotweed suggests that broad ecological tolerances and phenotypic plasticity, as opposed to high genetic variability, are responsible for its invasion success. Vegetative reproduction is common in both Europe and in North America, and a striking lack of genetic diversity has been reported in European populations (Hollingsworth and Bailey 2000; Hollingsworth et al. 1998). Although there were multiple North American introductions, these introductions were largely from horticultural specimens, suggesting that North American populations may have been founded by a limited number of genotypes (Barney 2006; Barney et al. 2006; Seiger and Merchant 1997). On the other hand, sexual reproduction, including hybridization and introgression, has lead to increased genetic diversity in Europe (Bailey et al. 2009) and may have increased the genetic diversity of Japanese knotweed in North America (Bram and McNair 2004; Forman and Kesseli 2003; Gammon et al. 2007). This would allow populations to respond to varying environmental pressures through adaptive evolution. Although earlier studies reported fairly low levels of genetic variation for European populations of Japanese knotweed, these studies only measured neutral variation. Such neutral variation may be a poor indicator of heritable variation for ecologically important traits (McKay and Latta 2002). For example, in roadside and salt marsh populations of Japanese knotweed on Long Island, Richards et al. (2008) found considerable phenotypic differentiation based on traits measured in a common garden, despite limited genetic variation using neutral amplified fragment length polymorphism (AFLP) markers.
To determine if hybridization in North America may have allowed for genetic diversification and adaptive evolution of Japanese knotweed populations on Long Island, I carried out a greenhouse common garden experiment and field surveys to look for variation among populations. If there is phenotypic similarity among populations in both the field and the common environment, then broad tolerances are likely to be a key factor in Japanese knotweed invasion, with a limited contribution from phenotypic plasticity. In contrast, if populations are distinct from each other in the field but similar in the common environment, then plasticity has likely played a role in invasion success. Finally, if populations are phenotypically distinct in the common environment and the field, then genetic differentiation could have played a role in invasion success. However, population differentiation does not necessarily support the presence of adaptive evolution. Such variation can arise from neutral sources such as founder effects and genetic drift, so that individual populations still depend on plasticity and broad tolerances to survive (Keller and Taylor 2008).
To determine if there has been adaptive evolution, I compared populations from beach sites with populations from wetland sites that represent more typical Japanese knotweed habitat. Because the low soil water availability at sandy beach sites could impose a strong selective force for drought tolerance (Kellman and Roulet 1990; Maun 1994; Salisbury 1952), I carried out a greenhouse experiment to test for variation in leaf traits and drought tolerance (measured as growth and biomass allocation) among populations and habitats. In its native range, Japanese knotweed generally inhabits riparian zones or other mesic areas, but F. japonica is also a primary colonist of volcanic soils in Japan (Zhou et al. 2003). As a result, there may be preexisting genetic variation in invasive populations for traits that allow plants to tolerate extremely well-drained, nutrient-poor soils, such as the ratio of root-to-shoot biomass (root/shoot) or specific leaf area (SLA; Knight and Ackerly 2003; Mokany et al. 2006; Ordonez et al. 2009). I analyzed variation in leaf size among populations because leaf size is associated with moisture and nutrient availability (Dudley 1996; Wilf et al. 1998) and correlates with many other leaf functional traits (Dunbar-Co et al. 2009; Niinemets et al. 2007). Furthermore, leaf size is known to vary between the two parent species and take on intermediate values in hybrids (Gammon et al. 2007). This taxonomic variation, coupled with the functional significance of leaf size, suggests that leaf size variation could be important for adaptive evolution. Salt tolerance could also be important for success in beach habitats, but, despite significant variation among individuals, there appears to be little variation for salt tolerance among Long Island salt marsh and roadside populations of Japanese knotweed (Richards et al. 2008). Significant phenotypic differences between beach and wetland populations in a common garden or significantly higher drought tolerance in beach populations would support the hypothesis of adaptive evolution to the edaphic conditions found in beach habitats.
Materials and Methods
Field Sites and Study Organisms
This study encompasses eight different field sites in Suffolk County, Long Island, NY, USA (Table 1). I chose sites that were geographically dispersed throughout the county and represented the range of beach and wetland habitats found in this area. On Long Island, beach habitat consists of sites along the north shore, adjacent to Long Island Sound, represented by HLB, HPL, and PJB, and sites along the south shore adjacent to the Atlantic Ocean, represented by ANW. Along the north shore, Japanese knotweed grows in the backshore and in sand at the foot of a bluff, with some plants extending up the bluff. Japanese knotweed is less common on south shore beaches, where it grows in interdunal areas. Japanese knotweed grows in a variety of wetland habitats on Long Island, from which I sampled riparian zones (GCP and PJP), a low-lying area surrounded by forest (SNW) and a salt marsh (WNW). Although these wetland areas are ecologically diverse, they all have consistently wet soil, which should reduce any selection for drought tolerance. Some populations occurred as contiguous stands, while others were comprised of multiple stands. Riparian and beach populations were roughly linear, with the remaining populations covering a roughly rectangular area. The approximate area occupied by each population is given in Table 1. Unfortunately, there are no historical records to indicate the age of these populations. I originally selected three beach (ANW, HPL, and PJB) and three wetland (GCP, SNW, and WNW) populations for the field and greenhouse studies. Two of the original sites, one wetland (SNW) and one beach (ANW), were mowed during the summer and were inappropriate for the collection of phenotypic data. They were replaced for field study by two additional sites (PJP and HLB).
At each site, I collected soil from three randomly selected locations directly under Japanese knotweed plants, from 10 cm below the surface. I measured gravimetric water content, the ratio of the mass of water in the soil to the mass of dry soil. I measured water content at the original six sites 1 day after rainfall and at all eight sites 1 week after rainfall. I compared gravimetric water content among sites and between habitats using the GLM procedure in SAS version 9.2 (SAS Institute Inc., Cary, North Carolina, USA), testing the effect of site nested within habitat [site (habitat)] over the error sum of squares mean square error (MSE) and the effect of habitat over the site (habitat) MSE.
I used morphological characteristics to identify the taxa at each field site (Bailey et al. 1995; Gammon et al. 2007; Zika and Jacobson 2003). This method has been used to identify Japanese knotweed taxa in Europe and North American and produces classifications that are consistent with molecular markers for each taxon (Bailey et al. 1995; Gammon et al. 2007). I only scored trait values in the range for F. japonica and F. × bohemica because I never observed F. sachalinensis during numerous surveys of potential field sites (personal observation), despite reports of its occurrence on Long Island (Weldy et al. 2005). I scored leaves from eight to ten plants at each site for length from base to tip (1:<18 cm, 2:18–20, 3: >20 cm), trichome size (1: glabrous, 2: scabrous or tuberculate, 3: muricate or palpillate), shape of base (1: acuminate, 2: truncate, 3: cordate), and shape of tip (1: cuspidate, 2: intermediate, 3: accuminate). In each case, the first character is indicative of F. japonica and the third character is indicative of F. × bohemica. A score of 2 for trichome size or shape of base is consistent with F. japonica, while a score of 2 for length or shape of tip is ambiguous. If all four characteristics for a leaf were consistent with one taxon, the plant was assigned to that taxon; otherwise, it was considered ambiguous. I used logistic regression to determine if there was a significant difference in the taxonomic composition among sites (Fit Model module in JMP v.5.1, SAS Institute Inc., Cary, NC, USA). Since there were no differences in taxonomic composition among populations (see “Results”), I did not consider taxon as an effect in other analyses.
