Abstract
The Pilosella alpicola group includes four species (P. alpicola s.str., P. ullepitschii, P. rhodopea and P. serbica) with allopatric distributions (Alps, Balkans, Carpathians) and contrasting cytotype patterns (diploid, diploid-polyploid and polyploid species). Whereas diploid taxa (P. ullepitschii and P. serbica) reproduce sexually, the mode of reproduction of polyploid cytotypes reflects their origin: autopolyploids of P. rhodopea reproduce sexually, while allopolyploid cytotypes of P. alpicola s.str. apomictically. We used allozymes to elucidate overall genetic variation within the group and to test their utility for taxon discrimination, assessment of polyploid origin and possible correlations with breeding systems. Variation of five allozyme systems encoded by eight polymorphic loci and 29 alleles was studied in 20 populations and 298 plants representing all taxa. Allozymes were proved to be only of limited usefulness for the taxonomic classification within the P. alpicola group. The Western Carpathian populations of P. ullepitschii formed the only genetically well-differentiated group. The same allele suite shared by all cytotypes of P. rhodopea and presence of both balanced and unbalanced heterozygotes in tetraploids was consistent with autopolyploid origins of polyploids and provided further evidence for a primary contact zone. An isolated relic population of P. rhodopea from the Southern Carpathians exhibited lowered values of genetic diversity when compared to the core area. Pronounced fixed heterozygosity was found in P. alpicola s.str., supporting its allopolyploid origin. In accordance with assumptions, genotypic variability was significantly higher in sexually reproducing diploid and diploid-polyploid taxa than in apomictic P. alpicola s.str.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the genus Pilosella Vaill. (syn. Hieracium subgenus Pilosella (Vaill.) Gray, Asteraceae, Lactuceae) are characterized by the presence of several ploidy levels in natural populations (2x–8x, based on x = 9), diverse breeding systems (self-incompatibility, facultative/obligate apomixis, vegetative reproduction via stolons) and widespread interspecific hybridization (reviewed in Krahulcová et al. 2000). The joint effect of these processes has led to a reticulate pattern of diversity reflected in contradictory taxonomic concepts (Nägeli and Peter 1885; Zahn 1922–1930; Tyler 2001; Bräutigam and Greuter 2007).
The Pilosella alpicola group encompasses four closely related, but morphologically distinct taxa distributed allopatrically (Šingliarová et al. 2011). Formerly, they were treated as subspecies of the Pilosella alpicola (Steud. & Hochst.) F.W. Schultz & Sch. Bip. (Zahn 1930). A cytogeographical survey of the group (Šingliarová and Mráz 2009; Šingliarová et al. 2011) revealed a taxon-specific pattern. Pilosella alpicola s.str. is a tetra- and pentaploid species (2n = 4x = 36 and 5x = 45) with an allopatric cytotype distribution in the Swiss and Italian Alps, respectively. The Carpathian endemics P. ullepitschii (Błocki) Szeląg and P. serbica (F.W. Schultz & Sch. Bip.) Szeląg from Serbia are exclusively diploid (2n = 2x = 18) and strictly self-incompatible (Šingliarová and Mráz 2009; Šingliarová et al. 2011). Populations of the Balkan subendemic P. rhodopea (Griseb.) Szeląg are diploid, diploid-polyploid or consist of several polyploid cytotypes (2n = 2x, 3x, 4x, 5x; Šingliarová and Mráz 2009; Šingliarová et al. 2011).
Our previous biosystematic study confirmed the close relatedness of the four recognized species of the P. alpicola group and indicated an autopolyploid as well as an allopolyploid origin of the polyploids (Šingliarová et al. 2011). Sexual reproduction has been proved in diploid-polyploid P. rhodopea, which, coupled with the absence of morphological and molecular (internal transcribed spacer) differentiation between cytotypes, suggests an autopolyploid origin of polyploids. On the contrary, an additive pattern of ITS polymorphism suggested hybridogeneous origin of polyploid P. alpicola s.str., putatively arising from P. rhodopea and Alpine P. glacialis (Reyn. ex Lachen.) F.W. Schultz & Sch. Bip. (= Hieracium angustifolium Hoppe). Moreover, an apomictic mode of reproduction was proved in this taxon (Šingliarová et al. 2011). Although the ITS sequences provided valuable data on the ‘rough’ phylogenetic relationship of the P. alpicola group to the other members of the genus (Fehrer et al. 2007; Šingliarová et al. 2011), as well as on the origin of polyploids (Šingliarová et al. 2011), there is no information about genetic differentiation within this group, and consequences of breeding systems and origins of polyploids for standing genetic variation.
Allozymes have been proved useful for establishing relationships at low systematic levels (Gottlieb 1984). As codominant markers, they can help elucidate origins of polyploids (Roose and Gottlieb 1976; Soltis and Rieseberg 1986; Soltis and Soltis 1989; Mahy et al. 2000; Rosquist and Prentice 2002; Crawford et al. 2006) or hybrids (Brochmann et al. 1992; Gauthier et al. 1998; Mráz et al. 2005) and indirectly estimate the breeding system within a population (Sipes and Wolf 1997; Sydes and Peakall 1998). Allozymes are especially suitable for detection of differences in population genetic structure between sexual and apomictic taxa (Bayer and Crawford 1986; Hughes and Richards 1988; Yahara 1990; Bayer 1991; Noyes and Soltis 1996; Hörandl et al. 2000, 2001; Hörandl and Greilhuber 2002; Kashin et al. 2005) due to different levels and patterns of heterozygosity and genotypic diversity. Allozymes have been used successfully also in several population genetic studies in the genus Pilosella (Krahulec et al. 2004; Kashin et al. 2005; Peckert et al. 2005; Tyler 2005; Bruun et al. 2007; Krahulcová et al. 2009a). In our previous paper, we used allozymes to compare the genetic diversity between the populations of P. ullepitschii from the core area (Western Carpathians) and isolated populations originated from Romanian Eastern and Southern Carpathians (Šingliarová et al. 2008).
The present study aimed to elucidate the amount and pattern of genetic variation within the Pilosella alpicola group using allozyme markers. The main objectives were to establish:
-
i)
the degree of genetic differentiation among four recognized species. Considering the clear morphological separation of species (Šingliarová et al. 2011), we would expect a similar pattern using allozymes.
