Abstract
Alkaline phosphatase expression by phytoplankton from two sites in Winyah Bay, SC, USA was investigated using nutrient-addition bioassays and cell-specific enzyme-labeled fluorescence (ELF) measurements. Our aim was to determine whether expression was group- or species-specific within the phytoplankton community. Diatoms dominated the riverine site in May, the coastal site in July, and both sites in August. Phytoplankton growth was limited by nitrogen (N) availability at the coastal site in May and the riverine site in August, but phosphate limitation was not observed. Alkaline phosphatase expression ranged from ∼30% of cells enumerated to less than 1% and was significantly reduced by inorganic phosphorus (P; 10 μM P) additions. Expression was restricted to species with low abundance, and there were no shifts in community composition consistent with alkaline phosphatase expression. Lack of phosphate limitation at higher-than-Redfield N/P ratios (up to 40:1), however, points to a potentially wider role of dissolved organic phosphorus in nutrition of Winyah Bay phytoplankton than indicated by the ELF assay.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phosphorus (P) is essential for the growth and reproduction of all aquatic organisms. In the water column, P exists in both particulate and dissolved forms, both of which may contain organic and inorganic constituents (Solórzano and Sharp 1980). The dissolved inorganic P fraction, operationally defined as soluble reactive phosphorus (SRP), is monitored routinely in rivers, lakes, and estuaries by water quality managers for regulation purposes. However, SRP is not always the dominant fraction of the total dissolved phosphorus (TDP) pool, nor is it the only fraction that is bioavailable (Kuenzler and Perras 1965; Cembella et al. 1984; Sakshaug et al. 1984). The dissolved organic phosphorus (DOP) component can comprise a significant portion of the TDP pool in a variety of aquatic environments (Meybeck 1993) and may serve as an alternative P source for bacteria and some phytoplankton (e.g., Cembella et al. 1984; Bentzen et al. 1992; Ruttenberg and Dyhrman 2005). DOP is a complex mixture of organic constituents such as monophosphate esters, nucleotides, polyphosphates, and phosphonates that have natural or anthropogenic origins (see reviews by Benitez-Nelson 2000; Karl and Björkman 2002). Currently, we have a limited understanding of the composition and biological utilization of DOP in marine systems (Björkman and Karl 1994; Karl and Yanagi 1997). In estuaries, the composition and biological role of DOP is even less well characterized. DOP can be a significant fraction of the P delivered from land to the marine environment (Meybeck 1993; Guildford and Hecky 2000), and river-transported terrigenous DOP has been recognized as a potential source of P for coastal marine organisms (Suzumura et al. 1998).
Bacteria have long been known to play an important role in DOP uptake and utilization (Kuenzler and Perras 1965; Ammerman and Azam 1985; Cotner and Wetzel 1992). More recently, attention has focused on the role of phytoplankton in DOP dynamics (Dyhrman and Palenik 1999; Rengefors et al. 2003; Dyhrman 2005). DOP utilization by both bacteria and phytoplankton is mediated by the expression of a number of enzymes including extracellular alkaline phosphatases (Kuenzler and Perras 1965; Ammerman 1991). Alkaline phosphatases specifically hydrolyze phosphomonoesters into bioavailable phosphate at alkaline pH (Kuenzler and Perras 1965; Jansson et al. 1988).
Alkaline phosphatase activity is thought to be triggered by low inorganic P availability (Kuenzler and Perras 1965) and has been used as an indicator of P status in a variety of phytoplankton communities including those in lakes (Rengefors et al. 2003; Dignum et al. 2004; Cao et al. 2007), coastal oceans (e.g., Labry et al. 2005; Dyhrman and Ruttenberg 2006; Nicholson et al. 2006), and open-ocean environments (Li et al. 1998; Lomas et al. 2004). More recently, alkaline phosphatase activity has been shown to be better correlated to intracellular P or total cellular P than to ambient concentrations (Vahtera et al. 2007; Litchman and Nguyen 2008), since phytoplankton are able to accumulate P beyond what is needed for immediate metabolic requirements (the “luxury consumption” concept of Droop 1973).
The traditional approach to measuring alkaline phosphatase activity relies on bulk assays of whole water samples, using a specific phosphomonoester substrate that is added to a sample and hydrolyzed in the presence of alkaline phosphatase (Perry 1972). While bulk assays yield the rate of potential alkaline phosphatase hydrolysis in a sample, they do not discriminate between the activities of the bacteria and the phytoplankton (unless samples are size-fractionated), nor do they distinguish species-specific responses within the phytoplankton community (Dyhrman and Palenik 1999; Rengefors et al. 2001; 2003). An alternative approach, developed more recently, is to use an enzyme-labeled fluorescence (ELF) technique that allows the visualization of alkaline phosphatase expression by individual cells (González-Gil et al. 1998). One specific assay known as ELF-97® (Molecular Probes, OR, USA) makes it possible to detect alkaline phosphatase expression in individual cells and thus phytoplankton species, while at the same time discriminating the activity of phytoplankton from bacteria in the same water sample (González-Gil et al. 1998; Dyhrman and Palenik 1999). The ELF assay has been used to study alkaline phosphatase expression by a variety of freshwater and marine phytoplankton taxa, including diatoms (Rengefors et al. 2001; Ruttenberg and Dyhrman 2005; Ou et al. 2006), dinoflagellates (González-Gil et al. 1998; Dyhrman and Palenik 1999; Nicholson et al. 2006), chlorophytes (Rengefors et al. 2001; Cao et al. 2007) and cyanobacteria (Dignum et al. 2004; Štrojsová et al. 2005; Mackey et al. 2007). Within these algal groups, alkaline phosphatase expression appears to be species-specific, and even within species, expression may vary both spatially and temporally (Ruttenberg and Dyhrman 2005; Mackey et al. 2007).
Temporal, spatial, and species-specific variability in alkaline phosphatase expression may be particularly important in dynamic estuarine ecosystems. Phytoplankton community composition will vary along an estuary as a function of many biological, chemical, and physical factors related to tidal forcing and riverine inflow. Phytoplankton communities are also subject to variable nitrogen (N) and P loading, and thus, productivity may be either N- or P-limited depending on season or location (Mallin et al. 1999; Klug 2006). While N is traditionally considered to be the limiting element in coastal marine habitats, reductions in inorganic P loading without concomitant reductions in N have resulted in periodic P limitation in some estuaries (Mallin et al. 1999; Murrell et al. 2002; Klug 2006). Evidence of P limitation of community production has been found in regimes ranging from restricted (Smith and Atkinson 1984; Krom et al. 1991) and shallow marine areas (Fourqurean et al. 1992; Glibert et al. 2004) to open-ocean regions of the North Atlantic and North Pacific (Cotner et al. 1997; Karl and Yanagi 1997). However, stoichiometric determinations of P limitation that assume inorganic P is the sole source for phytoplankton neglect to consider that DOP may provide an alternative source of P to phytoplankton capable of alkaline phosphatase expression. The ability to use both DOP and inorganic P may also confer a competitive advantage in terms of P acquisition and may contribute to the success of one species or taxonomic group of algae over another when inorganic P is low.
The extent of alkaline phosphatase expression by estuarine phytoplankton and associated impacts on community composition are poorly known. In this study, we focused on phytoplankton from a coastal plain estuary, Winyah Bay, SC, USA. We investigated whether phytoplankton in Winyah Bay showed alkaline phosphate expression, whether expression was group- or species-specific and whether there were accompanying shifts in phytoplankton community composition with nutrient manipulations.