Field Measurements
I measured stem density, aboveground biomass, lamina area, and SLA (lamina area divided by dry mass) at three wetland sites and three beach sites. Each site was harvested on a separate day during the last 2 weeks of August 2003, with the harvest dates chosen randomly. At each site, I randomly chose five 0.5 × 0.5-m plots and counted the number of stems per plot. This number was multiplied by four to get the stem density in stems per square meter. From the same five plots, I harvested all aboveground biomass. I sorted the biomass into support tissue (stems and petioles), laminas, and reproductive tissue. Reproductive tissue included the entire inflorescence. I dried all tissue to constant weight at 60°C and weighed it. To calculate lamina area and SLA, I collected approximately 50 sun-exposed leaves from each study site, using only leaves of the same developmental stage (seventh or eighth leaf from the tip of the stem). Each leaf was collected from a separate stem separated by at least 1 m. Since nearby stems may be ramets of the same genotype, this sampling method may have led to some pseudoreplication, so P values are interpreted conservatively. I scanned fresh leaves using a Canoscan 8000F flatbed scanner (Cannon USA Inc., Lake Success, NY, USA) and calculated lamina area using Scion Image Software (Scion Corporation, Frederick, MD, USA). After scanning, I dried leaves at 60°C, weighed them, and calculated SLA.
Greenhouse Experiment
I carried out a greenhouse experiment to examine the same traits I measured in the field under common environmental conditions, in order to determine if variation among sites had a genetic basis and to assess how plants responded to drought. I collected rhizomes from three beach sites and three wetland sites and grew the plants in a greenhouse at Stony Brook University in Stony Brook, NY, USA. At each site, I collected ten separate rhizome pieces from locations at least 10 m apart. I cut each rhizome into two pieces weighing 5 (±0.6) g and randomly assigned each half to control or dry treatment. I sprouted rhizomes in flats and then planted sprouted rhizome pieces into 4-l pots, in a mixture of six parts Turface (Profile Products, Buffalo Grove, IL, USA), two parts play sand, and one part vermiculite. Control plants were watered as needed to maintain the medium at field capacity. Dry-treatment plants were kept moist until the cuttings sprouted (6–12 day) and then watered only when the medium had dried out to at least 5 cm below the surface. Plants were arranged on the greenhouse bench in a random order. The rhizomes were planted on August 5, 2003 and maintained until harvest 8 weeks later.
On October 3, 2003, I collected two fully expanded leaves from each plant for analysis. Leaves were scanned, dried, and weighed, and the average lamina area and SLA of the two leaves per plant were calculated as described above. On October 5 and 6, 2003, I harvested the aboveground biomass of all greenhouse plants. Stems were cut at the soil level, and tissue was divided into leaves and stems. For the few plants that were flowering, reproductive tissue made up a very small part (<1%) of the plant biomass and was included with stem tissue. Leaves and stems were dried at 60°C and weighed. Pots were kept in a cool, dark location after harvest, and root tissue was harvested between October 7, 2003 and October 15, 2003. After rinsing with water, roots were dried and weighed as described for stems.
Data Analysis
Field Data
Since leaves were collected across whole sites rather than by plot, I analyzed leaf data and plot data separately. All transformations and exploratory statistical analyses were performed with JMP v. 5.1.2 (SAS Institute Inc., Cary, NC, USA). Lamina area and SLA were transformed with the Box–Cox algorithm to improve normality (Sokal and Rohlf 1995). The correlations between lamina area and SLA did not vary greatly or change signs between sites or habitats, suggesting that the assumption of multivariate normality was valid (Scheiner 2001). The data for stem density, stem mass, lamina mass, and reproductive biomass were transformed using the Box–Cox algorithm. Because field biomass data were collected on six separate days over the course of 2 weeks, with each site being harvested on a separate day, site may have been confounded with date of harvest. I used linear and quadratic regression to determine if there was a relationship between date of harvest and transformed biomass measurements. Only transformed reproductive biomass was significantly correlated with date of harvest (r = 0.50850, P = 0.004), since plants were in the reproductive phase and there was little vegetative growth during the period of harvest. To remove the effects of date of harvest on reproductive biomass, I used quadratic regression to fit the transformed reproductive biomass to the date of harvest and used the residuals from the regression in subsequent analyses. Untransformed stem and lamina biomass were strongly correlated (r > 0.95 for all sites), so I combined stem and lamina biomass into one measure (stem + leaf mass) by summing them and transforming the sum. I calculated the proportion of biomass in stems as the stem biomass divided by stem + leaf mass. This parameter evaluates investment in support versus assimilative tissue. I did not include reproductive biomass in the denominator because of the confounding effect of date. Both stem + leaf mass and proportion stem were transformed using the Box–Cox algorithm.
Greenhouse Data
For the greenhouse experiment, leaf data and biomass data were taken from the same plants and considered as one data set. There were strong correlations between the transformed stem, lamina, and root biomass (r > 0.85 for all contrasts for all sites), so I created a new variable total mass, by summing the stem, lamina, and root biomass. I calculated the proportion stem as the stem mass divided by the stem plus lamina mass. I calculated root/shoot as root biomass divided by stem and lamina biomass. Box–Cox transformations were applied as necessary to improve the normality of response variables.
Statistical Tests
For each data set (field leaf data, field plot data, and greenhouse data), I conducted a multivariate analysis of variance (MANOVA) using the GLM procedure in SAS version 9.2 (SAS Institute Inc., Cary, NC, USA). I conducted a univariate analysis of variance (ANOVA) for each response variable, for comparison with the MANOVA results. For field leaf data, I used lamina area and transformed SLA as response variables, with habitat and site (habitat) as factors. For the field plot data, I used stem density, stem + leaf mass, reproductive residuals, and proportion stem, with the transformations described above, as response variables, and habitat and site (habitat) as effects. For the greenhouse experiment, I used lamina area, SLA, total biomass, and root/shoot, with the transformations described above, as response variables and habitat, site (habitat), treatment, and two-way interactions between treatment and the other effects as effects. For both MANOVAs, the effect of site (habitat) was tested over the error sum of squares and cross-products (SSCP) matrix and the effect of habitat was tested over the site (habitat) SSCP matrix. Treatment and habitat × treatment were tested over the treatment × site (habitat) SSCP matrix, and treatment × site (habitat) was tested over the error SSCP matrix (Quinn and Keough 2002; Rencher 2002).
Results
Taxonomic Identification and Differences Among Field Sites
There were no differences in taxonomic composition among the populations or between habitats [site (habitat): Wald χ 2 = 4.3856, 8 df, P = 0.82; habitat: Wald χ 2 = 0.006, 2 df, P = 0.997]. Only two plants had all four characters consistent with F. japonica; 18% of the plants were scored as F. × bohemica, and the majority of the plants (79%) had a mix of characters from F. japonica and F. × bohemica, suggesting later-generation hybridization or introgression. Each population contained individuals with three to six different combinations of leaf characters. Since there is evidence for a molecular basis for these characters (Gammon et al. 2007), there appears to be multiple genotypes in each population. There do not appear to be any differences between beach and wetland populations in the presence of first- versus later-generation hybrids.