-
ii)
molecular evidence for auto- and allopolyploid origin of polyploids. Different criteria of genetic diversity are predicted for these two types of polyploids (Soltis and Soltis 2000; Ramsey and Schemske 2002). In the case of autopolyploids (P. rhodopea), polysomic inheritance resulting in multiallelic loci and the presence of both balanced and unbalanced heterozygotes is expected. Allopolyploid P. alpicola s.str., however, should be characterized by disomic inheritance mirrored in fixed heterozygosity. The distinction can be, however, imperfect in many instances, as auto- and allopolyploidy represent only opposite ends of a wide spectrum of intergenome differentiation (Stebbins 1980; Soltis and Soltis 2000; Obbard et al. 2006).
-
iii)
correlation between genetic diversity and breeding systems operating within the group. Breeding system was proved to crucially affect the patterns of genetic variation (Hamrick and Godt 1989; Karron 1991; Sun and Wong 2001). In general, sexual taxa (diploid P. serbica and P. ullepitschii and diploid-polyploid P. rhodopea) are expected to be more variable than their apomictic counterpart polyploid P. alpicola s.str.
Material and Methods
Sampling
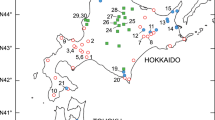
The plants were collected from their natural populations in 2005–2007 during extensive field survey across the whole range of the group (Table 1 , Fig. 1 ) and transferred into the experimental garden of the Institute of Botany, Slovak Academy of Sciences in Bratislava. If possible, the plants were collected in at least 4 m intervals. Altogether 298 plants originated from 20 populations from Switzerland, Italy, Slovakia, Romania, Serbia and Bulgaria were sampled (Table 1 , Fig. 1 ). Eleven populations (166 plants) of P. ullepitschii were allozymatically studied by Šingliarová et al. (2008), but only four systems (except LAP, see below) and six loci were scored. For the purposes of the present study we included these populations into the data set. Moreover, six plants of P. ullepitschii from population SMA were additionally analyzed for the first time. The number of plants analyzed per population ranged from 11 to 24 (Tables 1 and 2 ; Electronic Supplementary Material 1 ), except for population KOP (type locality of P. serbica) from which only four plants were studied because the remaining collected plants died during transport or in early stages of cultivation.
The ploidy level of all analyzed plants was assessed during a large-scale cytogeographical survey (Šingliarová et al. 2011), and populations were assigned to particular species according to the results of multivariate morphometric analyses (Šingliarová et al. 2011).
Electrophoresis
Young leaves were sampled from living cultivated plants and immediately used for allozyme extraction. Approximately 1 cm2 (ca. 40 mg) of fresh leaf tissue was ground in an ice-cold extraction buffer following Kato (1987): 0.1 M Tris-HCl (pH = 8.0), 70 mM mercaptoethanol, 26 mM sodium metabisulphite, 11 mM L-ascorbic acid, 4% (w/v) soluble PVP-40, the pH adjusted after the addition of the ascorbate, with Dowex Cl (1-8X). Crude homogenates were centrifuged at 15,000 rpm for 10 min. Supernatants were stored at –75°C. Polyacrylamid gel electrophoresis (PAGE) was carried out using 8.16% separation polyacrylamid gel with the buffer 1.82 M Tris-HCl, pH 8.9; 4% stacking gel with the buffer (0.069 M Tris-HCl, pH 6.9) and electrode buffer (0.02 M Tris, 0.24 glycine, pH 8.3).
PAGE was initially performed on six isozyme systems of which five turned out to be polymorphic and interpretable; their abbreviations and EC numbers are in parentheses: shikimate dehydrogenase SKDH (monomeric, EC 1.1.1.25), phosphoglucomutase PGM (monomeric, EC 5.4.2.2.), 6-phosphogluconate dehydrogenase 6-PGDH (dimeric, EC 1.1.1.44), superoxide dismutase SOD (dimeric, EC 1.15.1.1), leucine aminopeptidase LAP (monomeric, EC 3.4.11.1). Dihydrolipoamiddehydrogenase DIA (EC 1.6.4.3) was not interpretable.
Systems 6-PGDH and PGM displayed two zones of activity, and the LAP system showed three, interpreted as two or three putative loci respectively (e.g., 6-Pgdh-1, 6-Pgdh-2, with ‘1’ coding for the faster locus). Alleles were designated sequentially with the fastest one coded as ‘a’.
In total, nine polymorphic loci representing five enzyme systems were scored. The fastest locus of LAP (Lap1) had a clear and interpretable pattern in all plants from the Western Carpathians (P. ullepitschii), from the Alps (P. alpicola s.str.) and in 85% of P. rhodopea from the Balkans. Conversely, only a strong and consistent smear was observed in samples from the Eastern and Southern Carpathians (P. ullepitschii, P. rhodopea), Serbia (P. serbica) and in some polyploid plants from Bulgaria (P. rhodopea). This locus was therefore omitted, and only eight loci were subsequently used for evaluation of genetic diversity. Banding patterns of particular enzyme systems were interpreted with regard to their quaternary structure following Weeden and Wendel (1989). No deviations from expected patterns have been observed. Despite the generally complicated genetic interpretation in polyploids (Gottlieb 1981; Weeden and Wendel 1989; Obbard et al. 2006), the resolution of gels and staining intensity enable us to take relative band intensity into account. Phenotypes were compared to banding patterns of related diploids and interpreted as corresponding to genotypes of different allelic dosages (see e.g., Arft and Ranker 1998; Hörandl et al. 2000; Hardy and Vekemans 2001).
Genetic Analyses
To provide a measure of the level of genetic variation within populations of the Pilosella alpicola group, the following population genetics parameters were computed: P – proportion of polymorphic loci, where the frequency of the most common allele is less than or equal to 0.99 and 0.95 (including monomorphic loci with fixed heterozygotes), ΣA – sum of alleles, A – mean number of alleles per locus, H o – observed heterozygosity, H e – expected heterozygosity (computed for diploids and tetraploids only). The parameters were computed using POPGENE version 1.32 (Yeh et al. 1999) for diploids; for polyploids they were calculated by hand. Genotypic variation was estimated using standard parameters for clonal populations (Ellstrand and Roose 1987; Eckert and Barrett 1993): number of genotypes (G), proportion of distinguishable genotypes (i.e., number of distinguishable genotypes to the number of samples, G/N) and number of unique genotypes (G uni ). Multilocus genotype diversity D (modified Simpson diversity index) was calculated as \( D = 1 - \sum\nolimits_i {\left[ {{n_i}\left( {{n_i} - 1} \right)} \right]/N\left( {N - {1}} \right)]} \), where n i is the number of individuals of genotype i (Pielou 1969; but see also Noyes and Soltis 1996; Hörandl et al. 2000; Liston et al. 2003). The D value ranges from 0 to 1, 0 meaning that all individuals represent the same multilocus genotype and 1 meaning that each individual has a unique multilocus genotype. All parameters were generated for i) each population, ii) each of the four species analyzed and iii) particular cytotypes of P. rhodopea. Wright’s (1951) deficit or excess of heterozygotes (F IS ) in sexual populations (for diploid taxa and for diploids or tetraploids in P. rhodopea populations, where diploid or tetraploid plants prevailed) was calculated for each polymorphic locus and for overall loci as Weir and Cockerham’s (1984) estimates using the FSTAT software (Goudet 1995) for diploids and by hand for tetraploids.