Materials and Methods
Study Site
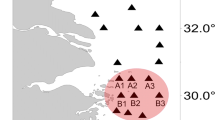
Winyah Bay is a 65 km2 partially mixed coastal plain estuary located near Georgetown County, SC, USA (Fig. 1). In terms of discharge, it is the fourth largest estuary on the eastern coast of the USA with a total drainage area of approximately 47,000 km2 (Patchineelam et al. 1999). Of the four rivers that flow into Winyah Bay, the Pee Dee River contributes approximately 90% of the total riverine discharge into the bay (Patchineelam et al. 1999). The Pee Dee, Black, and Waccamaw Rivers are influenced by agricultural runoff and flow from upstream swamps and forested land. The Sampit River, a tributary of Winyah Bay, is affected by effluent from a paper mill, a steel mill, and a municipal sewage treatment plant (South Carolina Sea Grant 1992).
Bioassays
We examined alkaline phosphatase expression and growth responses of phytoplankton communities from Winyah Bay to additions of inorganic P and N (as nitrate) using a bioassay approach. These experiments were designed to study alkaline phosphatase expression with varying inorganic P and excess N availability (the latter an experimentally forced P limitation treatment), but they also allowed us to address the question of whether N or P was limiting to phytoplankton growth in Winyah Bay.
Surface water was collected from two stations in Winyah Bay, designated “riverine” and “coastal” (Fig. 1) using a 1-m integrated water sampler on three dates (May 29, July 3, and August 14, 2006). The riverine site was at the confluence of the Black, Pee Dee, and Waccamaw Rivers; the coastal site was located between the jetties at the entrance to Winyah Bay (Fig. 1). Collection at these two sites allowed us to examine responses of phytoplankton communities under different relative influences of freshwater and the coastal ocean (Table 1). All water was prescreened using a 100-μm mesh to remove large grazers and was kept in 10-L acid-washed containers on ice in the dark until incubations began (for less than 1 h). A total of 90 L was collected from each site and was distributed into nine 10-L Cubitainers®. Triplicate cubitainers were amended daily with either nitrate (as KNO3; final concentration = 20 μM N) or phosphate (as KH2PO4; final concentration = 10 μM P). Controls, along with all containers, had silicate (Na2SiO3; final concentration = 200 μM Si) added to prevent possible silica limitation of diatoms. Silicate was the only nutrient added to the controls. The relatively high concentrations of N were added to prevent inorganic N limitation and to promote inorganic P limitation, while the relatively high P additions were used to suppress phosphatase activity.
Riverine samples were incubated in situ at the Georgetown Landing Marina close to the site of water collection. Since it was logistically difficult to incubate the coastal samples at the collection site, cubitainers were incubated in situ at Clam Bank Creek (Fig. 1), a site similar in temperature and light regime to the collection area (Table 1). Incubation of the samples in conditions similar to their site of collection was important because of the distinctly different spectral quality of available irradiance at the collection sites (Lawrenz et al., 2009). Irradiance at the riverine site is strongly influenced by the blackwater nature of the inflowing rivers where absorption of blue light by colored dissolved organic matter (CDOM) is high. The coastal collection site, in contrast, is much less strongly influenced by CDOM absorption (lower K 442 nmvalues, see Table 1). Vertical profiles of photosynthetically available radiation (PAR; 400–700 nm) at each collection site were measured using a submersible scalar quantum sensor (LI-COR, Lincoln, NE, USA) connected to a LI-COR data logger. At each incubation site, cubitainers were floated in a 1.1-m2 corral covered with two layers of neutral density screening to simulate average water column irradiance and to prevent photoinhibition. Samples were collected initially (day 0) and on days 1 and 2. Two-liter subsamples were removed from each container once per day (at 0900 hours for the riverine samples, 1300 hours for the coastal samples) for analyses.
Chemotaxonomic photosynthetic pigments, nutrient concentrations, and cell-specific alkaline phosphatase activity (by ELF assay) were determined on days 0, 1, and 2 of each experiment. Voucher samples for confirmation of pigment-based assessments of phytoplankton community structure by microscopy were collected on day 0, preserved in Lugol's iodine or 2% (final concentration) glutaraldehyde and were viewed using inverted microscopy (Uterhmöhl 1958).
Chemotaxonomic photosynthetic pigments were measured by high-performance liquid chromatography (HPLC; Pinckney et al. 1996) and were used to determine total phytoplankton biomass (as total chl a) and the relative and absolute contributions of major phytoplankton taxonomic groups (Jeffrey et al. 1997; Pinckney et al. 1998). Aliquots of water from the cubitainers (80–250 ml) were filtered through 25-mm GF/F filters under a gentle vacuum in dim light. Filters were immediately frozen and stored at −80°C. Frozen filters were freeze-dried overnight then placed in 90% acetone and extracted overnight at −20°C. Extracts were filtered through a 0.45-μm polytetrafluoroethylene filter (Pall Acrodisc). Ammonium acetate (1 M) was added as an ion-pairing agent in a ratio of three parts extract to one part ammonium acetate, and the extracts were then injected into a Shimadzu HPLC (LC10-AT) equipped with both a single monomeric and a polymeric reverse-phase C18 column in series. A nonlinear binary gradient was used for pigment separations (Pinckney et al. 1996). Solvent A consisted of 80% methanol:20% ammonium acetate (0.5 M adjusted to pH 7.2) and solvent B was 80% methanol:20% acetone. Each sample received 50 μl of the synthetic carotenoid β-apo-8′-carotenal (Sigma-Aldrich Chemical Company, No. 10810) that served as an internal standard. Absorption spectra and chromatograms (440 nm) were acquired using a Shimadzu SPD-M10av photodiode array detector. Pigment peaks were identified by comparing retention times and the absorbance spectra to certified pigment standards (DHI, Denmark). The contribution of each algal group to overall community composition was determined using ChemTax (CHEMical TAXonomy), a matrix factorization program (Mackey et al. 1996; Wright et al. 1996). The program uses steepest descent algorithms to determine the best fit based on an initial estimate of pigment ratios for algal classes. The absolute contribution of any algal group is the concentration of total chl a (in μg l−1) contributed by that group. Relative contributions were calculated as the proportion of total chl a accounted for by the group so that the sum of contributions from all groups equals one. Validated initial pigment ratio files were taken from Lewitus et al. (2005). Full discussions, validation, and sensitivity analyses for the ChemTax approach are found in Mackey et al. (1996), Wright et al. (1996), and Schlüter et al. (2000).
Inorganic nutrient (NO −3 + NO −2 , NH +4 , and silicate) concentrations were analyzed by a Technicon AutoAnalyzer II using standard protocols at the University of Maryland Center for Environmental Sciences Analytical Services Laboratory in Cambridge, MD, USA. Water samples were filtered through acid-washed (10% HCl) and precombusted 25-mm Whatman GF/F filters and were frozen at −20°C until analysis. Phosphorus analyses (TDP, SRP, and DOP) were done in the Marine Geochemistry Laboratory at the University of South Carolina. TDP was measured using the Monaghan and Ruttenberg (1999) high-temperature combustion and hydrolysis method. Measurements of SRP were made using a procedure based on Koroleff (1983) where samples were analyzed using a phosphomolybdate-blue colorimetric reaction and absorbance measured using a Beckman Coulter DU 640 spectrophotometer. Detection limits were ∼0.05 μM P (using a 10-cm spectrophotometer cell) at a wavelength of 880 nm. DOP concentrations were calculated as the difference between SRP and TDP measurements. It is important to note that due to the process of forming phosphomolybdate complex, SRP concentrations could include an unknown proportion of acid-labile organic compounds (e.g., simple sugars and monophosphate esters; Benitez-Nelson 2000).