There were significant differences in gravimetric water content among sites, and gravimetric water content was significantly higher at wetland sites (F = 7.5355, 4 df, P = 0.003 for site (habitat) and F = 27.418, 1 df, P < 0.001 for habitat, for measurements taken the day after rainfall, F = 3.298, 6 df, P = 0.026 for site(habitat) and F = 11.982, 1 df, P = 0.003 for habitat, for measurements taken 1 week after rainfall). There are different degrees of freedom for the two tests because the samples taken the day after rainfall included only the original sites, while the samples taken 1 week after rainfall included the two additional sites (PJP and PJB, Table 1).
Field Data
Statistical output for all tests is listed in “Appendix.” The MANOVA on field leaf data revealed a highly significant effect of site (habitat) (P < 0.001) but no effect of habitat (P = 0.587). Since the P values for this and other tests are either very high or very low, it is unlikely that any potential pseudoreplication due to unknowingly resampling the same genotype influenced the results. Lamina area had a greater effect on the differences among sites than SLA, based on the standardized canonical coefficients (“Appendix” Tables 4 and 5). However, the univariate ANOVAs suggest that both lamina area and SLA were significantly different among sites (Fig. 1, Table 2, and “Appendix” Table 6). There was also a highly significant effect of site (habitat) (P = 0.004) but no effect of habitat (P = 0.300) in the field plot MANOVA (Fig. 2, “Appendix” Table 7). Stem + leaf mass and the residuals of reproductive mass had the greatest influence on the differences among sites, based on the standardized canonical coefficients (“Appendix” Table 8). The opposite signs of the canonical coefficients imply that they were negatively correlated across sites (Scheiner 2001) and suggest a tradeoff between vegetative and reproductive biomass. For this analysis, the univariate ANOVAs contrasted with the MANOVA results, with none of the response variables showing a significant effect of site (habitat) (Table 2 and "Appendix" Table 9). The ANOVA for stem + leaf mass indicated that aboveground biomass was higher in beach sites (Table 2, Fig. 2).
Greenhouse Data
Site (habitat) had a significant effect on the outcome of the greenhouse experiment (P < 0.001, “Appendix” Table 10). Consistent with the field data, habitat did not affect the response variables when they were analyzed together (P = 0.292). Although treatment did not have a significant multivariate effect (P = 0.176), there was a significant interaction between treatment and site (habitat) (P = 0.001), suggesting that there was variation among sites in response to the drought treatment (Fig. 3). Based on the standardized canonical coefficients, lamina area had the greatest impact on the difference among sites, and root/shoot had the greatest impact on the treatment × site(habitat) interaction (“Appendix” Table 11). All univariate ANOVAs were significant overall (“Appendix” Table 12). Site (habitat) had a significant effect on lamina area, SLA, and total biomass (Table 3). Although there was no significant multivariate effect of treatment or habitat in the MANOVA, in univariate tests, treatment had a significant effect on lamina area, total biomass, and root/shoot, and habitat had a significant effect on SLA. There was a significant univariate treatment × habitat interaction for lamina area. The treatment × site (habitat) interaction was not significant for any individual traits, despite its multivariate significance. As with the field data, significant P values were well below 0.05, so it is unlikely that pseudoreplication affected the results.
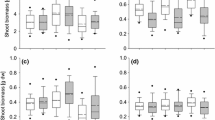
Reaction norms for a mean lamina area, b specific leaf area, c total mass, and d root/shoot, plus/minus SE, for Japanese knotweed grown from cuttings from six sites and two habitats, grown in the greenhouse under control or drought conditions (N ranged from 6 to 10, depending on the population and treatment). Filled symbols represent wetland populations and open symbols represent beach populations
Discussion
In a comparison of Japanese knotweed populations from beach and wetland habitats on Long Island, NY, USA, I found that all populations were composed almost entirely of first- and later-generation hybrids. Significant differences in the common garden provide evidence for genetic variation among populations in leaf morphology, growth, and biomass allocation patterns, and the significant treatment × site(habitat) interaction demonstrated that populations responded differently to imposed drought (“Appendix” Tables 10, 11, and 12). Despite the variation among populations in both the field and the greenhouse, the evidence for adaptation to beach or wetland habitats was weak. Habitat had no overall effect in any of the MANOVAs, indicating that both beach and wetland populations were equally sensitive to drought. There were significant univariate effects of habitat on several individual traits, suggesting that some differentiation according to habitat may have occurred. There was also considerable plasticity for ecologically important traits like lamina area, SLA, or root/shoot (Fig. 3). This may help Japanese knotweed establish and survive in beach habitats. The combination of plasticity and phenotypic variation is likely to increase the invasion potential of introduced populations via adaptive evolution.
Hybridization as a Source of Variation
Interspecific hybridization has long been recognized as a source of genetic diversity (Anderson 1949; Ellstrand and Schierenbeck 2000; Lewontin and Birch 1966), often resulting in a range of trait values for the offspring that exceeds that of the parent taxa (Burke and Arnold 2001; Parnell et al. 2008; Rieseberg et al. 1999). Extreme trait values or novel combinations of traits may allow hybrids to invade new habitat (Abbott 1992; Baumel et al. 2001; Facon et al. 2005; Lexer et al. 2003). For invasive species with low genetic diversity, hybridization can provide sufficient variation for adaptive evolution to proceed (Ellstrand and Schierenbeck 2000; O’Hanlon et al. 1999; Suehs et al. 2004). Previous studies of European populations of Japanese knotweed have reported very low genetic diversity (Hollingsworth and Bailey 2000). However, recent hybridization among the three species and F. × bohemica has led to increased genetic variation in Europe (Mandak et al. 2005; Pyšek et al. 2003; Tiebre et al. 2007). There is evidence of sexual reproduction and hybridization (including backcrosses) in American populations of Japanese knotweed, suggesting that genetic diversity could be higher in the USA (Bram and McNair 2004; Forman and Kesseli 2003; Gammon et al. 2007; Grimsby et al. 2007). In my survey of beach and wetland populations of Japanese knotweed, I found that all populations contained mostly individuals that were morphologically classified as F. × bohemica and either backcrosses or F2 hybrids (those scored as ambiguous), and even those scored as F. × bohemica could represent later-generation hybrids or backcrosses (Gammon et al. 2007). There were only two morphologically pure F. japonica genotypes. There was no evidence for transgressive trait expression in hybrid Japanese knotweed in the populations I surveyed or elsewhere in northeastern North America (Gammon et al. 2007). Instead, it appears that sexual reproduction and hybridization led to segregation of the variation present in the parent taxa, producing phenotypically variable offspring with intermediate forms. For example, F. japonica, F. × bohemica, and F. sachalinensis have small, medium, and large leaves, respectively (Gammon et al. 2007; Zika and Jacobson 2003), so hybrid offspring of these taxa should have highly variable leaf size. Consistent with this expectation, I found that lamina area was significantly different among populations both in the field and in the common garden (Tables 2 and 3). Since all populations were taxonomically similar, the differences among populations in this study were not due to differences in taxonomic composition but instead suggested idiosyncratic selection or founding events leading to a unique mix of genotypes in each population. Although I was careful to start with rhizome pieces of similar size for all populations, I cannot rule out maternal or other epigenetic effects that may have been carried over from the field sites (Rapp and Wendel 2005).