Hierarchical clustering (UPGMA), principal component (PCA) and principal coordinate analyses (PCoA) were performed to gain insight into the genetic relationships within the P. alpicola group. UPGMA was based on Nei's standard genetic distances calculated from allele frequencies (Nei 1972), PCA was based on allele frequencies, and PCoA was based on Jaccard's coefficient of dissimilarity computed from presence/absence of alleles. Based on Nei’s standard genetic distance, we computed genetic identity \( \left( {I = 1/\exp \left( {{D_a}} \right)} \right) \). Karyologically uniform populations and each ploidy level found in mixed-ploidy populations were either considered as separate OTUs (operational taxonomic units; PCA) or different ploidy levels from the same population were merged together (PCoA, UPGMA). Differences in genetic parameters (P95, P99, A, ∑A, H o , G and G/N) among three taxa (P. alpicola s.str., P. rhodopea and P. ullepitschii) were tested using the TukeyHSD post hoc comparison test. Because P. alpicola was represented by only two populations, we also performed separate two-sample t-tests with Welch approximation to test for the difference between P. rhodopea and P. ullepitschii with a higher number of populations analyzed. Genetic parameters violating the normality assumption (assessed by Shapiro-Wilk test) were tested using non-parametric Wilcoxon (two species) or Kruskal-Wallis sum rank tests when comparing three species. The correlation between pairwise Nei genetic distances and geographical distances was inferred using Mantel test with 999 permutations. All analyses were performed using various functions implemented in the ‘ade4’ and ‘adegenet’ packages (Chessel et al. 2004; Jombart 2008) and basic ‘stats’ package within R environment (R Development Core Team 2009).
Results
Eight loci yielded a total of 29 alleles. The allele frequencies are shown in the Electronic Supplementary Material 1 . Fifteen alleles were common to all taxa; however, they were not always present in all populations. Two alleles were shared by P. ullepitschii and P. rhodopea, two by P. ullepitschii and P. alpicola s.str., and one each by the P. alpicola s.str. and P. rhodopea pair, and P. rhodopea and P. serbica pair. Four alleles (Pgm2-c, 6Pgdh1-d, 6Pgdh2-b, 6Pgdh2-d) were unique to P. rhodopea, and one species-specific allele was detected in each of the diploid taxa P. ullepitschii (6Pgdh2-e) and P. serbica (Pgm1-c). Unique alleles were not evenly distributed in populations of particular species, but they were always found in plants of a single population (Electronic Supplementary Material 1 ). Only P. alpicola s.str. lacks any private alleles at either of the eight loci scored. However, if we included the Lap1 locus, which was absent in many individuals of other species (see Material and Methods), P. alpicola s.str. exhibited three unique alleles at this locus. Specifically, Lap1-a was moderately frequent in tetraploid plants from the Simplon Pass (SIM), and alleles Lap1-b and Lap1-c were present in all pentaploid plants studied from the Val de Duron valley (DUR).
In most of the plants, at most one or two alleles per locus were recorded. Only in five polyploid plants of P. rhodopea, all from the BOT population (representing 8% of all P. rhodopea polyploids), were found three alleles at the Pgm2 locus. Similarly, loci Lap1 and Lap2 in pentaploid P. alpicola s.str. (DUR) consistently possessed three alleles.
Within particular species, the highest genetic identity (I = 0.944) was found between two P. alpicola s.str. populations (Table 3 ). Slightly lower values of genetic identity were observed between P. rhodopea populations (mean = 0.894, ranging from 0.808 to 0.975). The lowest values (mean = 0.852) of the parameter along with the widest range (from 0.646 to 0.989) were detected between P. ullepitschii populations. As for the genetic identities between taxa, the highest values were observed between P. alpicola s.str. and P. rhodopea (from 0.809 to 0.965, mean = 0.903), while pair of taxa P. serbica and P. ullepitschii displays the lowest values (from 0.678 to 0.837, mean = 0.781). Remaining pairs of taxa show moderate values of genetic identity.
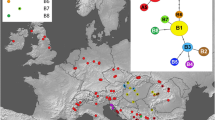
Cluster and ordination analyses based on allele frequencies (UPGMA, PCA) and presence/absence of alleles (PCoA) were fairly congruent. All analyses revealed clear separation of the Western Carpathian populations of P. ullepitschii from the rest (Figs. 2 , 3 and 4 , Table 3 ). In UPGMA (Fig. 2 ), the populations of P. ullepitschii from the Western Carpathians were separated at the highest level of dissimilarity. The second cluster consisted of two sub-clusters. The first subcluster was formed by populations of P. ullepitschii from the Eastern Carpathians (NMA, SMA) and P. rhodopea from the Southern Carpathians (COZ), indicating a rather geographical grouping. The second subcluster contained a mixture of P. alpicola s.str., P. rhodopea, P. serbica and P. ullepitschii populations from the Southern Carpathians (BUC).
