Abstract
Among the applications of DNA barcoding for plant conservation is the identification of illegally traded endangered species from small samples or vegetative specimens. DNA barcoding offers an important tool for the phytosanitary authorities to identify species belonging to groups such as the bamboos and orchids, which command high prices in the horticultural trade. In this study we created a DNA barcode library for 20 endangered Orchidaceae species and 36 species of bamboo (Bambusoideae, Poaceae) distributed in Mexico. We applied several metrics to evaluate the efficiency of the barcodes matK and rbcL and, for bamboos, that of the plastid spacer psbI-K. Our results coincide with those of previous barcoding projects in which alone matK allowed for the identification of the most orchid species. For bamboos, the psbI-K spacer retrieved more polymorphic sites and in combination with matK we were able to identify bamboos to at least the generic level.
Resumen
Una de las aplicaciones del código de barras genético en la conservación de plantas es la de identificar especies amenazadas e ilegalmente comerciadas. El código de barras genético puede constituir una herramienta importante para las autoridades fitosanitarias para determinar especímenes de grupos como bambúes y orquídeas, que alcanzan precios elevados en horticultura. En este trabajo se creó una biblioteca de ADN para 20 especies amenazadas de Orchidaceae y 36 especies de bambúes de la subfamilia Bambusoideae distribuidas en México. Se aplicaron varios métodos para evaluar la eficiencia de los códigos de barras matK y rbcL y para los bambúes también se secuenció el espaciador psbI-K. Los resultados coinciden con proyectos previos, en que utilizando únicamente matK es posible identificar más especies de orquídeas. Se encontró una gran variación en las secuencias de psbI-K para bambúes y en combinación con matK fue posible identificar un mayor número de géneros.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
At present, DNA is commonly used as a source of evidence in comparative biology. It has brought important changes to molecular biology, molecular systematics, genomics, bioinformatics and even to alpha-taxonomy (Miller, 2007; Goldstein & DeSalle, 2011). However, nucleotide sequences are also applied to identify organisms (Hebert et al., 2003a). This use is known as DNA barcoding, as a nod to the application of unique combinations of variable-width vertical marks to identify products in commercial activities (Hebert et al., 2003b; Stoeckle & Hebert, 2008; reviewed by Brower, 2006 and Meier, 2008; Cräutlein et al., 2011). The use of the DNA barcode is not only intended for taxonomic work to identify overlooked species, but also has agricultural, clinical, ecological, forensic, illegal trade-related and even recreational applications (Janzen, 2004; Hebert & Gregory, 2005; Schindel & Miller, 2005, Stoeckle & Hebert, 2008, Alacs et al., 2010).
DNA barcoding in land plants is based on standardized regions of the chloroplast genome. Following a debate and a long period of testing, a 2-core-locus consisting of the coding regions matK and rbcL was suggested by the Consortium for the Barcode of Life (CBOL) as the valid alternative for DNA barcoding (CBOL Plant Working Group, 2009). Prior to this, both coding regions were widely used in plant phylogenetic studies (reviewed by Palmer et al., 2004). To reach this conclusion, the Plant Working Group compared the performance of seven leading candidate plastid DNA regions: atpF-atphH spacer, matK gene, rbcL gene, rpoB gene, rpoC1 gene, psbK-psbI spacer, and trnH-psbA spacer (CBOL Plant Working Group, 2009).
Among the applications of DNA barcoding for plant conservation is the identification of illegally traded endangered species from small samples or vegetative specimens. DNA barcoding offers an important tool for the phytosanitary authorities to identify species in groups such as bamboos, cacti, cycads and orchids, all of which fetch high prices in the horticultural trade, and in many cases are regulated by CITES agreement for the international trade of endangered species.
Orchids are highly sought after by collectors and nurseries worldwide and are in high demand. In Mexico, 188 species of Orchidaceae are classified as endangered; 60 % are critically endangered and moreover Laelia gouldiana Rchb.f. is considered extinct (SEMARNAT, 2001). Populations are being decimated by habitat destruction and because plants are extracted from natural habitats for illegal export and sale in local markets (Flores-Palacios & Valencia-Díaz, 2007; Chow, 2010). Another plant group whose species are also heavily collected from native vegetation is the woody bamboos (Bambusoideae, Poaceae). These plants have large, showy stems, which are used for constructing buildings and for making handicrafts. At present, bamboo cane is a good substitute for timber and an increasing number of nurseries are becoming involved in their trade (Lobokivov, 2003). Of the 41 bamboo species found in Mexico, 38 are endemic and restricted to a few localities; most of the species in two genera, Olmeca Soderstr. and Otatea (McClure & W.W. Sm.) C.E. Calderón & Soderstr. are endangered. Particularly, Otatea glauca L.G. Clark & G. Cortés, O. ximenae Ruiz-Sanchez & L. G. Clark and Olmeca zapotecorum Ruiz-Sanchez, Sosa & Mejía-Saulés are critically endangered (Ruiz-Sanchez et al., 2008, 2011).