My experimental design did not allow for a statistical test of phenotypic or genetic variation among genotypes within each population. My observations suggest that it was present but limited. In the common environment, there were clear differences in leaf color, leaf shape, growth habit, flowering time, and other traits that made it easy to distinguish plants from different populations, and those traits did not vary much within populations. The significant effect of population on plant phenotype also requires lower within than among population variation. Nonetheless, there was some variation within populations, especially for root/shoot (see error bars in Fig. 3). Each population contained individuals with at least three different combinations of taxonomic traits, suggesting a minimum of three genotypes per populations. In their recent study of Long Island Japanese knotweed populations, Richards et al. (2008) found little or no AFLP variation within populations but still observed significant phenotypic variation among individuals within populations in a common garden, for traits such as biomass and succulence. Although I cannot rule out the possibility of having sampled multiple ramets from the same clone within each population, my sampling design was similar to Richards et al. (2008) and thus likely to have similar levels of intrapopulation variation.
Unlike this study, Richards et al. (2008) did not find significant differences among sites for plants grown in a common garden. However, they did report that adjacent Japanese knotweed populations had the same AFLP genotype regardless of habitat, while geographically separated populations had different AFLP genotypes. This is consistent with the pattern I found since all of my populations were geographically distant. The significant variation among but apparently limited variation within populations suggests that establishment by seed was probably important for the founding of each population, with both clonal spread and sexual reproduction contributing to population growth. Grimsby et al. (2007) recently found the same pattern in a survey of Massachusetts populations of Japanese knotweed, using simple sequence repeat markers.
In the common garden, I found significant variation among sites for lamina area, SLA, and total biomass. I found significant variation in reproductive biomass in the field, but I could not measure this trait in the greenhouse to determine if that variation was genetic. These traits illustrate the many ways that hybrid offspring can differ to provide the phenotypic and genetic variation on which selection could act. Traits such as lamina size and SLA have known functional significance and suggest that individual populations are functionally different from each other. Both within and among species, lamina size correlates with water availability, and SLA is an indicator of physiological functions such as photosynthesis and respiration (Dudley 1996; Reich et al. 1998; Wilf et al. 1998). Differences among populations in total biomass and reproductive biomass suggest a high potential for variation in reproductive output and fitness. If that variation is heritable, this could eventually lead to a shift in dominance of particularly fit genotypes. The functional variation present in Long Island Japanese knotweed suggests not only high potential for evolutionary change but also the possibility of variable impacts among populations. Since Japanese knotweed can dominate plant communities in the areas it invades, differences in functions like gas exchange or growth rate could translate into significant difference in ecosystem-level properties such as net primary production or transpiration.
Natural Selection and Response to Drought Treatment
The high variation among sites and the lack of any significant effect of habitat suggest that site-specific factors—either variable selection at each site or chance events during the establishment of each population—are driving the divergence of populations. Given the small population sizes and putatively young age of the populations, selection would have to have been very strong and occurred during initial establishment to result in such divergence. Selection of this sort has been demonstrated for other clonal plants. For example, North American coastal populations of Spartina patens displayed adaptive genetic divergence among adjacent stands on dunes, swales, and marsh sites (Silander 1979). Likewise, invasive populations of Spartina alterniflora on the Chinese coast contain molecular markers specific to each of three habitat types (Deng et al. 2007). Deng et al. suggest that selection acted on S. alterniflora seedlings during establishment and led to habitat specialization. Those studies found variation among habitats, however, and I found variation within each habitat type. Divergent natural selection for individual populations within a habitat could have occurred if there were divergent selection pressures among sites within each habitat. This is plausible for the wetland sites that included a salt marsh, a riparian zone, and a wet forest clearing but seems less likely for the beach sites. Beach sites were ecologically very similar yet showed as much among population variation as wetland sites. If there were habitat-based selection occurring at the population level, I would expect beach populations to show some convergence, at least in the phenotypes of plants growing in the field. Rather than site-specific selection, the pattern of high variation among and low variation within sites suggests that populations were founded by a few seeds and/or clonal propagules, followed by clonal spread within each population.
Despite the lack of an overall effect of habitat in either the field or the greenhouse, there were responses in some individual traits. In the greenhouse, SLA was significantly different among habitats (Table 3), suggesting that this trait may have diverged genetically between habitats. Two of the three beach populations had lower SLA than wetland populations when grown in the common environment (Fig. 3), and, although the effect was not significant, SLA of beach populations measured in the field was equal to or lower than that of all wetland populations (Fig. 2). Lower SLA, which indicates thicker or denser leaves, can confer drought tolerance because those leaves have a higher proportion of their mass in cell wall tissue and are able to withstand lower water potentials (Cano et al. 2008; Martinez et al. 2007; Mitchell et al. 2008). Lower SLA in beach habitats could also be a response to generally higher light levels (Evans and Poorter 2001) since the beach habitats were very open, while the wetland habitats (excluding the salt marsh) experienced partial shade. Nonetheless, the population with the highest SLA in the common environment was a beach population (ANW), so clearly there has been no uniform selection for lower SLA in beach environments.
The response to drought treatment in the greenhouse experiment suggests that both beach and wetland populations are equally sensitive to drought. Different populations responded differently to drought, as indicated by the significant treatment × site (habitat) interaction, but there was no overall habitat effect or treatment × habitat interaction (“Appendix” Table 10). All populations had reduced lamina area, lower total biomass, and increased root/shoot under drought (Fig. 3). There was a significant univariate habitat × treatment interaction for lamina area (Table 3), but this appears to be driven primarily by the responses of two populations. WNW, a wetland population, had an exceptionally large decrease in lamina area, while ANW, a beach population, had a smaller than average reduction (Fig. 3). Although this result is consistent with an increased sensitivity to drought in one wetland population and a decreased sensitivity in one beach population, most of the populations had almost identical changes in lamina area, regardless of their habitat. As with other traits in this study, it appears that any selective evolution for drought tolerance has occurred at the level of the population and not the habitat.
Although my results demonstrated that Japanese knotweed plants from beach and wetland sites are equally sensitive to drought when grown in pots, field-grown plants may have mechanisms that allow them to avoid drought stress rather than tolerate it. The lack of a difference in lamina area between beach and wetland populations in the field supports the idea that beach-grown plants do not need to tolerate drought because they are avoiding it. Field-grown plants have extensive rhizomes and large, succulent stems that may be used to buffer the plant from water loss during periods of rapid transpiration (Meinzer et al. 2001), whereas greenhouse-grown plants had no substantial rhizome development and limited stem growth during the course of the experiment. Leaf succulence could also be important since Richards et al. (2008) found significant variation for leaf succulence among individuals in a common garden. Plants at beach sites may also have deeper roots and greater belowground biomass than those at wetland sites, further helping field-grown beach plants to avoid drought. The greenhouse experiment suggests that there is no genetic variation among sites or habitats for root/shoot, but there was significant plasticity in this trait (see below), which could be important in the field.