Cluster analysis (UPGMA) of 20 populations of the Pilosella alpicola group based on Nei’s (1972) genetic distance. Population codes are given in Table 1 . Pilosella alpicola s.str. (N = 2, black circles), P. rhodopea (N = 6, white circles with dot), P. serbica (N = 1, open triangle), P. ullepitschii (N = 11, black pentagons). Only bootstrap values (based on 999 permutations) above 50% are shown
Principal component analysis (PCA) of populations of the Pilosella alpicola group based on allelic frequencies of karyologically uniform populations and particular cytotypes from ploidy-mixed populations. Pilosella alpicola s.str. (N = 2, black circles), P. rhodopea (N = 12, circles with dot), P. serbica (N = 1, open triangle), P. ullepitschii (N = 11, black pentagons) as OTUs. For population codes see Table 1
Principal coordinates analysis (PCoA) of populations of the Pilosella alpicola group based on Jaccard’s coefficient of dissimilarity. Pilosella alpicola s.str. (N = 2, black circles), P. rhodopea (N = 6, white circles with dot), P. serbica (N = 1, open triangle), P. ullepitschii (N = 11, black pentagons) as OTUs. For population codes see Table 1
In the ordination analyses (Figs. 3 and 4 ), only populations of the Western Carpathian P. ullepitschii (PCA, PCoA) and population of P. serbica (KOP) (PCA) can be distinguished, while the Romanian populations of P. ullepitschii were only slightly differentiated (PCoA) or intermingled (PCA) with the populations of P. alpicola s.str., P. rhodopea and P. serbica.
There was no significant association between Nei’s genetic distances and geographical distances (Mantel test, r = 0.152, P = 0.064). However, when two P. alpicola s.str. populations were removed from the analysis, the correlation was highly significant (Mantel test, r = 0.657, P < 0.001; Fig. 5 ).
Correlations between pairwise genetic distances (according to Nei 1972) among populations of the Pilosella alpicola group and their pairwise geographical distances. Regression line including all populations is represented by the full line, while the dashed line shows the correlation after removing two P. alpicola s.str. populations. Black dots denote pairwise comparisons between populations of P. ullepitschii, P. serbica and P. rhodopea, while gray dots denote comparisons between populations of P. alpicola s.str. and populations of three remaining taxa
Genetic diversity values at both species and population level are given in Table 2 . The highest values for most of genetic diversity parameters (P99, P95, A, ΣA) were found in diploid P. serbica; however, the small sample size (four plants) could significantly bias the values. Therefore, the values of genetic parameters for this species are given in all tables but they are not further discussed. Mean genetic diversity estimates per population and species of P. rhodopea were slightly higher (P99 = 70.8, P95 = 62.5, ΣA = 25, A = 2) than those of P. alpicola s.str. (P99 = 75, P95 = 56.3, ΣA = 18, A = 2) and P. ullepitschii (P99 = 57.3, P95 = 56.5, ΣA = 22, A = 1.72). Values of observed heterozygosity (H o ) averaged per population were similar in P. ullepitschii (0.232) and P. rhodopea (0.294), while those observed in P. alpicola s.str. were approximately two fold higher (0.53). Similarly, the highest levels of expected heterozygosity (H e ) was recorded in tetraploid populations of P. alpicola s.str. (H e = 0.540) and P. rhodopea (H e = 0.571 and 0.419), while values observed in diploid populations were considerably lower − P. ullepitschii (H e = 0.263), P. serbica (H e = 0.371), P. rhodopea (H e = 0.212 and 0.349). In P. rhodopea, H o and H e of particular populations increased from pure diploid (COZ, H o = 0.205, H e = 0.212) through diploid-polyploid (BOT, H o = 0.301, H e for tetraploids = 0.419 and DOD, H o = 0.330, H e of diploids = 0.349) to polyploid populations (BEZ, H o = 0.344 and MUS, H o = 0.491, H e of tetraploids = 0.571). The only exception was polyploid population VICH (3x+4x) with the lowest proportion of heterozygotes among all analyzed populations (H o = 0.108). In sexual taxa (P. ullepitschii, P. serbica and all cytotypes of P. rhodopea), values of observed heterozygosity averaged for particular loci covered a whole spectrum between zero and one (Table 4 ). On the contrary, mean observed heterozygosity for particular loci in apomictic P. alpicola s.str. was equal or close to zero (homozygous) or one (heterozygous, see Table 4 ). No significant differences in parameters P95, P99, A, ∑A, H o , G and G/N were found among tested taxa (TukeyHDS test when comparing P. alpicola s.str., P. rhodopea and P. ullepitschii; or two-sample t-test, when comparing P. rhodopea and P. ullepitschii; results not shown).
The most striking difference between sexual taxa (P. ullepitschii, P. rhodopea) and apomictic P. alpicola s.str. was found in genotypic variability (G, G/N and D, see Table 2 ). P. alpicola str. showed significantly reduced clonal variation when compared to both sexual taxa (G: TukeyHSD test, P < 0.05 for both comparisons; G/N: Kruskal-Wallis test, P = 0.064; D: Kruskal-Wallis test, P = 0.048).
Pooled values of genetic and genotypic diversity for particular cytotypes of P. rhodopea (Table 5 ) revealed that both genetic diversity (D) and genotypic variation (G/N) were highest in diploids and tetraploids, while in triploids they were lower. The mean heterozygosity of P. rhodopea tetraploids (H o = 0.458) was nearly double that of diploids and triploids (H o = 0.255 and 0.240, respectively).
Inbreeding coefficients calculated for sexual taxa (Table 6 ) markedly varied among loci and populations of particular taxa. Except P. serbica, populations of all taxa displayed both excess and deficit of heterozygotes of rather moderate character.
Discussion
Correspondence between Allozyme and Morphological Differentiation
Although many studies showed allozymes to be useful taxonomic marker (e.g., Samuel et al. 1990; Bayer 1991; Hörandl 2004; Ruiz et al. 2004), our allozymic results were rather incongruent with previously detected morphological variation of the Pilosella alpicola group (Šingliarová et al. 2011). We propose two non-exclusive explanations for the low resolution of allozymes in the P. alpicola group: i) the group has diversified quite recently and the morphologically well differentiated taxa had insufficient time to accumulate taxon-specific mutations at the protein level. The low number of taxon-specific alleles detected in the present study supports this hypothesis. Furthermore, no parsimoniously informative ITS site was found in 21 sequenced accessions and this supports a very recent origin of the P. alpicola member species (Šingliarová et al. 2011). In such a situation, low allozymic variation and high level of shared ancestral polymorphism is not surprising. ii) As suggested by Tyler (2005), frequent interspecific hybridization observed in the genus Pilosella could potentially blur genetic differentiation between species. Indeed, we observed some interspecific hybrids between the members of the P. alpicola group and other Pilosella species, but they were less frequent when compared to the lowland taxa. In addition to their intermediate morphology, these interspecific hybrids showed an additive pattern of ITS polymorphism corroborating their hybrid origin (Šingliarová and Mráz, unpubl.). However, no such ITS pattern, neither conspicuous morphology (indicating hybrid origin), nor introgression were recorded in the P. alpicola plants included in our study. Based on this, interspecific hybridization seems not to be a primary reason of weak allozymic differentiation of the P. alpicola group.