Cell-specific alkaline phosphatase activity was assayed using the ELF-97 approach of González-Gil et al. (1998) as modified by Dyhrman and Palenik (1999). To concentrate the samples, aliquots (250–500 ml) of each sample were filtered through a 0.8 μm Gelman SUPOR filter until the filter was nearly dry. Cells were transferred to a microcentrifuge tube, spun at 4000 rpm at room temperature in a Beckman-Coulter Microfuge 18 centrifuge to form a pellet and were stored in 1 ml of 70% ethanol at 4°C in the dark prior to analysis. Before analysis, samples were recentrifuged (4,000 rpm for 2 min), and the supernatant was discarded. The pellet was rinsed with 100 μL of filtered sterilized seawater and was then resuspended in 100 μL of ELF reagent. After 1 h of incubation in the dark at 4°C, the sample was centrifuged, decanted, and rinsed twice with sterile sea water. The pellet was resuspended in 20 μL of the sterile seawater, from which a 7–10 μL aliquot was mounted on a slide and analyzed using a Nikon TE300 inverted microscope. In the presence of alkaline phosphatase, the soluble colorless substrate (2-(5′-chloro-2′-phosphoryloxyphenyl)-6-chloro-4-(3H)-quinazolinone) is converted to an insoluble product (2-(5′chloro-2′-hydroxyphenyl)-6-chloro-4-(3H)-quinazolinone) that fluoresces green under UV excitation (365 nm; Huang et al. 1993; Dyhrman and Palenik 1999). For the purposes of this study, a cell was scored positive for alkaline phosphatase expression if 75% or more of the cell surface appeared green (see “Discussion” for the implications of this choice of criterion). At least 400 cells on each slide were counted and scored and were identified as a diatom, dinoflagellate (to genus), or “other” taxa (see “Alkaline Phosphatase Expression” section in “Results” below). Results are expressed as percentages of total cells counted. This scoring approach differs from that of some researchers, who first identify taxa that can express alkaline phosphatase then count the number of labeled vs. unlabeled cells in that taxa (see “Discussion”). Note also that we did not specifically enumerate the picoplankton component of the community, so “total cells” refers only to phytoplankton visible under light microscopy (up to ×400).
Statistics
Differences in total phytoplankton biomass and in the relative and absolute abundance of phytoplankton taxonomic groups were analyzed using a two-factor repeated measures analysis of variance (RM-ANOVA) (Scheiner and Gurevich 1993) using SPSS 14.0 for Windows. Factors were nutrient treatment and collection site. Absolute concentrations of algal groups and total community biomass (as total chl a) were ln-transformed before analysis to satisfy the normality assumption. Relative abundance data were analyzed similarly but were arcsine square root-transformed. Homogeneity of error variances was checked using Cochran's Test, and because homogeneity was satisfied, a Bonferroni test (α = 0.01) was used for post hoc comparison of means.
Results
Inorganic Nutrient and DOP Concentrations
Ambient DOP concentrations ranged from 0.7 to ∼1 μM at the riverine station and 0.3 to 0.7 μM at the coastal site (Table 2; using concentrations on day 0 for control and +N treatments only for each month). These concentrations represent 40–67% of the TDP pool at the riverine location and 50–89% at the coastal site (Table 2). DOP concentrations were higher at the riverine site than at the coastal site in all months. SRP was highest at both sites in July (Table 2). SRP, nitrate + nitrite and silicate concentrations at the coastal sampling location were consistently lower than at the riverine site in all months (Table 2). The unusually high DOP concentrations in the +P treatments at both riverine and coastal stations in May (∼3–5 μM) were traced to the stock of KH2PO4 used for the P additions. A new bottle of KH2PO4 was purchased for subsequent experiments.
Phytoplankton Biomass and Community Composition
Initial phytoplankton community composition and total biomass differed between collection site and with month of collection. In May, the phytoplankton community at the riverine site was dominated by diatoms (55% or more of the total chl a) with smaller varying contributions from the cryptophytes, prasinophytes, cyanobacteria, and haptophytes (Fig. 2a). The coastal phytoplankton community in May was initially composed primarily of haptophytes (59% or more of total chl a) and prasinophytes, but community composition in the containers shifted by day 2 to dominance by diatoms (Fig. 2a). The addition of N significantly increased total phytoplankton biomass and the absolute abundance of diatoms and prasinophytes in May (Table 3). As indicated by the significant nutrient × site interaction terms for total chl a, diatoms, and prasinophytes, this was true only for the coastal site, that is, site of collection had a significant influence on whether N additions resulted in significantly increased biomass (Fig. 2a, Table 3). N additions also resulted in a significant shift in the relative abundance of cyanobacteria, but overall, this group accounted for a relatively low percentage of the total phytoplankton biomass at both sites (Table 3). P additions had no significant effect on total phytoplankton biomass or on the relative or absolute abundance of phytoplankton taxonomic groups in May (Fig. 2a, Table 3).
Phytoplankton community composition and total biomass (as chl a) from HPLC/ChemTax data collected during experiments in May (a), July (b), and August (c) 2006 in Winyah Bay, SC, USA. Treatments were daily additions of nitrate (+N; 20 μM N) or phosphate (+P; 10 μM P). Error bars indicate standard deviation of the mean. Note the differences in y-axis scale between a, b, and c
In July, there were no significant effects of N or P additions on total chl a or on the absolute or relative abundance of the different phytoplankton groups, except for a higher relative abundance of haptophytes in the N-amended cubitainers as compared to controls (Table 3).
In August, total phytoplankton biomass was significantly higher at the riverine site than at the coastal site (Fig. 2c, Table 3). Diatoms dominated the phytoplankton community at both locations contributing 70% or more of the total chl a (Fig. 2c). The addition of N significantly increased total phytoplankton biomass at both riverine and coastal sites (Fig. 2c, Table 3) as compared to controls. Diatoms, dinoflagellates, and prasinophytes all increased in absolute (but not relative) abundance particularly at the riverine site (see significant nutrient × site interaction terms for dinoflagellates and prasinophytes in Table 3). P additions had no significant effect on total chl a or on the absolute or relative abundance of algal groups in August, except diatoms were relatively less abundant at the riverine station in the P addition treatment (Fig. 2c, Table 3).
Alkaline Phosphatase Expression
Alkaline phosphatase expression by phytoplankton from Winyah Bay varied with site and month of collection. Overall, the highest percentages of cells labeled as positive for alkaline phosphatase expression were found in communities from the coastal site in May (up to 32% of cells labeled; Fig. 3a) with the lowest observed in phytoplankton communities collected in August at both riverine and coastal sampling locations (less than 2% of cells labeled; Fig. 3c). In May, alkaline phosphatase expression was significantly higher in phytoplankton communities collected from the coastal site as compared to the river site (Fig. 3a, Table 4), whereas the opposite relationship was observed in August (Fig. 3c, Table 4).