The Role of Phenotypic Plasticity and Broad Ecological Tolerances
In contrast to genetic diversity and adaptive evolution, some invasions may succeed due to a preponderance of general-purpose genotypes (Baker 1965; Parker et al. 2003). Earlier selection may give rise to species or populations that have limited genetic variation but are able to tolerate a broad range of ecological conditions through phenotypic plasticity. Given that many invasive species enter their new range with low genetic diversity, plasticity can facilitate invasion by allowing organisms to tolerate harsh or novel conditions (Freeman and Byers 2006; Funk 2008; Muth and Pigliucci 2007; Parker et al. 2003; Richards et al. 2006). In the case of Japanese knotweed, phenotypic plasticity may facilitate the spread into coastal habitats since there is no evidence to adaptive evolution for this novel habitat. All populations showed plasticity in response to drought, although the amount was variable (Fig. 3). Some plastic responses to drought, such as reduced biomass and lamina area, arise because of the high sensitivity of stomatal conductance, cell elongation, and protein synthesis to low water potential (Lambers et al. 1998). This may reduce reproductive output but help plants survive under drought conditions. Greater sensitivity of aboveground than belowground tissue leads to an increased root/shoot and increases the ability to take up water relative to the amount of transpiring tissue (Lambers et al. 1998).
The phenotypic breadth of characters measured in the field may arise from genetic variation, or it could be an expression of phenotypic plasticity. There was a marginal effect of habitat on proportion stem in the field (P = 0.064, Table 2), with no effect in the greenhouse (P = 0.518 in a univariate ANOVA, data not shown). This suggests that beach plants may be able to plastically allocate less biomass to support tissue, possibly because of less competition for light. This characteristic would allow them to invest more of their resources in light capture, reproductive, or root tissue, increasing their overall resource use efficiency. There was a significant effect of habitat on stem + lamina mass (equal to aboveground biomass minus reproductive tissue) in the field, but no differences in total biomass in the greenhouse, either at the population or the habitat level (Tables 2 and 3). This suggests that the difference in biomass in the field is a plastic response. Since stem + lamina mass was higher in beach habitats than wetland habitats, beach habitat actually may be better for Japanese knotweed growth than wetland habitat. Plasticity in response to habitat variation may compliment the genetic variation arising from hybridization, with both characteristics contributing to invasion success.
Conclusions
The invasive species literature contains many examples of previously reproductively isolated species or populations that interbreed in their invasive range, leading to highly successful invasive populations, either through the generation of genetic novelty or fixed heterosis (Abbott et al. 2003; Bleeker 2003; Ellstrand and Schierenbeck 2000; Facon et al. 2005; Gaskin and Schaal 2002; Lexer et al. 2003; Pooler et al. 2002). Earlier studies of invasive Japanese knotweed reported evidence of low genetic diversity and clonal reproduction (Hollingsworth and Bailey 2000; Hollingsworth et al. 1998). While some particularly widespread genotypes continue to be found in both Europe and North America (Mandak et al. 2004; Pyšek et al. 2003; Richards et al. 2008), more and more studies have reported later-generation hybridization, sexual reproduction, and phenotypic diversity (Bailey et al. 2007; Bram and McNair 2004; Forman and Kesseli 2003; Gammon et al. 2007; Grimsby et al. 2007; Mandak et al. 2003; Richards et al. 2008; Tiebre et al. 2007). I have observed extensive seed production throughout Long Island and seedling establishment at many sites (personal observation). Whatever barriers to sexual reproduction were originally present, such as the lack of male fertile plants (Hollingsworth et al. 1998), those barriers appear to have been overcome. Despite the evidence for sexual reproduction, clonal spread continues to be important for Japanese knotweed invasion on Long Island, as evidenced by the presence of a single, widespread F. japonica genotype in both roadside and salt marsh habitats (Richards et al. 2008). Significant among-population variation in a common garden for ecologically important traits such as lamina size, SLA, and growth rate, and habitat differentiation for SLA suggest that sexual reproduction and adaptive evolution may be contributing to Japanese knotweed’s spread into beaches. Future studies involving reciprocal transplants and the measurement of correlations between traits and fitness in specific environments will help to clarify the importance of local adaptation in this system.
Phenotypic plasticity also seems to be playing an important role in Japanese knotweed invasion. Plasticity in traits like lamina size, root/shoot, and succulence may help Japanese knotweed to survive in habitats ranging from roadsides and riparian zones to salt marshes and beaches. Plasticity, coupled with sexual reproduction and the potential for adaptive evolution, could increase the already high rate of spread of Japanese knotweed in North America, particularly in coastal areas. Invasion of coastal ecosystems by nonnative plants can have significant impacts on ecosystem-level processes (Dukes and Mooney 2004; Grosholz et al. 2000; Levin et al. 2006; Windham 2001) and presents a serious challenge to managers of these systems. Information on the genetic and phenotypic makeup of invasive species can aid managers in assessing the threat of future spread and help them to develop appropriate control strategies.
References
Abbott, R.J. 1992. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends in Ecology & Evolution 7: 401–405.
Abbott, R.J., J.K. James, R.I. Milne, and A.C.M. Gillies. 2003. Plant introductions, hybridization and gene flow. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 358: 1123–1132.
Allendorf, F.W. and L.L. Lundquist. 2003. Introduction: Population biology, evolution, and control of invasive species. Conservation Biology 17: 24–30.
Amsberry, L., M.A. Baker, P.J. Ewanchuk, and M.D. Bertness. 2000. Clonal integration and the expansion of Phragmites australis. Ecological Applications 10: 1110–1118.
Anderson, E. 1949. Introgressive hybridization. New York: Wiley.
Bailey, J.P. 2003. Japanese knotweed s.l. at home and abroad. In Plant invasions: ecological threats and management solutions, ed. L.E. Child, J.H. Brock, G. Brundu, K. Prach, P. Pyšek, P.M. Wade, and M. Williamson. Leiden: Backhuys.
Bailey, J., L.E. Child, and M. Wade. 1995. Assessment of the genetics variation of British populations of Fallopia japonica and its hybrid Fallopia x bohemica. In Plant invasions: general aspects and special problems, ed. P. Pyšek, K. Prach, P. Rejmánek, and M. Wade, 141–150. Amsterdam: SPB Academic.
Bailey, J.P., K. Bimova, and B. Mandak. 2007. The potential role of polyploidy and hybridisation in the further evolution of the highly invasive Fallopia taxa in Europe. Ecological Research 22: 920–928.
Bailey, J.P., K. Bimova, and B. Mandak. 2009. Asexual spread versus sexual reproduction and evolution in Japanese knotweed s.l. sets the stage for the “Battle of the Clones”. Biological Invasions 11: 1189–1203.
Baker, H.G. 1965. Characteristics and modes of origins of weeds. In The genetics of colonizing species, ed. H.G. Baker and G.L. Stebbins, 147–168. New York: Academic.