In this paper we set out to create a DNA barcode library for 20 endangered Orchidaceae species and 36 species of bamboo distributed in Mexico. We apply several metrics to evaluate the efficiency of the matK and rbcL barcodes and, for the bamboos, that of the plastid spacer psbI-K, which was one of the candidate barcode loci evaluated by CBOL.
Materials and Methods
Sampling
A total of 20 endangered species of Orchidaceae were included in the study. They were selected from the Mexican Red List (SEMARNAT, 2001) and moreover they are found in cloud forests of the mountains of the Gulf of Mexico, one of the most endangered habitats in the country. Tissues were obtained mostly from the living collection of the Botanical Garden Clavijero or in the field (Appendix 1). Of the 41 species of bamboo found in Mexico, 36 were analysed. They were collected in the field as part of the project of establishing the collection of Mexican native bamboos at the Botanical Garden Clavijero. In order to determine variation of matK and rbcL among individuals of the same species, for Arthrostylidium excelsum, Olmeca fulgor, Olmeca reflexa and Rhipidocladum martinezii we obtained nucleotide sequences for four or five individuals (Table 1). For orchids we were not able to get several individuals of same species. Vouchers are indicated in Appendix 1.
DNA Sequencing
DNA was isolated using either the modified 2× CTAB method (Rogers & Bendich, 1985; Doyle & Doyle, 1987; Cota-Sánchez et al., 2006) or the DNeasy Plant Mini Kit (Quiagen, Valencia, California), following the manufacturer’s instructions. For bamboos the rbcL and matk genes were amplified and sequenced using the primer pairs rbcLaf - bcLajf634R for the former and matK Xf - matk 5r for the latter, and following the protocols outlined by Erickson et al. (2008), CBOL Plant Working Group (2009), and Dunning & Savolainen (2010). The psbK-psbI spacer was amplified and sequenced using primers and protocols by Lahaye et al. (2008). For orchids, the primers rbcLa_f (Erickson et al., 2008; CBOL Plant Working Group, 2009) and rbcLajf-634R (Fazekas et al., 2008, CBOL Plant Working Group, 2009) were utilized to amplify rbcL. For matK the primer pais Xf - 5r was utilized (Ford et al., 2009; CBOL Plant Working Group, 2009). For both the bamboo and the orchid sequences, amplified double-stranded DNA fragments were purified using QIAquick columns (Qiagen) following the protocols provided by the manufacturer and subsequently were sequenced at a commercial facility (Macrogen INC. Seoul, Korea). GenBank accession numbers are recorded in Appendix 1. Electropherograms were edited and assembled using Sequencher 4.1 (Gene Codes, Ann. Arbor, MI). Sequences were aligned manually using the program Se-Al v. 2.0a11 (Rambaut, 2002).
Evaluation of Barcodes
Analyses of nucleotide polymorphisms were performed for aligned matrices of every barcode with the program eDnaSP (DNA Sequence Polymorphism, v. 3.99, Rozas et al., 2003). Inter- and intra-specific genetic divergences were calculated using every DNA barcode locus. Average pairwise distances were estimated between every species, as well as for the four or five accessions in the selected bamboo species. For each single barcode and in combination, pairwise distances were calculated with the simplest K2P model following Lahaye et al. (2008) with the software MEGA 5.0 (Tamura et al., 2011). This model also utilizes the CBOL recommendations for distance calculations (barcoding.si.edu/). Phylogenetic analyses were carried out to evaluate if species or genera were retrieved as monophyletic with each barcode following Lahaye et al. (2008) and thus able to identify species or genera. Parsimony analyses were run in TNT (Goloboff et al., 2003), under a traditional search, with equal weights and 10,000 iterations saving 100 trees per run. Clade support was estimated by bootstrap (BS), with 1,000 replicates. In addition, UPGMA trees were inferred from K2P distances with MEGA 5.0 (Tamura et al., 2011) and evaluated by bootstrap.