The percentage of total cells and HPLC/ChemTax-derived Phytoplankton community taxa (days 0–2) that were scored positive for expression of alkaline phosphatase (“AP positive”) during time-course experiments in May (a), July (b), and August(c), 2006. Treatments were daily additions of nitrate (+N; 20 μM N) or phosphate (+P; 10 μM P). Data for group-specific alkaline phosphatase expression are not available for control and + N treatments on days 0 and 1 at the coastal station in May. Error bars indicate standard deviation of the mean. Note the differences in y-axis scale between a, b, and c
Alkaline phosphatase expression was significantly lower in bioassay containers amended with inorganic P in all experiments (Table 4). This is shown clearly by results from both sampling sites in the July experiment (Fig. 3b) and also by the response of phytoplankton from the coastal site in May (Fig. 3a) and the riverine site in August (Fig. 3c). Note the significant interaction terms in Table 3 for May and August indicating that the effect of P addition on alkaline phosphatase expression was site-specific.
Diatoms and dinoflagellates were the only phytoplankton taxa that could be positively identified among the cells that were scored positive for alkaline phosphatase expression. Species commonly found to express alkaline phosphatase in all experiments included those of the dinoflagellate genera Protoperidinium and Gymnodinium and of the diatom genera Pleurosigma and Cylindrotheca (Fig. 4) with less common expression by the diatoms Thalassionema and Asterionellopsis. All other cells that were scored positive for alkaline phosphatase expression could not be readily identified because they did not preserve well using standard ELF protocols; therefore, our results are presented as contributions from diatoms, dinoflagellates, and the unidentified “other” cells (Fig. 3).
Taxon-specific alkaline phosphatase expression varied with site and month of collection. In May at the riverine site, diatoms dominated the phytoplankton community, and alkaline phosphatase expression was due mainly to the diatoms (27–87% of all cells scored positive depending on day and treatment; Fig. 3a). Dinoflagellates represented 0–29% of all cells that expressed alkaline phosphatase, but it is important to note that dinoflagellates were low enough in abundance in the overall community composition as to be included in the “other” category based on ChemTax (see Fig. 2a). “Other” cells scored as positive (7–45%) likely included species of prasinophytes, haptophytes, or cryptophytes as illustrated by ChemTax results (Fig. 2a).
At the coastal site in May, diatoms again dominated the cells that were scored as positive for alkaline phosphatase expression as well as community composition, accounting for 59–74% of all cells that expressed alkaline phosphatase (Fig. 3a). Dinoflagellates and other cells contributed less than 25% to cells scored alkaline phosphatase positive. Unfortunately, we do not have taxon-specific counts for days 0 and 1 in this experiment (only for day 1 in the P treatment). It would have been informative to see what taxa contributed to our highest observed percentages of alkaline phosphatase expression (day 0, control) before the shift in community composition from haptophytes to diatoms (shown in Fig. 2a) and the drop in the percentage of cells scored as positive for expression.
In the July experiment, phytoplankton taxa from the riverine site that expressed alkaline phosphatase were mostly unidentified “others” (26–85%; Fig. 3b) which agrees well with the relatively diverse overall community composition at this site (Fig. 2b). Surprisingly, alkaline phosphatase expression by others also accounted for a high percentage (26–58%) of total expression at the coastal sampling location, where diatoms dominated the community composition (Fig. 2b), though diatoms also accounted for a substantial proportion of the expression (Fig. 3b).
In August, dinoflagellates of the genus Gymnodinium accounted for most of the taxon-specific alkaline phosphatase expression (Fig. 3b), but overall expression was lowest in August. Diatoms dominated the phytoplankton community composition at both the riverine (Navicula and Cyclotella) and coastal (Thalassiosira and Chaetoceros) sampling sites; however, these diatoms were not species that were routinely scored positive for alkaline phosphatase expression.
The day 0 samples in our bioassay experiments gave us a snapshot of what taxa could express alkaline phosphatase under the prevailing environmental conditions. The time-course nature of these experiments then allowed us to look at how alkaline phosphatase expression changed with ambient N and inorganic P availability. From day 0 through to day 2 in these bioassays, we often saw shifts in the overall percentage of cells that expressed alkaline phosphatase (increasing or decreasing trends depending on site and month of collection). For example, in samples from the riverine station in July, the percentage of cells scored as positive for alkaline phosphatase expression increased from day 0 to day 2 in the control and +N treatments (Fig. 3b). These increases were coincident with a decreasing availability of SRP (Table 2). The opposite trend was observed in samples from the coastal station in both May and July: the percentage of cells scored as positive decreased through the incubation period in both control and +N treatments. In this case, however, there was no clear relationship to either SRP concentrations or DOP availability (Table 2).
Discussion
Summer phytoplankton communities from surface waters of Winyah Bay varied in community composition with site and month of collection. The most diverse community composition was observed at the riverine site in July and likely reflected a strong freshwater influence. Salinities along the estuary in July were the lowest of all months sampled and followed a 3-week period of high discharge from the Pee Dee River (USGS; data not shown). Phytoplankton community composition varied in response to nitrate and inorganic P additions. Bioassays showed that phytoplankton growth was limited by N availability only at the coastal station in May and at the riverine site in August. There was no evidence of inorganic P limitation at either site in any month. These results lend support to the contention that both P and N should be controlled to reduce eutrophication in estuaries (Conley et al. 2009; but see Schindler et al. 2008). There was some evidence that silicate might have been co-limiting to diatoms in May at the coastal site as shown by the shift in phytoplankton community composition in control containers after silicate additions on day 0. The repeated measures approach of our statistical analyses accounts for such shifts in the control bottles. Ambient nitrate + nitrite and silicate concentrations at the coastal station in May were the lowest of all months sampled. Nutrient concentrations were highest in July, following the period of high discharge from the Pee Dee River. This could account for the observation that neither N nor inorganic P was limiting to phytoplankton growth in July.
DOP concentrations measured at our two study sites (∼0.3 to 1 μM) fall within the range measured in other estuaries and river-influenced coastal oceans. Concentrations up to 0.6 μM were measured in the Delaware Estuary (Lebo and Sharp 1993), Chesapeake Bay (Conley et al. 1995), on the Eel River Shelf (Monaghan and Ruttenberg 1999), and in the Bay of Biscay (Labry et al. 2005). Slightly higher values (up to ∼1 μM) were reported for the Tanshui Estuary (Fang 2000), Apalachicola Bay (Mortazavi et al. 2000), and the Scheldt Estuary (van der Zee et al. 2007), while concentrations in the Mississippi River often exceeded 1 μM and were sometimes >2 μM (Rinker and Powell 2006). In terms of percent contribution to the TDP pool, our overall ranges are similar to those of Monaghan and Ruttenberg (1999) for two stations on the Eel River Shelf, where DOP comprised 28–86% of the TDP pool depending on season. DOP values in their study were highest in the summer and were higher than SRP concentrations, as we observed often at our coastal station.