Barney, J.N. 2006. North American history of two invasive plant species: phytogeographic distribution, dispersal vectors, and multiple introductions. Biological Invasions 8: 703–717.
Barney, J.N., N. Tharayil, A. DiTommaso, and P.C. Bhowmik. 2006. The biology of invasive alien plants in Canada. 5. Polygonum cuspidatum Sieb. & Zucc. [= Fallopia japonica (Houtt.) Ronse Decr.]. Canadian Journal of Plant Science 86: 887–905.
Baumel, A., M.L. Ainouche, and J.E. Levasseur. 2001. Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France). Molecular Ecology 10: 1689–1701.
Blair, A.C. and L.M. Wolfe. 2004. The evolution of an invasive plant: an experimental study with Silene latifolia. Ecology 85: 3035–3042.
Bleeker, W. 2003. Hybridization and Rorippa austriaca (Brassicaceae) invasion in Germany. Molecular Ecology 12: 1831–1841.
Bram, M.R. and J.N. McNair. 2004. Seed germinability and its seasonal onset of Japanese knotweed (Polygonum cuspidatum). Weed Science 52: 759–767.
Burke, J.M. and M.L. Arnold. 2001. Genetics and the fitness of hybrids. Annual Review of Genetics 35: 31–52.
Cano, L., J. Escarre, I. Fleck, J.M. Blanco-Moreno, and F.X. Sans. 2008. Increased fitness and plasticity of an invasive species in its introduced range: a study using Senecio pterophorus. Journal of Ecology 96: 468–476.
Chambers, R.M., L.A. Meyerson, and K. Saltonstall. 1999. Expansion of Phragmites australis into tidal wetlands of North America. Aquatic Botany 64: 261–273.
Collyer, M.L., C.A. Stockwell, C.A. Dean, and M.H. Reiser. 2007. Phenotypic plasticity and contemporary evolution in introduced populations: Evidence from translocated populations of white sands pupfish (Cyprinodon tularosa). Ecological Research 22: 902–910.
Daehler, C.C. and D.R. Strong. 1997. Hybridization between introduced smooth cordgrass (Spartina alterniflora; Poaceae) and native California cordgrass (S. foliosa) in San Francisco Bay, California, USA. American Journal of Botany 84: 607–611.
Deng, Z.F., S.Q. An, C.F. Zhou, Z.S. Wang, Y.B. Zhi, Y.J. Wang, S.H. Shi, et al. 2007. Genetic structure and habitat selection of the tall form Spartina alterniflora Loisel. in China. Hydrobiologia 583: 195–204.
Dudley, S.A. 1996. Differing selection on plant physiological traits in response to environmental water availability: a test of adaptive hypotheses. Evolution 50: 92–102.
Dukes, J.S. and H.A. Mooney. 2004. Disruption of ecosystem processes in western North America by invasive species. Revista Chilena De Historia Natural 77: 411–437.
Dunbar-Co, S., M.J. Sporck, and L. Sack. 2009. Leaf trait diversification and design in seven rare taxa of the Hawaiian Plantago radiation. International Journal of Plant Sciences 170: 61–75.
Dybdahl, M.F. and S.L. Kane. 2005. Adaptation vs. phenotypic plasticity in the success of a clonal invader. Ecology 86: 1592–1601.
Ellstrand, N.C. and K.A. Schierenbeck. 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the United States of America 97: 7043–7050.
Evans, J.R. and H. Poorter. 2001. Photosynthetic acclimation of plants to growth irradiance: the relative importance of specific leaf area and nitrogen partitioning in maximizing carbon gain. Plant Cell and Environment 24: 755–767.
Facon, B., P. Jarne, J.P. Pointier, and P. David. 2005. Hybridization and invasiveness in the freshwater snail Melanoides tuberculata: hybrid vigour is more important than increase in genetic variance. Journal of Evolutionary Biology 18: 524–535.
Fitzpatrick, B.M. and H.B. Shaffer. 2007. Hybrid vigor between native and introduced salamanders raises new challenges for conservation. Proceedings of the National Academy of Sciences of the United States of America 104: 15793–15798.
Forman, J. and R.V. Kesseli. 2003. Sexual reproduction in the invasive species Fallopia japonica (Polygonaceae). American Journal of Botany 90: 586–592.
Freeman, A.S. and J.E. Byers. 2006. Divergent induced responses to an invasive predator in marine mussel populations. Science 313: 831–833.
Funk, J.L. 2008. Differences in plasticity between invasive and native plants from a low resource environment. Journal of Ecology 96: 1162–1173.
Galatowitsch, S.M., N.O. Anderson, and P.D. Ascher. 1999. Invasiveness in wetland plants in temperate North America. Wetlands 19: 733–755.
Gammon, M.A., J.L. Grimsby, D. Tsfrelson, and R. Kesseli. 2007. Molecular and morphological evidence reveals introgression in swarms of the invasive taxa Fallopia japonica, F. sachalinensis, and F. x bohemica (Polygonaceae) in the United States. American Journal of Botany 94: 948–956.
Gaskin, J.F. and B.A. Schaal. 2002. Hybrid Tamarix widespread in US invasion and undetected in native Asian range. Proceedings of the National Academy of Sciences of the United States of America 99: 11256–11259.
Grimsby, J.L., D. Tsirelson, M.A. Gammon, and R. Kesseli. 2007. Genetic diversity and clonal vs. sexual reproduction in Fallopia spp. (Polygonaceae). American Journal of Botany 94: 957–964.
Grosholz, E.D., G.M. Ruiz, C.A. Dean, K.A. Shirley, J.L. Maron, and P.G. Connors. 2000. The impacts of a nonindigenous marine predator in a California bay. Ecology 81: 1206–1224.
Hollingsworth, M.L. and J.P. Bailey. 2000. Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese knotweed). Botanical Journal of the Linnean Society 133: 463–472.
Hollingsworth, M.L., P.M. Hollingsworth, G.I. Jenkins, J.P. Bailey, and C. Ferris. 1998. The use of molecular markers to study patterns of genotypic diversity in some invasive alien Fallopia spp. (Polygonaceae). Molecular Ecology 7: 1681–1691.
Hollingsworth, M.L., J.P. Bailey, P.M. Hollingsworth, and C. Ferris. 1999. Chloroplast DNA variation and hybridization between invasive populations of Japanese knotweed and giant knotweed (Fallopia, Polygonaceae). Botanical Journal of the Linnean Society 129: 139–154.
Huebner, C.D., C. Olson, and H.C. Smith. 2004. Invasive plants field and reference guide: An ecological perspective of plant invaders of forests and woodlands. Washington, DC: USDA Forest Service.
Inamura, A., Y. Ohashi, E. Sato, Y. Yoda, T. Masuzawa, M. Ito, and K. Yoshinaga. 2000. Intraspecific sequence variation of chloroplast DNA reflecting variety and geographical distribution of Polygonum cuspidatum (Polygonaceae) in Japan. Journal of Plant Research 113: 419–426.
Johnston, J.A., D.J. Grise, L.A. Donovan, and M.L. Arnold. 2001. Environment-dependent performance and fitness of Iris brevicaulis, I. fulva (Iridaceae), and hybrids. American Journal of Botany 88: 933–938.