The Western Carpathian populations of P. ullepitschii formed the only well-differentiated group, while the Romanian populations of P. ullepitschii were clustered with the populations of P. alpicola s.str. from the Alps and P. rhodopea from the Balkan Peninsula (Figs. 2 , 3 and 4 ). Such pattern could result from longer geographical separation of the Western Carpathian populations from the remaining populations. Indeed, several phylogeographic studies of high mountain plants confirmed the existence of a strong genetic barrier between the Western Carpathians and Southeastern Carpathians (e.g., Mráz et al. 2007; Ronikier et al. 2008; Těšitel et al. 2009). Alternatively, such separation could be caused by other processes such as genetic drift due to population size reduction or recent founder event(s) of isolated Romanian populations (cf. Šingliarová et al. 2008).
Genetic relatedness of the Romanian P. ullepitschii and P. rhodopea populations could be tentatively explained by recent contact of both taxa in this area. Although extremely rare and allopatrically distributed, both taxa are still present in the Southern Carpathians (see above). Indeed, the distributional and genetic pattern of many mountain species suggested biogeographical connections between the Bulgarian mountains and the Southern Carpathians (Reed et al. 2004; Puşcaş et al. 2008; Varga and Schmitt 2008; Schmitt 2009). It has been demonstrated that, despite the assumed selective neutrality of allozyme markers (e.g., Kimura 1968), environmental factors (temperature, humidity) can promote a shift in allele frequencies in populations through directional selection mechanisms (Koehn and Hilbish 1987; Karl and Avise 1992; Prentice et al. 2000; Karl et al. 2009). Thus, more similar environmental conditions between the southeastern Carpathians and the Balkans might be another reason of grouping of southeastern Carpathian P. ullepitschii and P. rhodopea populations.
The intermingled position of P. alpicola s.str. populations within populations of other species (Figs. 2 and 3 ) might be surprising considering the geographical distance, striking morphological differences and allopolyploid origin (Šingliarová et al. 2011). The grouping with P. rhodopea populations can nonetheless be explained because this species was suggested to be one of the putative parental species of the allopolyploid P. alpicola s.str. (Šingliarová et al. 2011). A different level of genomic dosage of both putative parental taxa, with putatively greater influence of P. rhodopea (P. alpicola str. is morphologically more similar to P. rhodopea than to P. glacialis), could have shaped the allozyme pattern of P. alpicola s.str. In spite of its hybrid origin, no unique alleles were found in P. alpicola s.str. in eight loci that were interpretable for all taxa and regions. If the three unique alleles in the Lap1 locus, however, had been included in the analyses, they would have contributed to better separation of P. alpicola s.str. from the Balkan populations. Moreover, different alleles recorded in two subareas (Lap1-a in the tetraploids from SIM and Lap1-b and c in the pentaploids from DUR) indicate a polytopic origin of allopatric cytotypes.
Allozyme Variation in Diploid-Polyploid Pilosella rhodopea
Allozyme variation ascertained for diploid-polyploid P. rhodopea in the present study is consistent with the hypothesis of an autopolyploid origin (Šingliarová et al. 2011). All cytotypes possessed almost identical suites of alleles (Electronic Supplementary Material 1 ). The only two unique alleles in P. rhodopea missing in diploids (6Pgdh2-b, 6Pgdh2-d) were present in one triploid plant from the BEZ population, where we found only 3x and 4x plants (Šingliarová et al. 2011). Considering that also diploids were previously detected from the same locality by Krahulcová et al. (2009b), these unique alleles could be a part of common allele suite of unsampled diploids. The third unique allele found in P. rhodopea (Pgm2-c) was frequent in all cytotypes at the BOT locality (2x, 3x, 4x), the only population from the Stara planina Mts analyzed. Because all cytotypes shared this unique allele, we consider it evidence of in situ origin of polyploid cytotypes. Alternatively, the triploid and tetraploid cytotypes might still be later immigrants to the already established diploid population and could have acquired this allele through frequent intercytotype gene-flow. Independent autopolyploid origin of polyploids within diploid populations is supported also by previously established commonness of mixed ploidy populations (75%) and absence of morphological and ITS differentiation (Šingliarová et al. 2011). While there are numerous examples of secondary contact zones of previously allopatric cytotypes (e.g., Thompson and Lumaret 1992; Hardy et al. 2000; Husband and Sabara 2004; Schlaepfer et al. 2008; Kolář et al. 2009), evidence for primary contact zones is still quite sparse (Soltis and Soltis 1989; Weiss et al. 2002; Stuessy et al. 2004; Kolář et al. 2009; Trávníček et al. 2011).
Observed heterozygosity in polyploids of P. rhodopea is in agreement with the expectation that polyploids exhibit higher heterozygosity when compared to diploid relatives (Soltis and Rieseberg 1986; Soltis and Soltis 1989, 1993). Values detected in tetraploids (0.458, Table 5 ) resembled those published for other autopolyploid taxa (e.g., 0.43 for Dactylis, Soltis and Soltis 1993; 0.474 for Lotus, Gauthier et al. 1998; 0.54 for Centaurea, Hardy and Vekemans 2001; 0.472 for Thymus, López-Pujol et al. 2004). Heterozygous tetraploids of P. rhodopea exhibited both balanced and unbalanced allele dosages, H o across loci was variable (Table 4 ), and there is no evidence for fixed heterozygozity. These findings indicate polysomic inheritance and thus an autopolyploid origin of P. rhodopea polyploids (Soltis and Soltis 1993). Conversely, estimates of genetic diversity (P, A) are lower for polyploids of P. rhodopea when compared to the diploids.
Although the polyploid state allows for the presence of three or more alleles at a single locus (Soltis and Rieseberg 1986; Soltis and Soltis 1989; Mahy et al. 2000), more than two alleles per locus was recorded only in small portion of P. rhodopea polyploids (8%) when compared to other autopolyploids (e.g., 37% for Thymus loscosii, López-Pujol et al. 2004). Nevertheless, similar values were also reported in the literature (e.g., 12% for Vaccinium oxycoccos, Mahy et al. 2000).