Results of our ELF assays showed that some phytoplankton taxa from Winyah Bay can express alkaline phosphatase. We saw a significant reduction in ELF labeling of phytoplankton in bioassay containers with inorganic P added, indicating that the expression of alkaline phosphatase was repressed when inorganic P was freely available. This observation is in agreement with previous reports of reduced labeling upon re-introduction of phosphate (e.g., Dyhrman and Palenik 1999, 2003; Labry et al. 2005; Dyhrman and Ruttenberg 2006; Mackey et al. 2007). We did not, however, see a significant increase in ELF labeling in containers with added nitrate (our scenario of “forced” phosphate limitation), even though, by day 2, SRP was reduced to extremely low concentrations (<0.1 μM in most cases). The reason for this lack of up-regulation of alkaline phosphatase expression is not immediately apparent. Previous studies have shown that nitrate amendments result in both increased ELF labeling for specific taxa (e.g., chlorophytes (Rengefors et al. 2003) and diatoms (Dyhrman and Ruttenberg 2006)) and increased alkaline phosphatase activity in the bulk microbial community (Dyhrman and Ruttenberg 2006). However, Rengefors et al. (2003) also found that N additions did not lead to increased alkaline phosphatase expression in freshwater species of Chlorococcales (Dictyosphaerium and Actinastrum). They attributed the lack of increased ELF labeling to the possibility of either luxury uptake of phosphate prior to the incubations (and thus the presence of internal phosphate stores), innately low phosphate requirements, and/or the lack of growth of these species in the incubation containers (and therefore the absence of “P stress”). In our case, daily measurements of algal biomass showed step-wise increases in total chl a in most containers, indicating that the populations were growing; however, we cannot rule out intracellular stores of phosphate or innately low (taxon-specific) P requirements as possible reasons behind our observed lack of up-regulation of alkaline phosphatase expression.
Our experimental design (daily sampling) also allowed us to observe step-wise changes in the percentage of labeled cells with incubation time. Samples from the control and +N treatment containers from the riverine site in July, for example, showed a step-wise increase in the percentage of cells that expressed alkaline phosphatase that followed progressive declines in ambient inorganic P. More difficult to explain are the step-wise decreases in ELF labeling shown in coastal samples from the May and July sampling times. Decreasing concentrations of total chl a with time in the control samples, along with observations of grazers in fixed samples, indicate that, despite our prescreening with 100 μm mesh, small grazers were still present in the containers. We therefore speculate that either grazers were removing taxa that tended to be ELF-labeled (dinoflagellates and some taxa in the “others” category), or grazers were regenerating enough inorganic P to meet phytoplankton demands, thus reducing alkaline phosphatase expression. These observations illustrate the importance of examining the temporal dynamics of phytoplankton community composition and ELF labeling during nutrient addition experiments, rather than making a single measurement at the end of the incubation period.
Alkaline phosphatase expression by phytoplankton from Winyah Bay was species-specific. Expression was commonly observed in dinoflagellate species of the genera Protoperidinium and Gymnodinium and diatoms, including Pleurosigma and Cylindrotheca, consistent with previous reports of ELF labeling in these algal taxa (e.g., Rengefors et al. 2003; Dyhrman and Ruttenberg 2006; Ou et al. 2006). Expression was not limited to diatoms and dinoflagellates, however, as many ELF-positive species could not be readily identified as they did not preserve well using the standard protocol. Similar problems were experienced by Rengefors et al. (2001), who found some phytoplankton, such as chrysophytes, cryptophytes, and haptophytes, were too small and/or fragile to label with the ELF reagent. While we had no problems with labeling, the 70% ethanol preservation resulted in sufficient cell shrinking and related morphological changes to render them unidentifiable. These “other” cells were most important in our July bioassay at the riverine site, where haptophytes, prasinophytes, and chlorophytes made substantial contributions to the total phytoplankton biomass. We expect that the chlorophytes and haptophytes are key contributors to the ELF activity of the “others” at the riverine location. Previous studies have shown that some species of both freshwater chlorophytes and haptophytes can express alkaline phosphatase (Rengefors et al. 2001, 2003; Cao et al. 2007). Cyanobacteria have also been shown to express alkaline phosphatase, though expression appears to be more common in larger species of the genera Anabaena, Microcystis, or Trichodesmium (as examples), than in the picoplanktonic cyanobacteria (Lomas et al. 2004; Štrojsová et al. 2005; Mackey et al. 2007). Even within Trichodesmium, however, species differences exist: one species of Trichodesmium (Trichodesmium erythraeum) has the gene for alkaline phosphatase (phoA), but a co-occurring species, Trichodesmium thiebautii, does not (Orchard et al. 2003).
The percentages of total cells counted that showed alkaline phosphatase expression were generally less than 15%, except for one experiment (May) where up to 32% of all cells counted were labeled. Our results can be directly compared to those of Lomas et al. (2004), who investigated taxonomic variability of phosphorus stress in Sargasso Sea phytoplankton and found that 30% of the enumerated autotrophic eukaryotes collected from surface waters in the summer season were ELF-labeled, and nearly 70% of cells were labeled in the fall season. All of their values are substantially higher than our observations of ELF-labeling, as might be expected for an open-ocean environment where inorganic P was below detection (Lomas et al. 2004). Lomas et al. (2004) used similar taxonomic discriminations as ours: autotrophic flagellates displayed higher ELF labeling in summer than in fall, whereas diatoms displayed higher labeling in fall than in summer. In their study, dinoflagellates did not contribute substantially to ELF labeling (Lomas et al. 2004).
Our approach and the Lomas et al. (2004) approach to assessing community-wide alkaline phosphatase expression differ somewhat from other studies, where the term “percent labeled” refers to the percentage of each individual species or taxa that is labeled (e.g., Rengefors et al. 2001, 2003; Dyhrman and Ruttenberg 2006), as opposed to the percentage of the total phytoplankton cells counted. The former approach assesses the degree of variability in ELF labeling within a species or taxa. Variability can be high due to differences in the extent of cellular alkaline phosphatase expression or the specific activity of individual alkaline phosphatase enzyme complexes (Lomas et al. 2004). We used a relatively conservative criterion (∼75% of total cell surface labeled with ELF precipitate) for scoring cells as positive for alkaline phosphatase expression. This might explain, in part, why our percentages are lower than those of Lomas et al. (2004), who used a less stringent criterion (∼25% of cell surface area covered; Lomas, personal communication). The use of 75% cell surface coverage for a score of positive for alkaline phosphatase expression was an arbitrary decision, and we had no specific criterion for choosing 75% coverage, except the goal of ensuring that a cell scored positive was truly positive. In hindsight, we should have chosen a percentage more comparable to studies already published (e.g., Rengefors et al. 2001, 2003; Dyhrman and Ruttenberg 2006). We have now implemented a binning system that scores ELF labeling as 0–33%, 33–66%, or 66–100% of the cell surface covered, which should help us compare future results to those of other investigators (or at least those with defined criteria). Unfortunately, these data are not available for these experiments, and it is not possible to re-analyze the samples. We attempted to calculate a correction factor based on subsequent work on alkaline phosphatase expression in the dinoflagellate Prorocentrum minimum and the cryptophyte Rhodomonas salina (Ranhofer et al. in prep). For P. minimum, scoring a cell as positive with 75% or more of the cell surface labeled resulted in a 56% underestimate as compared to using 33% or more of the cell surface labeled as the criterion for “positive”. For R. salina, however, there was only a 4% underestimate, and there were similarly low values for cultures of Emiliania huxleyi and Thalassiosira weissflogii (although both these species had very low (∼1%) labeling overall). Thus, it appears that the choice of percent coverage criterion matters for some species (like dinoflagellates) but not for others.