Results
Molecular Characteristics
Amplification was successful for each barcode tested, for bamboos as well for orchids. Therefore, nucleotide sequences from all the taxa were successfully amplified and sequenced. Table 1 includes DNA sequence statistics for every DNA barcode and possible combinations. In the orchids, matK was the plastid sequence with the highest number of sequence polymorphic sites, while for the bamboos it was psbI-K. The second barcode locus proposed by CBOL, rbcL, retrieved the lowest polymorphisms. For bamboos the combination of matK + psbI-K retrieved more polymorphic sites than the combination of matk + rbcL.
Inter- and Intra-Specific Diversity
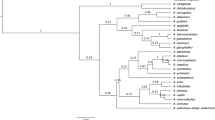
The performance of each barcode and all their combinations was assessed by means of the genetic distance from the K2P (Kimura’s two parameters) pairwise distance matrices (Table 2, Fig. 1). The highest inter-specific diversity was reached by matK for Orchidaceae and for intra-specific variation in bamboos, and the combination of matK with psbI-K for Bambusoideae. For orchids and bamboos (intra- and inter-specific variation) rbcL had the lowest distances.
Species Identification
The performance of each DNA barcode in identifying and delineating species was assessed by the percentage of monophyletic groups recovered by Parsimony and UPGMA analyses (Table 3, Fig. 1). The highest values of species monophyly were obtained from UPGMA reconstruction in comparison with parsimony reconstruction. One hundred percent of monophyly was retreived with matrices based on matK alone in both orchids and individuals of same species in bamboos and for inter-specific monophyly of genera in bamboos. Percentage of monophyly retrieved by inter-specific matrices with matK was identical to the retrieved by the combination of matK-psbI-K for species of bamboos. The lowest percentage of monophyly in both bamboos and orchids was reached by rbcL.
Discussion
For orchids our results are concordant with those reported by Asahina et al. (2010), in that with matK alone it was possible to identify more taxa than with rbcL. These authors obtained sequences of the two markers for five medicinal species in Dendrobium Sw. Moreover, our results also match those obtained Gigot et al. (2007) who found small genetic distances among Mesoamerican orchid species (intra- and inter-specific variation) utilizing plastid regions proposed in phase 2 of the DNA barcode protocol for plants, including rbcL and matK. The Mesoamerican orchids we studied can be identified to at least the genus level by just sequencing matK. Lahaye et al. (2009) also concluded that the most variable DNA barcode is matK for a broader array of species in several families found at Kruger National Park, South Africa. By contrast, to identify tropical plant species in Panama and tropical tree species in Puerto Rico Kress et al. (2009, 2010) utilized a three-locus barcode.
There is still no DNA barcoding project for species in Subfamily Bambusoideae. The only record for Poaceae is the detection of a cryptic species in Tripogon Roem. & Schult. (subfamily Chloridoideae) based on matK and trnH-psbA sequences (Ragupathy et al., 2009). In our study we further estimated variation of matK, psbI-K and rbcL in 4 or 5 individuals of the same bamboo species. We found that variation is very low and thus with the combination of the three barcodes is possible to identify species just sequencing only one sample.
In Mesoamerica the cycads comprise another group of plants that is highly threatened and vulnerable to illegal trade. Of the approximately 55 species of cycads from Mexico, 80 % are endangered (Vovides, 2000; SEMARNAT, 2001). Nicolalde-Morejón et al. (2011) examined the performance of seven plastid DNA regions considered in phase 2 by CBOL and the nuclear ITS region as barcodes for the cycad genera Ceratozamia, Dioon and Zamia. They concluded that for the successful identification of the species, it would be necessary to sequence four or five markers. Thus in comparison with bamboos and orchids, in which a two- locus barcode is useful to identify at the level of species, cycads require two or three additional barcodes to recognize genera.
Our results indicate that the identification of endangered Mesoamerican orchids is achieved at the generic level using only matK. Moreover, the failure to identify species of bamboos with the 2-core locus barcode system indicates that the proposed barcode combination needs further refinement, as indicated by a number of previous studies (reviewed by Vijayan & Tsou, 2011). However, considering that it is noteworthy that the 20 orchid species included in here are not as closely related as the studied bamboos. We sample species from most of the Orchidaceae clades; while, for the bamboos our sampling was restricted to subfamily Bambusoideae. Bamboos in Mesoamerica are closely related and belong to the subtribes Guaduinae, Arthrostyliidinae and Chusqueinae grouped in this subfamily (Ruiz-Sanchez et al., 2008, 2011).