Keller, S.R. and D.R. Taylor. 2008. History, chance and adaptation during biological invasion: Separating stochastic phenotypic evolution from response to selection. Ecology Letters 11: 852–866.
Kellman, M. and N. Roulet. 1990. Nutrient flux and retention in a tropical sand-dune succession. Journal of Ecology 78: 664–676.
Knight, C.A. and D.D. Ackerly. 2003. Evolution and plasticity of photosynthetic thermal tolerance, specific leaf area and leaf size: Congeneric species from desert and coastal environments. New Phytologist 160: 337–347.
Lambers, H., F.S. Chapin III, and T.L. Pons. 1998. Plant physiological ecology. New York: Springer.
Lavergne, S. and J. Molofsky. 2007. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences of the United States of America 104: 3883–3888.
Lecerf, A., D. Patfield, A. Boiche, M. P. Riipinen, E. Chauvet, and M. Dobson. 2007. Stream ecosystems respond to riparian invasion by Japanese knotweed (Fallopia japonica). Canadian Journal of Fisheries and Aquatic Sciences 64:1273–1283.
Lee, C.E. 2002. Evolutionary genetics of invasive species. Trends in Ecology & Evolution 17: 386–391.
Leger, E.A. and K.J. Rice. 2003. Invasive California poppies (Eschscholzia californica Cham.) grow larger than native individuals under reduced competition. Ecology Letters 6: 257–264.
Levin, L.A., C. Neira, and E.D. Grosholz. 2006. Invasive cordgrass modifies wetland trophic function. Ecology 87: 419–432.
Lewontin, R.C. and L.C. Birch. 1966. Hybridization as a source of variation for adaptation to new environments. Evolution 20: 315.
Lexer, C., M.E. Welch, O. Raymond, and L.H. Rieseberg. 2003. The origin of ecological divergence in Helianthus paradoxus (Asteraceae): Selection on transgressive characters in a novel hybrid habitat. Evolution 57: 1989–2000.
Maerz, J. C., B. Blossey, and V. Nuzzo. 2005. Green frogs show reduced foraging success in habitats invaded by Japanese knotweed. Biodiversity and Conservation 14:2901–2911.
Mandak, B., P. Pysek, M. Lysak, J. Suda, A. Krahulcova, and K. Bimova. 2003. Variation in DNA-ploidy levels of Reynoutria taxa in the Czech Republic. Annals of Botany 92: 265–272.
Mandak, B., P. Pyšek, and K. Bimova. 2004. History of the invasion and distribution of Reynoutria taxa in the Czech Republic: A hybrid spreading faster than its parents. Preslia 76: 15–64.
Mandak, B., K. Bimova, P. Pysek, J. Stepanek, and I. Plackova. 2005. Isoenzyme diversity in Reynoutria (Polygonaceae) taxa: Escape from sterility by hybridization. Plant Systematics and Evolution 253: 219–230.
Maron, J.L., M. Vila, R. Bommarco, S. Elmendorf, and P. Beardsley. 2004. Rapid evolution of an invasive plant. Ecological Monographs 74: 261–280.
Martinez, J.P., H. Silva, J.F. Ledent, and M. Pinto. 2007. Effect of drought stress on the osmotic adjustment, cell wall elasticity and cell volume of six cultivars of common beans (Phaseolus vulgaris L.). European Journal of Agronomy 26: 30–38.
Maun, M.A. 1994. Adaptations enhancing survival and establishment of seedlings on coastal dune systems. Vegetatio 111: 59–70.
McKay, J.K. and R.G. Latta. 2002. Adaptive population divergence: Markers, QTL and traits. Trends in Ecology & Evolution 17: 285–291.
Meinzer, F.C., M.J. Clearwater, and G. Goldstein. 2001. Water transport in trees: Current perspectives, new insights and some controversies. Environmental and Experimental Botany 45: 239–262.
Minchinton, T.E., J.C. Simpson, and M.D. Bertness. 2006. Mechanisms of exclusion of native coastal marsh plants by an invasive grass. Journal of Ecology 94: 342–354.
Mitchell, P., E. Veneklaas, H. Lambers, and S. Burgess. 2008. Using multiple trait associations to define hydraulic functional types in plant communities of south-western Australia. Oecologia 158: 385–397.
Mokany, K., R.J. Raison, and A.S. Prokushkin. 2006. Critical analysis of root: shoot ratios in terrestrial biomes. Global Change Biology 12: 84–96.
Mooney, H.A. and E.E. Cleland. 2001. The evolutionary impact of invasive species. Proceedings of the National Academy of Sciences of the United States of America 98: 5446–5451.
Muth, N.Z. and M. Pigliucci. 2007. Implementation of a novel framework for assessing species plasticity in biological invasions: responses of Centaurea and Crepis to phosphorus and water availability. Journal of Ecology 95: 1001–1013.
Niinemets, U., A. Portsmuth, D. Tena, M. Tobias, S. Matesanz, and F. Valladares. 2007. Do we underestimate the importance of leaf size in plant economics? Disproportional scaling of support costs within the spectrum of leaf physiognomy. Annals of Botany 100: 283–303.
O’Hanlon, P.C., R. Peakall, and D.T. Briese. 1999. Amplified fragment length polymorphism (AFLP) reveals introgression in weedy Onopordum thistles: Hybridization and invasion. Molecular Ecology 8: 1239–1246.
Ordonez, J.C., P.M. van Bodegom, J.P.M. Witte, I.J. Wright, P.B. Reich, and R. Aerts. 2009. A global study of relationships between leaf traits, climate and soil measures of nutrient fertility. Global Ecology and Biogeography 18: 137–149.
Parker, I.M., J. Rodriguez, and M.E. Loik. 2003. An evolutionary approach to understanding the biology of invasions: Local adaptation and general-purpose genotypes in the weed Verbascum thapsus. Conservation Biology 17: 59–72.
Parnell, N.F., C.D. Hulsey, and J.T. Streelman. 2008. Hybridization produces novelty when the mapping of form to function is many to one. BMC Evolutionary Biology 8: 122.
Pooler, M.R., R.L. Dix, and J. Feely. 2002. Interspecific hybridizations between the native bittersweet, Celastrus scandens, and the introduced invasive species, C. orbiculatus. Southeastern Naturalist 1: 69–76.
Pyšek, P., B. Mandak, Francirkova, and K. Prach. 2001. Persistence of stout clonal herbs as invaders in the landscape: A field test of historical records. In Plant invasions: Species ecology and ecosystem management, ed. G. Brundu, J. Brock, I. Camarda, L. Child, and M. Wade. Leiden: Backhuys.
Pyšek, P., J.H. Brock, K. Bimova, B. Mandak, V. Jarosik, I. Koukolikova, J. Pergl, et al. 2003. Vegetative regeneration in invasive Reynoutria (Polygonaceae) taxa: The determinant of invasibility at the genotype level. American Journal of Botany 90: 1487–1495.
Quinn, G.P. and M.J. Keough. 2002. Experimental design and data analysis for biologists. New York: Cambridge University Press.