Significant loss of genetic diversity was found in isolated Romanian populations of P. ullepitschii (Šingliarová et al. 2008). Because of secondarily formed habitats, we assumed that these populations experienced a strong bottleneck probably due to a founder effect mediated by human activities. A similar pattern was found in the remote diploid P. rhodopea population in the Cozia Mts (Southern Carpathians). When compared with diploids from the main range in the Balkans (DOD population), COZ population displayed markedly lower values in all genetic parameters (Table 2 ). Considering relic habitat of P. rhodopea in the Cozia Mts (crevices in steep rock cliffs) and presence of several endemic and relic taxa (Nyárády 1955; Popescu et al. 1970), long-term vicariance scenario with genetic depauperation of small isolated population appears the most plausible.
Allozyme Variation in Allopolyploid Pilosella alpicola s.str.
Contrary to the previous assumption of a combination of two divergent genomes (Šingliarová et al. 2011), allozyme phenotypes of exclusively polyploid P. alpicola s.str. lacked unique alleles in eight shared loci. Three unique alleles, however, were found at the locus Lap1, which has not been scorable for all analyzed plants of the P. alpicola group (see Material and Methods). These unique alleles could originate from the second parental taxon – P. glacialis; unfortunately this species has not been included in the analyses. Nevertheless, the pronounced fixed heterozygosity observed at several loci (three for tetraploids and four for pentaploids, see Table 4 and Electronic Supplementary Material 1 ) is convincing evidence of allopolyploid origin of the species. The values of observed heterozygosity of P. alpicola s.str. at individual loci were either close to zero or close to one, as usual in allopolyploids with disomic inheritance (Roose and Gottlieb 1976; Soltis and Soltis 1990; Arft and Ranker 1998).
Allozyme Variation and Mode of Reproduction
The patterns of allozyme variation within the Pilosella alpicola group agree with the mode of reproduction detected by Šingliarová and Mráz (2009) and Šingliarová et al. (2011). The populations of sexually reproducing species were genetically more variable than apomictic populations of P. alpicola s.str. (Table 2 ).
Despite significant loss of genetic diversity in isolated Romanian populations of P. ullepitschii (Table 2 , and Šingliarová et al. 2008), mean genetic variation found in diploid P. ullepitschii was even higher than average values published for other sexual plants (P and A, Hamrick and Godt 1989). Values of averaged observed heterozygosity were similar or slightly exceeding those in other diploid sexual taxa (Tolmiea, Soltis and Rieseberg 1986; Taraxacum, Hughes and Richards 1988; Lotus, Gauthier et al. 1998; Hemerocallis, Kang and Chung 2000; Ranunculus, Hörandl et al. 2000). However, even higher values are known for sexual diploid taxa from apomictic complexes (e.g., Kashin et al. 2005 for Taraxacum and Pilosella; Hoebee et al. 2006 for Sorbus). Multilocus genotypic measures (G/N and D, Table 2 ) reached values close to maximum, which fits well the assumption for self-incompatible species (Ellstrand and Roose 1987).
Diploid and diploid-polyploid populations of sexual P. rhodopea (COZ, DOD, BOT) displayed genotypic diversity comparable to diploid P. ullepitschii. Decreased genotype variability, however, was found in polyploid populations (BEZ, VICH, MUS). Though the lower values of G/N for the MUS population were mainly due to genotypically uniform pentaploids, in two populations from the Pirin Mts, they were apparently caused by higher clonality of triploids (Tables 2 and 5 ). The triploid cytotype, not frequent in the genus Pilosella (Schuhwerk 1996 and references therein), predominates in most mixed-ploidy populations of P. rhodopea (Šingliarová et al. 2011). A similar shift of proportion of cytotypes in the field in favour of triploids was observed also in Pilosella echioides (Peckert and Chrtek 2006; Trávníček et al. 2011). All populations with decreased genotype variability were formed only by polyploids (Šingliarová et al. 2011 and this study). We therefore hypothesize that these populations could have been founded by a small number of polyploid plants representing only a fraction of variability present in the source population(s). Moreover, the significantly decreased value of mean observed heterozygosity in the VICH population indicated a higher level of inbreeding. According to G/N values, vegetative reproduction could also play a role in the success of triploids. However, we did not observe increased formation of accessory rosettes in triploids, which could form independent ramets (Šingliarová, unpubl.). Alternatively, it cannot be ruled out that lower genotypic diversity may be a result of apomixis, but no asexually formed seeds were detected in polyploids of P. rhodopea so far (Šingliarová et al. 2011).
Averaged values of genotypic diversity (G/N, D) of apomictic P. alpicola s.str. (G/N = 0.292, D = 0.467, Table 2 ) are consistent with values detected in other apomictic taxa (Pilosella echioides G/N = 0.05–0.5, D = 0–0.55, Peckert et al. 2005; Hieracium alpinum G/N = 0.1–0.133, D = 0.453–0.66, Štorchová et al. 2002; Chrtek and Plačková 2005; G/N = 0.06, D = 0.87 in Erigeron annuus, Ellstrand and Roose 1987; G/N = 0.1, D = 0.49 in E. compositus, Noyes and Soltis 1996; G/N = 0.12–0.13, D = 0.2–0.35 in Ranunculus variabilis, Hörandl et al. 2001).
When considering two P. alpicola s.str. populations separately, the tetraploid population (SIM) showed a lower value of genotypic diversity when compared to the other taxa (see above) but much higher when compared to the pentaploid DUR population, where only two multilocus genotypes were detected. This might indicate relatively frequent sexual recombination and thus facultative character of apomixis in tetraploids. Indeed, facultative apomixis is quite common in polyploid Pilosella taxa, especially with even ploidies, whereas it is more obligate in odd-ploid Pilosella including pentaploids (Koltunow et al. 1998; Krahulcová et al. 2000, 2004; Krahulec et al. 2004; Fehrer et al. 2005; for rare exceptions see Pogan and Wcisło 1995; Rotreklová et al. 2002).
Conclusion
Our study showed that genetic differentiation among morphologically well-defined species of the P. alpicola group inferred by allozymes was rather low. We assume that recent diversification of the group shaped this pattern and more discriminative markers should be used in the future to achieve possibly better taxonomic separation. The type of polyploidy (auto- vs allo-) and reproduction modes, however, were proved to have significant impact on genetic and genotypic diversity observed in particular taxa. Close genetic relatedness of cytotypes found in ploidy-mixed populations of the P. rhodopea suggests the existence of primary contact zones of diploid and polyploid cytotypes. In this light, the P. rhodopea populations represent a promising system to explore evolutionary processes involved in coexistence of conspecific cytotypes.