We can compare our results to studies that provide data on both the percent of species labeled and the relative abundance of that species in the phytoplankton community. Nicholson et al. (2006), for example, examined alkaline phosphatase expression in diatoms and dinoflagellates that comprised the bulk of the eukaryotic phytoplankton collected from Monterey Bay, CA, USA. Using data from their Table 1, we calculated that the percentages of labeled cells of those enumerated (diatoms and dinoflagellates only) ranged from 0% to 6.5%. These numbers are within range of our observations but are generally lower even considering the relatively stringent criteria we used in scoring. Similarly, less than 6% of the total cells in phytoplankton communities studied by Mackey et al. (2007) were scored positive for ELF labeling (calculated from data presented in their Fig. 7). Their study included an assessment of the picophytoplankton, which comprised a large fraction of the total phytoplankton cell abundance, yet less than 4% of the picophytoplankton cells were scored as positive for ELF labeling. Generally, it appears that the phytoplankton species that are routinely ELF labeled tend to be those that are relatively less abundant in the community. In the Nicholson et al. (2006) study, for example, the highest ELF labeling was for the dinoflagellate P. minimum, yet the relative abundance of this dinoflagellate in the community was only 8.8% of the total cells counted (Table 1 in Nicholson et al. 2006). In our study, the diatoms and dinoflagellates that were routinely scored as ELF positive (Gymnodinium, Protoperidinium, Cylindrotheca, and Pleurosigma) also made relatively low contributions to the total overall abundance, and this is likely why we saw no significant shifts in phytoplankton community composition that we could associate with alkaline phosphatase expression, even at low inorganic P concentrations.
The relatively low percentage of cells that expressed alkaline phosphatase in our study, together with the lack of any significant shifts in phytoplankton community composition, indicate that utilization of DOP may not be of broad-scale importance to nutrient dynamics and phytoplankton growth in spring/summer assemblages in Winyah Bay. However, we should note that blooms of Gymnodinium-like dinoflagellates (e.g., Kryptoperidinium) and diatoms (like Cylindrotheca) are common in North Inlet, a nearby estuary (Kawaguchi et al. 1997; Lewitus and Holland 2003), and diatoms dominate the community composition of Winyah Bay in winter (Lawrenz et al., 2009). Thus, their overall importance to DOP dynamics in Winyah Bay may be underestimated. Also, the ELF assay detects activity of the alkaline phosphatase enzymes only. These specifically target DOP compounds with ester bonds, yet “DOP” is a bulk characterization of a variety of different compounds, so utilization of DOP compounds that require diesterases, phytases, C-P lyases, or 5′nucleotidases cannot be detected by the ELF approach. We also observed no P limitation of phytoplankton communities at either site in any month, even at dissolved inorganic N/P ratios that well exceed the Redfield ratio of 16:1 (calculated from data in our Table 1). While the Redfield ratio of dissolved N/P may not necessarily be a good indicator of phytoplankton nutrient limitation (see review by Geider and LaRoche 2002), the combined evidence points towards a potentially wider role of DOP in the nutrition of phytoplankton from Winyah Bay than that indicated by our ELF-labeling approach. This study highlights the need for continued work towards molecular characterization of the bulk DOP pool and development of cell-specific assays for enzymes (other than alkaline phosphatases) involved in DOP degradation.
References
Ammerman, J.W. 1991. Role of ecto-phosphohydrolases in phosphorus regeneration in estuarine and coastal ecosystems. In Microbial Enzymes in Aquatic Environments, ed. R.J. Chrost, 165–186. New York: Springer.
Ammerman, J.W. and F. Azam. 1985. Bacterial 5′-nucleotidase in aquatic ecosystems: A novel mechanism of phosphorus regeneration. Science 227: 1338–1340. doi:10.1126/science.227.4692.1338.
Benitez-Nelson, C.R. 2000. The biogeochemical cycling of phosphorus in marine systems. Earth Science Reviews 51: 109–135. doi:10.1016/S0012-8252(00)00018-0.
Bentzen, E., W.D. Taylor, and E.S. Millard. 1992. The importance of dissolved organic phosphorus to phosphorus uptake by limnetic plankton. Limnology and Oceanography 37: 217–231.
Björkman, K. and D.M. Karl. 1994. Bioavailability of inorganic and organic phosphorus compounds to natural assemblages of microorganisms in Hawaiian coastal waters. Marine Ecology Progress Series 111: 265–273. doi:10.3354/meps111265.
Cao, X., C. Song, Q. Li, and Y. Zhou. 2007. Dredging effects on P status and phytoplankton density and composition during winter and spring in Lake Taihu, China. Hydrobiologia 581: 287–295. doi:10.1007/s10750-006-0516-2.
Cembella, A.D., N.J. Antia, and P.J. Harrison. 1984. The utilization of inorganic and organic phosphorus compounds as nutrients by eukaryotic microalgae: A multidisciplinary perspective: Part 1. CRC Critical Reviews of Microbiology 10: 317–391. doi:10.3109/10408418209113567.
Conley, D.J., W.M. Smith, J.C. Cornwell, and T.R. Fisher. 1995. Transformation of particle-bound phosphorus at the land-sea interface. Estuarine, Coastal and Shelf Science 40: 161–176. doi:10.1016/S0272-7714(05)80003-4.
Conley, D.J., H.W. Paerl, R.W. Howarth, D.F. Boesch, S.P. Seitzinger, K.E. Havens, C. Lancelot, and G.E. Likens. 2009. Controlling eutrophication: Nitrogen and phosphorus. Science 323: 1014–1015. doi:10.1126/science.1167755.
Cotner, J. and R. Wetzel. 1992. Uptake of dissolved inorganic and organic phosphorus compounds by phytoplankton and bacterioplankton. Limnology and Oceanography 37: 232–243.
Cotner, J.B., J.W. Ammerman, E.R. Pelle, and E. Bentzen. 1997. Phosphorus-limited bacterioplankton growth in the Sargasso Sea. Aquatic Microbial Ecology 13: 141–149. doi:10.3354/ame013141.
Dignum, M., H.L. Hoogveld, H.C.P. Matthijs, H.J. Laanbroek, and R. Pel. 2004. Detecting the phosphate status of phytoplankton by enzyme-labelled fluorescence and flow cytometry. FEMS Microbial Ecology 48: 29–38. doi:10.1016/j.femsec.2003.12.007.
Droop, M.R. 1973. Some thoughts on nutrient limitation in algae. Journal of Phycology 9: 264–272.
Dyhrman, S. 2005. Ectoenzymes in Prorocentrum minimum. Harmful Algae 4: 619–627. doi:10.1016/j.hal.2004.08.011.
Dyhrman, S.T. and B. Palenik. 1999. Phosphate stress in cultures and field populations of the dinoflagellate Prorocentrum minimum detected by a single-cell alkaline phosphatase assay. Applied and Environmental Microbiology 65: 3205–3212.
Dyhrman, S.T. and B. Palenik. 2003. Characterization of ectoenzyme activity and phosphate-regulated proteins in the coccolithophorid Emiliania huxleyi. Journal of Plankton Research 25: 1215–1225.
Dyhrman, S.T. and K.C. Ruttenberg. 2006. Presence and regulation of alkaline phosphatase activity in eukaryotic phytoplankton from the coastal ocean: Implications for dissolved organic phosphorus remineralization. Limnology and Oceanography 51: 1381–1390.
Fang, T.H. 2000. Partitioning and behavior of different forms of phosphorus in the Tanshui Estuary and one of its tributaries, Northern Taiwan. Estuarine, Coastal and Shelf Science 50: 689–70. doi:10.1006/ecss.1999.0604.