Our results concord with other barcoding projects, in which matK retrieved more variation in land plant groups. In orchids, the identification of more species was achieved utilizing only matK. For bamboos we found that the spacer psbI-K retrieved more polymorphic sites and in combination with matK we were able to identify bamboos to at least the generic level.
Literature Cited
Alacs, E. A., A. Georges, N. N. FitzSimmons & J. Robertson. 2010. DNA detective: a review of molecular approaches to wildlife forensics. Forensic Science Medicine and Pathology 6: 180–194.
Asahina, H., J. Shinozaki, K. Masuda, Y. Morimitsu & M. Satake. 2010. Identification of medicinal Dendrobium species by phylogenetic analysis using matK and rbcL sequences. Journal of Natural Medicines 64: 133–138.
Brower, A. V. Z. 2006. Problems with DNA barcodes for species delimitation: ‘ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae). Systematics and Biodiversity 4: 127–132.
CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences 106: 12794–12797.
Chow, Y. N. 2010. Orchid smugglers and the use of biotechnology to combat them. Asia-Pacific Journal of Molecular Biology and Biotechnology 18: 175–179.
Cota-Sánchez, J. H., K. Remrchuk & K. Ubayasena. 2006. Ready to use DNA extracted with CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Molecular Biology Reporter 24: 161–167.
Cräutlein, M. V., H. Korpelainen, M. Pietiläinen & J. Rikkinen. 2011. DNA barcoding: a tool for improved taxon identification and detection of species diversity. Biodiversity and Conservation 20: 373–389.
Doyle, J. J. & J. L. Doyle. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
Dunning, L. T. & V. Savolainen. 2010. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Botanical Journal of the Linnean Society 164: 1–9.
Erickson, D. L., J. Spouge, A. Resch, L. A. Weigt & W. J. Kress. 2008. DNA barcoding in land plants: developing standards to quantify and maximize success. Taxon 57: 1304–1316.
Fazekas, A. J., K. S. Burgess, P. R. Kesanakurti, S. W. Graham, S. G. Newmaster, B. C. Husband, D. M. Percy, M. Hajibabaei & S. C. H. Barrett. 2008. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3: 1–12.
Flores-Palacios, A. & S. Valencia-Díaz. 2007. Local illegal trade reveals unknown diversity and involves a high species richness of wild vascular epiphytes. Biological Conservation 136: 372–387.
Ford, C. S., K. L. Ayres, N. Toomey, N. Haider, J. V. Alphen-Stahl, L. J. Kelly, N. Wikström, P. M. Hollingsworth, R. J. Duff, S. B. Hoot, R. S. Cowan, M. W. Chase & M. J. Wilkinson. 2009. Selection of candidate coding DNA barcoding regions for use on land plants. Botanical Journal of the Linnean Society 159: 1–11.
Gigot, G., J. V. Alphen-Stahl, D. Bogarin, J. Warner, M. W. Chase & V. Savolainen. 2007. Finding a suitable DNA barcode for Mesoamerican orchids. Lankesteriana 7: 200–203.
Goldstein, P. Z. & R. DeSalle. 2011. Integrating DNA barcode data and taxonomic practice: determination, discovery and description. Bioessays 33: 135–147.
Goloboff, P. A., J. S. Farris & K. Nixon. 2003. TNT: tree analysis using new technology. Version 1.0, Beta test v. 0.2. Systematic Biology 54: 176–178.
Hebert, P. D. N. & T. R. Gregory. 2005. The promise of DNA barcoding for taxonomy. Systematic Biology 54: 852–859.
———, S. Ratnasingham & J. R. deWaard. 2003a. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B (suppl.) 270: 96–99.
———, A. Cywinska, S. L. Ball & J. R. de Waard. 2003b. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B 270: 313–32-1.
Janzen, D. H. 2004. Now is the time. Philosophical Transactions of the Royal Society of London B 359: 731–732.
Kress, W. J., D. L. Erickson, F. A. Jones, N. G. Swenson, R. Perez, O. Sanjur & E. Bermingham. 2009. Plant DNA barcodes and community phylogeny of a tropical forest dynamics plot in Panama. PNAS 106: 18621–18626.
———, ———, N. G. Swenson, J. Thompson, M. Uriarte & J. K. Zimmerman. 2010. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. Plos One 5: e15409.
Lahaye, R., V. Savolainen, S. Duthoit, O. Maurin & M. van der Bank. 2008. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park as a model system (South Africa). Nature Precedings hd1:10101/npre.2008.1896.1. Posted 16 May 2008.