References
Arft AM, Ranker TA (1998) Allopolyploid origin and population genetics of the rare orchid Spiranthes diluvialis. Amer J Bot 85:110–122
Bayer RJ (1991) Allozymic and morphological variation in Antennaria (Asteraceae: Inuleae) from the low arctic of Northwestern North America. Syst Bot 16:492–506
Bayer RJ, Crawford DJ (1986) Allozyme divergence among five diploid species of Antennaria (Asteraceae, Inuleae) and their allopolyploid derivatives. Amer J Bot 73:287–296
Bräutigam S, Greuter W (2007) A new treatment of Pilosella for the Euro-Mediterranean flora [Notulae ad floram euro-mediterraneam pertinentes No. 24]. Willdenowia 37:123–137
Brochmann C, Soltis DE, Soltis PS (1992) Electrophoretic relationships and phylogeny of Nordic polyploids in Draba (Brassicaceae). Pl Syst Evol 182:35–70
Bruun HH, Scheepens JF, Tyler T (2007) An allozyme study of sexual and vegetative regeneration in Hieracium pilosella L. Canad J Bot 85:10–15
Chessel D, Dufour AB, Thioulouse J (2004) The ade4 package-I- One-table methods. R News 4:5–10
Chrtek J Jr, Plačková I (2005) Genetic variation in Hieracium alpinum (Asteraceae) in the Krkonoše Mts (West Sudeten Mts, Czech Republic). Biologia, Bratislava 60:387–391
Crawford DJ, Archibald JK, Santos-Guerra A, Mort ME (2006) Allozyme diversity within and divergence among species of Tolpis (Asteraceae-Lactuceae) in the Canary Islands: Systematic, evolutionary, and biogeographical implications. Amer J Bot 93:656–664
R Development Core Team (2009) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Eckert CG, Barrett SCH (1993) Clonal reproduction and patterns of genotypic diversity in Decodon verticillatus (Lythraceae). Amer J Bot 80:1175–1182
Ellstrand NC, Roose ML (1987) Patterns of genotypic diversity in clonal plant species. Amer J Bot 74:121–131
Fehrer J, Šimek R, Krahulcová A, Krahulec F, Chrtek J Jr, Bräutigam E, Bräutigam S (2005) Evolution, hybridisation, and clonal distribution of apo- and amphimictic species of Hieracium subgen. Pilosella (Asteraceae: Lactuceae) in a Central European mountain range. In Bakker FT, Chatrou LW, Gravendeel B, Pelser P (eds) Plant species-level systematics: new perspectives on pattern & process. Regnum Veg 143:175–201
Fehrer J, Gemeinholzer B, Chrtek J Jr, Bräutigam S (2007) Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Molec Phylogen Evol 42:347–361
Gauthier P, Lumaret R, Bédécarrats A (1998) Genetic variation and gene flow in Alpine diploid and tetraploid populations of Lotus (L. alpinus (D.C.) Schleicher/L. corniculatus L.). I. Insights from morphological and allozyme markers. Heredity 80:683–693
Gottlieb LD (1981) Electrophoretic evidence and plant populations. Progr Phytochem 7:1–46
Gottlieb LD (1984) Isozyme evidence and problem solving in plant systematics. In Grant WF (ed) Plant biosystematics. Academic Press, London, pp 343–357
Goudet J (1995) FSTAT: a computer program to calculate F-statistics (version 2.9.3). J Heredity 86:485–486
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In Brown AHD, Clegg MT, Kahler AL, Wier BS (eds) Plant population genetics. Sinauer, Sunderland, Massachusetts, pp 43–63
Hardy OJ, Vekemans X (2001) Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evolution 55:943–954
Hardy OJ, Vanderhoeven S, De Loose M, Meerts P (2000) Ecological, morphological and allozymic differentiation between diploid and tetraploid knapweeds (Centaurea jacea) from a contact zone in the Belgian Ardennes. New Phytol 146:281–290
Hoebee SE, Menn C, Rotach P, Finkeldey R, Holderegger R (2006) Spatial genetic structure of Sorbus torminalis: The extent of clonal reproduction in natural stands of a rare tree species with a scattered distribution. Forest Ecol Managem 226:1–8
Hörandl E (2004) Comparative analysis of genetic divergence among sexual ancestors of apomictic complexes using isozyme data. Int J Pl Sci 165:615–622
Hörandl E, Greilhuber J (2002) Diploid and autotetraploid sexuals and their relationships to apomicts in the Ranunculus cassubicus group: insights from DNA content and isozyme variation. Pl Syst Evol 234:85–100
Hörandl E, Greilhuber J, Dobeš C (2000) Isozyme variation within the apomictic Ranunculus auricomus complex: evidence for a sexual progenitor species in southeastern Austria. Pl Biol 2:1–10
Hörandl E, Jakubowsky G, Dobeš C (2001) Isozyme and morphological diversity within apomictic and sexual taxa of the Ranunculus auricomus complex. Pl Syst Evol 226:165–185
Hughes J and Richards AJ (1988) The genetic structure of populations of sexual and asexual Taraxacum (dandelions). Heredity 60:161–171
Husband BC, Sabara HA (2004) Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytol 161:703–713
Jombart T. (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kang SS, Chung MG (2000) High levels of allozyme variations and low allozyme divergence within and among species of Hemerocallis (Liliaceae). Amer J Bot 87:1634–1646
Karl SA, Avise JC (1992) Balancing selection at allozyme loci in oysters: implications from nuclear RFLPs. Science 256:100–102
Karl I, Schmitt T, Fischer K (2009) Genetic differentiation between alpine and lowland populations of a butterfly is related to PGI enzyme genotype. Ecography 32:488–496
Karron JD (1991) Patterns of genetic variation and breeding systems in rare plant species. In Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, Oxford, pp 87–98
Kashin AS, Anfalov VE, Demochko YuA (2005) Studying allozyme variation in sexual and apomictic Taraxacum and Pilosella (Asteraceae) populations. Russ J Genet 41:144–154