Fourqurean, J.W., J.C. Zieman, and V.N. Powell. 1992. Phosphorus limitation of primary production in Florida Bay: Evidence from C:N:P ratios of the dominant seagrass Thalassia testudinum. Limnology and Oceanography 37: 162–171.
Geider, R.J. and J. LaRoche. 2002. Redfield revisited: Variability of C:N:P in marine microalgae and its biochemical basis. European Journal of Phycology 37: 1–17. doi:10.1017/S0967026201003456.
Glibert, P.M., C.A. Heil, D. Hollander, M. Revilla, A. Hoare, J. Alexander, and S. Murasko. 2004. Evidence for dissolved organic nitrogen and phosphorus uptake during a cyanobacterial bloom in Florida Bay. Marine Ecology Progress Series 280: 73–83. doi:10.3354/meps280073.
González-Gil, S., B.A. Keafer, R.V.M. Jovine, A. Aguilera, S. Lu, and D.M. Anderson. 1998. Detection and quantification of alkaline phosphatase in single cells of phosphorus-starved marine phytoplankton. Marine Ecology Progress Series 164: 21–35. doi:10.3354/meps164021.
Guildford, S.J. and R.E. Hecky. 2000. Total nitrogen, total phosphorus, and nutrient limitation in lakes and oceans: Is there a common relationship? Limnology and Oceanography 45: 1213–1223.
Huang, Z., W. You, R.P. Haugland, V.B. Paragas, N.A. Olson, and R.P. Haugland. 1993. A novel fluorogenic substrate for detecting alkaline phosphatase activity in situ. Journal of Histochemistry and Cytochemistry 41: 313–317.
Jansson, M., H. Olsson, and K. Pettersson. 1988. Phosphatases: Origin, characteristics and function in lakes. Hydrobiologia 170: 157–175.
Jeffrey, S., R. Mantoura, and S. Wright. 1997. Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods. Paris: United Nations Educational, Scientific and Cultural Organization Publishing.
Karl, D.M. and K. Björkman. 2002. Dynamics of DOP. In Biogeochemistry of Marine Dissolved Organic Matter, ed. C.A. Carlson, 249–366. San Diego, California: Academic.
Karl, D.M. and K. Yanagi. 1997. Partial characterization of the dissolved organic phosphorus pool in the oligotrophic North Pacific Ocean. Limnology and Oceanography 42: 1398–1405.
Kawaguchi, T., A.J. Lewitus, C.M. Aelion and H.N. McKellar. 1997. Can urbanization limit iron availability to estuarine algae? Journal of Experimental Marine Biology and Ecology 213: 53–69.
Klug, J. 2006. Nutrient limitation in the lower Housatonic River estuary. Estuaries and Coasts 29: 831–840.
Koroleff, F. 1983. Determination of phosphorus. In Methods of Seawater Analysis, ed. K. Grasshoff, M. Ehrhardt, and K. Kremling, 117–156. New York: Verlag-Chemie.
Krom, M.D., N. Kress, S. Benner, and L.I. Gordon. 1991. Phosphorus limitation of primary productivity in the eastern Mediterranean Sea. Limnology and Oceanography 36: 424–432.
Kuenzler, E.J. and J.P. Perras. 1965. Phosphatases of marine algae. Biological Bulletin 128: 271–284. doi:10.2307/1539555.
Labry, C., D. Delmas, and A. Herbland. 2005. Phytoplankton and bacterial alkaline phosphatase activities in relation to phosphate and DOP availability within the Gironde plume waters (Bay of Biscay). Journal of Experimental Marine Biology and Ecology 318: 213–225. doi:10.1016/j.jembe.2004.12.017.
Lawrenz, E., J.L. Pinckney, M.L. Ranhofer and T.L. Richardson. 2009. Spectral irradiance and phytoplankton community composition in a blackwater-dominated estuary, Winyah Bay, South Carolina, USA. Estuarine, Coastal and Shelf Science, in review.
Lebo, M.E. and J.H. Sharp. 1993. Distribution of phosphorus along the Delaware, an urbanized coastal plane estuary. Estuaries 16: 290–301. doi:10.2307/1352502.
Lewitus, A.J. and A.F. Holland. 2003. Initial results from a multi-institutional collaboration to monitor harmful algal blooms in South Carolina. Proceedings of the EMAP Symposium 2001: Coastal Monitoring Through Partnership, Environmental Monitoring and Assessment 81: 361-371. doi:10.1023/A:1021362032676
Lewitus, A.J., D.L. White, R.G. Tymowski, M.E. Geesey, S.N. Hymel, and P.A. Noble. 2005. Adapting the CHEMTAX method for assessing phytoplankton taxonomic composition in southeastern U.S. estuaries. Estuaries 28: 160–172. doi:10.1007/BF02732761.
Li, H., M.J.W. Veldhuis, and A.F. Post. 1998. Alkaline phosphatase activities among planktonic communities in the northern Red Sea. Marine Ecology Progress Series 173: 107–115. doi:10.3354/meps173107.
Litchman, E. and B.L.V. Nguyen. 2008. Alkaline phosphatase activity as a function of internal phosphorus concentration in freshwater phytoplankton. Journal of Phycology 44: 1379–1383. doi:10.1111/j.1529-8817.2008.00598.x.
Lomas, M.W., A. Swain, K. Shelton, and J. Ammerman. 2004. Taxonomic variability of phosphorus stress in Sargasso Sea plankton. Limnology and Oceanography 49: 2303–2310.
Mackey, M.D., D.J. Mackey, H.W. Higgins, and S.W. Wright. 1996. CHEMTAX- A program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Marine Ecology Progress Series 11: 265–283. doi:10.3354/meps144265.
Mackey, K.R.M., R.G. Labiosa, M. Calhoun, J.H. Street, A.F. Post, and A. Paytan. 2007. Phosphorus availability, phytoplankton community dynamics and taxon-specific phosphorus status in the Gulf of Aqaba, Red Sea. Limnology and Oceanography 52: 873–885.
Mallin, M.A., L.B. Cahoon, M.R. McIver, D.C. Parsons, and G.C. Shank. 1999. Alternation of factors limiting phytoplankton production in the Cape Fear River Estuary. Estuaries 22: 825–836. doi:10.2307/1353064.
Meybeck, M. 1993. C, N, P and S in rivers: from sources to global inputs. In Interactions of C, N, P, and S Biogeochemical Cycles and Global Change, ed. R. Wollast, F. MacKenzie, and L. Chou, 163–193. Berlin: Springer.
Monaghan, E.J. and K.C. Ruttenberg. 1999. Dissolved organic phosphorus in the coastal ocean: Reassessment of available methods and seasonal phosphorus profiles from the Eel River Shelf. Limnology and Oceanography 44: 1702–1714.
Mortazavi, B., R.L. Iverson, W.M. Landing, and W. Huang. 2000. Phosphorus budget of Apalachicola Bay: A river-dominated estuary in the northeastern Gulf of Mexico. Marine Ecology Progress Series 198: 33–42. doi:10.3354/meps198033.
Murrell, M.C., R.S. Stanley, E.M. Lores, G.T. Didonato, L.M. Smith, and D.A. Flemer. 2002. Evidence that phosphorus limits phytoplankton growth in a Gulf of Mexico estuary: Pensacola Bay, Florida, U.S.A. Bulletin of Marine Science 70: 155–167.