———, M. van der Bank, D. Bogarin, J. Warner, F. Pupulin, G. Gigot, O. Maurin, S. Duthoit, T. G. Barraclough & V. Savolainen. 2009. DNA barcoding the floras of biodiversity hotspots. Proceedings of the National Academy of Sciences of U.S.A. 105: 2923–2928.
Lobokivov, M. 2003. Bamboo and rattan products and trade. Journal of Bamboo and Rattan 2: 397–406.
Meier, R. 2008. DNA sequences in taxonomy, opportunities and challenges in Wheeler, Q. D. (ed.), The new taxonomy systematics. The Systematics Association Special Volume Series 76: 65–127.
Miller, S. E. 2007. DNA barcoding and the renaissance of taxonomy. Proceedings of the National Academy of Sciences 104: 4775–4776.
Nicolalde-Morejón, F., F. Vergara-Silva, J. González-Astorga, D. W. Stevenson, A. P. Vovides & V. Sosa. 2011. A character-based approach in the Mexican cycads supports diverse multigene combinations for DNA barcoding. Cladistics 27: 150–164.
Palmer, J. D., D. E. Soltis & M. W. Chase. 2004. The plant tree of life: an overview and some points of view. American Journal of Botany 91: 1437–1455.
Ragupathy, S., S. G. Newmaster, M. Murugesan & V. Balasubramaniam. 2009. DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the Hill tribes of the Western Ghats in southern India. Molecular Ecology Resources 9: 164–171.
Rambaut, A. 2002. Se-Al sequence alignment Editor, V2. 0a11. Department of Zoology. University of Oxford, Oxford.
Rogers, S. O. & A. J. Bendich. 1985. Extraction of DNA from milligram amounts of fresh, herbarium, and mummified plant tissues. Plant Molecular Biology 5: 69–76.
Rozas, J., J. C. Sánchez-Del Barrio, X. Messeguer & R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Ruiz-Sanchez, E., V. Sosa & M. T. Mejia-Saules. 2008. Phylogenetics of Otatea inferred from morphology and chloroplast DNA sequence data and recircumscription of Guaduinae (Poaceae: Bambusoideae). Systematic Botany 33: 277–283.
———, ——— & ———. 2011. Molecular phylogenetics of the Mesoamerican bamboo Olmeca (Poaceae, Bambuseae): Implications for taxonomy. Taxon 60: 89–98.
Schindel, D. E. & S. E. Miller. 2005. DNA barcoding a useful tool for taxonomists. Nature 435: 17.
SEMARNAT. 2001. Norma Oficial Mexicana NOM-059-ECOL-2001. Protección ambiental – Especies nativas de México de flora y fauna silvestres – Categorías de riesgo y especificaciones para su inclusión, exclusión o cambio – Lista de especies en riesgo. Diario Oficial de la Federación. México.
Stoeckle, M. Y. & P. D. Hebert. 2008. Bar code of life: DNA tags help classify life. Scientific American 299: 82–88.
Tamura, K., D. Peterson, N. Peterson, G. Stecher, M. Nei & S. Kumar. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739.
Vijayan, K. & C. H. Tsou. 2011. DNA barcoding in plants: taxonomy and a new perspective. Current Science 99: 1530–1541.
Vovides, A. P. 2000. México: segundo lugar mundial en diversidad de cícadas. Biodiversitas 6: 6–10.
Acknowledgments
We thank Javier Francisco Ortega and Angélica Cibrián for inviting us to the “ADN y la conservación de la diversidad vegetal en América Latina” symposium, part of the “X Congreso Latinoamericano de Botánica” in La Serena, Chile (October, 2010). We acknowledge funding from CONABIO (HB004 “Barcode de bambúes nativos de México”). We thank Arith Pérez and Claudia Navarro for assisting with the lab work. Bianca Delfosse improved the English. Ismael Guzmán-Valdivieso and José M. Pech helped with the analyses. Eduardo Ruiz Sanchez helped collect the bamboos and Milton Díaz helped collect the orchids. Sequencing the Orchidaceae was part of the B.Sc. thesis project carried out at the Universidad Veracruzana by the third author of this paper.
Author information
Authors and Affiliations
Corresponding author
Appendix 1
Appendix 1
Rights and permissions
About this article
Cite this article
Sosa, V., Mejía-Saules, T., Cuéllar, M.A. et al. DNA Barcoding in Endangered Mesoamerican Groups of Plants. Bot. Rev. 79, 469–482 (2013). https://doi.org/10.1007/s12229-013-9129-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12229-013-9129-4