Rapp, R.A. and J.F. Wendel. 2005. Epigenetics and plant evolution. New Phytologist 168: 81–91.
Rapson, G.L. and J.B. Wilson. 1992. Genecology of Agrostis capillaris L (Poaceae)—an invader into New Zealand. 2. Responses to light, soil fertility, and water availability. New Zealand Journal of Botany 30: 13–24.
Reich, P.B., M.B. Walters, D.S. Ellsworth, J.M. Vose, J.C. Volin, C. Gresham, and W.D. Bowman. 1998. Relationships of leaf dark respiration to leaf nitrogen, specific leaf area and leaf life-span: A test across biomes and functional groups. Oecologia 114: 471–482.
Rencher, A.C. 2002. Methods of multivariate analysis: Wiley series in probability and statistics. New York: Wiley-Interscience.
Richards, C.L., O. Bossdorf, N.Z. Muth, J. Gurevitch, and M. Pigliucci. 2006. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecology Letters 9: 981–993.
Richards, C.L., R.L. Walls, J.P. Bailey, R. Parameswaran, T. George, and M. Pigliucci. 2008. Plasticity in salt tolerance traits allows for invasion of novel habitat by Japanese knotweed s. l. (Fallopia japonica and F. bohemica, Polygonaceae). American Journal of Botany 95: 931–942.
Rieseberg, L.H., M.A. Archer, and R.K. Wayne. 1999. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372.
Rieseberg, L.H., S.C. Kim, R.A. Randell, K.D. Whitney, B.L. Gross, C. Lexer, and K. Clay. 2007. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129: 149–165.
Sakai, A.K., F.W. Allendorf, J.S. Holt, D.M. Lodge, J. Molofsky, K.A. With, S. Baughman, et al. 2001. The population biology of invasive species. Annual Review of Ecology and Systematics 32: 305–332.
Salisbury, E. 1952. Downs and dunes: Their plant life and environment. London: G. Bell and Sons.
Scheiner, S.M. 2001. MANOVA, multiple response variables and multispecies interactions. In Design and analysis of ecological experiments, ed. S.M. Scheiner and J.G. Gurevitch, 99–115. New York: Oxford University Press.
Schierenbeck, K.A. and N.C. Ellstrand. 2009. Hybridization and the evolution of invasiveness in plants and other organisms. Biological Invasions 11: 1093–1105.
Schweitzer, J.A. and K.C. Larson. 1999. Greater morphological plasticity of exotic honeysuckle species may make them better invaders than native species. Journal of the Torrey Botanical Society 126: 15–23.
Seiger, L.A. and H.C. Merchant. 1997. Mechanical control of Japanese knotweed (Fallopia japonica [Houtt.] Ronse Decraene): Effects of cutting regime on rhizomatous reserves. Natural Areas Journal 17: 341–345.
Siemens, T. J., and B. Blossey. 2007. An evaluation of mechanisms preventing growth and survival of two native species in invasive bohemian knotweed (Fallopia x bohemica, Polygonaceae). American Journal of Botany 94:776–783.
Silander, J.A. 1979. Micro-evolution and clone structure in Spartina patens. Science 203: 658–660.
Sokal, R.R. and F.J. Rohlf. 1995. Biometry. New York: Freeman.
Stockwell, C.A., A.P. Hendry, and M.T. Kinnison. 2003. Contemporary evolution meets conservation biology. Trends in Ecology & Evolution 18: 94–101.
Suehs, C.M., L. Affre, and F. Medail. 2004. Invasion dynamics of two alien Carpobrotus (Aizoaceae) taxa on a Mediterranean island: I. Genetic diversity and introgression. Heredity 92: 31–40.
Tickner, D.P., P.G. Angold, A.M. Gurnell, and J.O. Mountford. 2001. Riparian plant invasions: Hydrogeomorphological control and ecological impacts. Progress in Physical Geography 25: 22–52.
Tiebre, M.S., J.P. Bizoux, O.J. Hardy, J.P. Bailey, and G. Mahy. 2007. Hybridization and morphogenetic variation in the invasive alien Fallopia (Polygonaceae) complex in Belgium. American Journal of Botany 94: 1900–1910.
Topp, W., H. Kappes, and F. Rogers. 2008. Response of ground-dwelling beetle (Coleoptera) assemblages to giant knotweed (Reynoutria spp.) invasion. Biological Invasions 10:381–390.
Weldy, T., R. Mitchell, and R. Ingalls. 2005. New York flora atlas. Albany: New York Flora Association, New York State Museum.
Wiedemann, A.M. and A. Pickart. 1996. The Ammophila problem on the northwest coast of North America. Landscape and Urban Planning 34: 287–299.
Wilf, P., S.L. Wing, D.R. Greenwood, and C.L. Greenwood. 1998. Using fossil leaves as paleoprecipitation indicators: An Eocene example. Geology 26: 203–206.
Williams, S.L. and E.D. Grosholz. 2008. The invasive species challenge in estuarine and coastal environments: Marrying management and science. Estuaries and Coasts 31: 3–20.
Windham, L. 2001. Comparison of biomass production and decomposition between Phragmites australis (common reed) and Spartina patens (salt hay grass) in brackish tidal marshes of New Jersey, USA. Wetlands 21: 179–188.
Wolfe, L.M., J.A. Elzinga, and A. Biere. 2004. Increased susceptibility to enemies following introduction in the invasive plant Silene latifolia. Ecology Letters 7: 813–820.
Xu, C.Y., W.J. Zhang, C.Z. Fu, and B.R. Lu. 2003. Genetic diversity of alligator weed in China by RAPD analysis. Biodiversity and Conservation 12: 637–645.
Zhou, Z.H., M. Miwa, K. Nara, B.Y. Wu, H. Nakaya, C.L. Lian, N. Miyashita, et al. 2003. Patch establishment and development of a clonal plant, Polygonum cuspidatum, on Mount Fuji. Molecular Ecology 12: 1361–1373.
Zika, P.F. and A.L. Jacobson. 2003. An overlooked hybrid Japanese knotweed (Polygonum cuspidatum x Sachalinense; Polygonaceae) in North America. Rhodora 105: 143–152.
Acknowledgments
The author thanks Manuel Lerdau, Christina Richards, Elizabeth Leger, Jennifer Verdolin, Jessica Gurevitch, R. Geeta, the Lerdau laboratory group, and two anonymous reviewers for discussions on this research and comments on earlier versions of this manuscript. Thanks to Ashley Frye, Anna Chaldesheva, and James Meany for assistance in the field and greenhouse and the Long Island National Wildlife Refuge, the Village of Port Jefferson, and the Town of Southold for access to field sites. This study was funded by a Budweiser Environmental Scholarship/National Wildlife Refuge Centennial Scholarship from Anheiser Busch and The National Fish and Wildlife Foundation.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Rights and permissions
About this article
Cite this article
Walls, R.L. Hybridization and Plasticity Contribute to Divergence Among Coastal and Wetland Populations of Invasive Hybrid Japanese Knotweed s.l. (Fallopia spp.). Estuaries and Coasts 33, 902–918 (2010). https://doi.org/10.1007/s12237-009-9190-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12237-009-9190-8