Nicholson, D., S. Dyhrman, F. Chavez, and A. Payton. 2006. Alkaline phosphatase activity in the phytoplankton communities of Monterey Bay and San Francisco Bay. Limnology and Oceanography 51: 874–883.
Orchard, E., E. Webb, and S.T. Dyhrman. 2003. Characterization of phosphorus-regulated genes in Trichodesmium spp. Biological Bulletin 205: 230–231. doi:10.2307/1543268.
Ou, L., B. Huang, L. Lin, H. Hong, F. Zhang, and Z. Chen. 2006. Phosphorus stress of phytoplankton in the Taiwan Strait determined by bulk and single-cell alkaline phosphatase activity assays. Marine Ecology Progress Series 327: 95–106. doi:10.3354/meps327095.
Patchineelam, S.M., B. Kjerfve, and L.R. Gardner. 1999. A preliminary sediment budget for the Winyah Bay Estuary, South Carolina, USA. Marine Geology 162: 133–144. doi:10.1016/S0025-3227(99)00059-6.
Perry, M.J. 1972. Alkaline phosphatase activity in subtropical Central North Pacific waters using a sensitive fluorometric method. Marine Biology 15: 113–119. doi:10.1007/BF00353639.
Pinckney, J.L., D.F. Millie, K.E. Howe, H.W. Paerl, and J.P. Hurley. 1996. Flow scintillation counting of 14C-labled microalgal photosynthetic pigments. Journal of Plankton Research 18: 1867–1880. doi:10.1093/plankt/18.10.1867.
Pinckney, J.L., H.W. Paerl, M.B. Harrington, and K.E. Howe. 1998. Annual cycles of phytoplankton community structure and bloom dynamics in the Neuse River estuary, North Carolina. Marine Biology 131: 371–381. doi:10.1007/s002270050330.
Rengefors, K., K. Petterson, T. Blenchner, and D.M. Anderson. 2001. Species-specific alkaline phosphatase activity in freshwater spring phytoplankton: Application of a novel method. Journal of Plankton Research 23: 435–443. doi:10.1093/plankt/23.4.435.
Rengefors, K., K. Rutttenberg, C.L. Haupert, C. Taylor, B.L. Howes, and D.M. Anderson. 2003. Experimental investigation of taxon-specific response of alkaline phosphatase activity in natural freshwater phytoplankton. Limnology and Oceanography 48: 1167–1175.
Rinker, K.R. and R.T. Powell. 2006. Dissolved organic phosphorus in the Mississippi River plume during spring and fall 2002. Marine Chemistry 102: 170–179. doi:10.1016/j.marchem.2005.09.013.
Ruttenberg, K.C. and S.T. Dyhrman. 2005. Temporal and spatial variability of dissolved organic and inorganic phosphorus, and metrics of phosphorus bioavailability in an upwelling-dominated coastal system. Journal of Geophysical Research 110: 1–22. doi:10.1029/2004JC002837.
Sakshaug, E., E. Granéli, M. Elbrächter, and H. Kayser. 1984. Chemical composition and alkaline phosphatase activity of nutrient-saturated and P-deficient cells of four marine dinoflagellates. Journal of Experimental Marine Biology and Ecology 77: 241–254. doi:10.1016/0022-0981(84)90122-9.
Scheiner, S. and J. Gurevich. 1993. Design and Analysis of Ecological Experiments. New York: Chapman and Hall.
Schindler, D.W., R.E. Hecky, D.L. Findlay, M.P. Stainton, B.R. Parker, M.J. Paterson, K.G. Beaty, M. Lyng, and S.E.M. Kaisan. 2008. Eutrophication of lakes cannot be controlled by reducing nitrogen input: Results of a 37-year whole-ecosystem experiment. Proceedings of the National Academy of Sciences 105: 11254–11258. doi:10.1073/pnas.0805108105.
Schlüter, L., F. Mǿhlenberg, H. Havskum, and S. Larsen. 2000. The use of phytoplankton pigments for identifying and quantifying phytoplankton groups in coastal areas: Testing the influence of light and nutrients on pigment/chlorophyll a ratios. Marine Ecology Progress Series 192: 49–63. doi:10.3354/meps192049.
Smith, S.V. and M.J. Atkinson. 1984. Phosphorus limitation of net production in a confined ecosystem. Nature 307: 626–627. doi:10.1038/307626a0.
Solórzano, L. and J.H. Sharp. 1980. Determination of total dissolved phosphorus and particulate phosphorus in natural waters. Limnology and Oceanography 25: 754–758.
South Carolina Sea Grant Consortium, 1992. Characterization of the Physical, Chemical and Biological Conditions and Trends in Three South Carolina Estuaries: 1970–1985, Vol. 2. Winyah Bay and North Inlet Estuaries. Charleston, South Carolina: South Carolina Sea Grant Consortium.
Štrojsová, A., J. Vrba, J. Nedoma, and K. Šimek. 2005. Extracellular phosphatase activity of freshwater phytoplankton exposed to different in situ phosphorus concentrations. Marine and Freshwater Research 56: 417–424. doi:10.1071/MF04283.
Suzumura, M., K. Ishikawa, and H. Ogawa. 1998. Characterization of dissolved organic phosphorus in coastal seawater using ultrafiltration and phosphohydrolytic enzymes. Limnology and Oceanography 43: 1553–1564.
Uterhmöhl, H. 1958. Zur Vervollkommung der quantitativen Phytoplankton-Methodik. Mitteilungen der Internationalen Vereinigung für Limnologie 9: 1–38.
Vahtera, E., M. Laamanen, and J.-M. Rintala. 2007. Use of different phosphorus sources by the bloom-forming cyanobacteria Aphanizomenon flos-aquae and Nodularia spumigena. Aquatic Microbial Ecology 46: 225–237. doi:10.3354/ame046225.
van der Zee, C., N. Roevros, and L. Chou. 2007. Phosphorus speciation, transformation and retention in the Scheldt estuary (Belgium/The Netherlands) from the freshwater tidal limits to the North Sea. Marine Chemistry 106: 76–91. doi:10.1016/j.marchem.2007.01.003.
Wright, S.W., D.P. Thomas, H.J. Marchant, H.W. Higgins, M.D. Mackey, and D.J. Mackey. 1996. Analysis of phytoplankton of the Australian sector of the Southern Ocean: Comparisons of microscopy and size frequency data with interpretation of pigment HPLC data using the ‘CHEMTAX’ matrix factorization program. Marine Ecology Progress Series 11: 285–298. doi:10.3354/meps144285.
Acknowledgements
We gratefully acknowledge the hard work of Steven Schmidt, Kent Ray, Jean-Marie Buschur, Elyse Walker, Max Bangs, and Dale Soblo who collected and filtered many liters of water. Thanks to Renée Styles and Lois Lane for technical assistance. Special thanks to Sonya Dyhrman and Karin Rengefors for help with the ELF assay and for answering multiple questions from TLR. We are grateful to two anonymous reviewers for their constructive comments. This work was funded in part by the South Carolina Sea Grant Program (grant numbers P/M-2 J-V410 and R/ER 29), and the Slocum-Lunz Foundation (to MLR).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ranhofer, M.L., Lawrenz, E., Pinckney, J.L. et al. Cell-Specific Alkaline Phosphatase Expression by Phytoplankton from Winyah Bay, South Carolina, USA. Estuaries and Coasts 32, 943–957 (2009). https://doi.org/10.1007/s12237-009-9180-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12237-009-9180-x