Kato T (1987) Hybridization between Dianthus superbus var. longicalycinus and D. shinanensis evidenced by resolvable esterase isozymes from herbarium specimens. Ann Tsukuba Bot Gard 6:9–18
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626
Koehn RK, Hilbish TJ (1987) The adaptative importance of genetic variation. Amer Sci 75:134–141
Kolář F, Štech M, Trávníček P, Rauchová J, Urfus T, Vít P, Kubešová M, Suda J (2009) Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Ann Bot (Oxford) 103:963–974
Koltunow AM, Johnson SD, Bicknell RA (1998) Sexual and apomictic development in Hieracium. Sex Pl Reprod 11:213–230
Krahulcová A, Krahulec F, Chapman HM (2000) Variation in Hieracium subgen. Pilosella (Asteraceae): What do we know about its sources? Folia Geobot 35:319–338
Krahulcová A, Papoušková S, Krahulec F (2004) Reproduction mode in the allopolyploid facultatively apomictic hawkweed Hieracium rubrum (Asteraceae, H. subgen. Pilosella). Hereditas 141:19–30
Krahulcová A, Rotreklová O, Krahulec F, Rosenbaumová R, Plačková I (2009a) Enriching ploidy level diversity: the role of apomictic and sexual biotypes of Hieracium subgen. Pilosella (Asteraceae) that coexist in polyploid populations. Folia Geobot 44:281–306
Krahulcová A, Vladimirov V, Krahulec F, Bräutigam S (2009b) The agamic complex of Pilosella (Asteraceae) in Bulgaria and SW Romania: variation in ploidy level and breeding systems. Phytol Balcan 15:377–384
Krahulec F, Krahulcová A, Fehrer J, Bräutigam S, Plačková I, Chrtek J Jr (2004) The Sudetic group of Hieracium subgen. Pilosella from the Krkonoše Mts: a synthetic view. Preslia 76:223–243
Liston A, Wilson BL, Doescher PS, Robinson WA, Harris N, Svejcar T (2003) Genetic evidence for sexual and clonal reproduction in a 59-year old population of Festuca idahoensis (Poaceae). Oecologia 137:216–225
López-Pujol J, Bosch M, Simon J, Blanche C (2004) Allozyme diversity in the tetraploid endemic Thymus loscossi (Lamiaceae). Ann Bot (Oxford) 93:323–332
Mahy G, Bruederle LP, Connors B, Van Hofwegen M, Vorsa N (2000) Allozyme evidence for genetic autopolyploidy and high genetic diversity in tetraploid cranberry, Vaccinium oxycoccos (Ericaceae). Amer J Bot 87:1882–1889
Mráz P, Chrtek J Jr, Fehrer J, Plačková I (2005) Rare recent natural hybridization in Hieracium s.str. – evidence from morphology, allozymes and chloroplast DNA. Pl Syst Evol 255:177–192
Mráz P, Gaudeul M, Rioux D, Gielly L, Choler P, Taberlet P, IntraBioDiv Consortium (2007) Genetic structure of Hypochaeris uniflora (Asteraceae) suggests vicariance in the Carpathians and rapid post-glacial colonisation of the Alps from an Eastern Alpine refugium. J Biogeogr 34:2100–2114
Nägeli C, Peter A (1885) Die Hieracien Mittel-Europas. Monographishe Bearbeitung der Piloselloiden mit besonderer Berücksichtigung der mitteleuropäischen Sippen I. R. Oldenburg, München.
Nei M (1972) Genetic distances between populations. Amer Naturalist 106:283–292
Noyes RD, Soltis DE (1996) Genotypic variation in agamospermous Erigeron compositus (Asteraceae). Amer J Bot 83:1292–1303
Nyárády EI (1955) Vegetaţia muntelui Cozia şi cîteva plante noi pentru flora Olteniei, Moldovei şi Transilvaniei (Vegetation of the Cozia Mts and some new plants for the flora of Oltenia, Moldova and Transylvania). Bul Şti Secţ Sţi Biol, Agron, Geol Geogr 7:209–246
Obbard DJ, Harris SA, Pannell JR (2006) Simple allelic-phenotype diversity and differentiation statistics for allopolyploids. Heredity 97:296–303
Peckert T, Chrtek J Jr (2006) Mating interactions among coexisting diploid, triploid and tetraploid cytotypes of Hieracium echioides (Asteraceae). Folia Geobot 41:323–334
Peckert T, Chrtek J Jr, Plačková I (2005) Genetic variation in agamospermous populations of Hieracium echioides in southern Slovakia and northern Hungary (Danube Basin). Preslia 77:307–315
Pielou EC (1969) An introduction to mathematical ecology. John Wiley, New York.
Pogan E, Wcisło H (1995) Embryological analysis of Hieracium pilosella L. from Poland. Acta Biol Cracov, Ser Bot 37:53–61
Popescu A, Sanda V, Roman N, Şerbănescu GH, Doniţă N (1970) Investigations on the Olt gorge flora. Rev Roum Biol 15:259–269
Prentice HC, Lönn M, Lager H, Rosén E, van der Maarel E (2000) Changes in allozyme frequencies in Festuca ovina populations after a nine-year nutrient/water experiment. J Ecol 88:331–347
Puşcaş M, Choler P, Tribsch A, Gielly L, Rioux D, Gaudeul M, Taberlet P (2008) Post-glacial history of the dominant alpine sedge Carex curvula in the European Alpine System inferred from nuclear and chloroplast markers. Molec Ecol 17:2417–2429
Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annual Rev Ecol Syst 33:589–639
Reed JM, Kryštufek B, Eastwood WJ (2004) The physical geography of the Balkans and nomenclature of place names. In Griffths HI, Kryštufek B, Reed JM (eds) Balkan biodiversity pattern and process in the European hotspot. Kluwer Academic Publishers, Dordrecht, pp 9–22
Ronikier M, Cieślak E, Korbecka G (2008) High genetic differentiation in the alpine plant Campanula alpina Jacq. (Campanulaceae): evidence for glacial survival in several Carpathian regions and long isolation between the Carpathians and the Eastern Alps. Molec Ecol 17:1763–1775
Roose ML, Gottlieb LD (1976) Genetic and biochemical consequences of polyploidy in Tragopogon. Evolution 30:818–830
Rosquist G, Prentice HC (2002) Genetic variation in Scandinavian Anthericum liliago (Anthericaceae): allopolyploidy, hybridization and immigration history. Pl Syst Evol 236:55–72
Rotreklová O, Krahulcová A, Vaňková D, Peckert T, Mráz P (2002) Chromosome numbers and breeding systems in some species of Hieracium subgen. Pilosella from Central Europe. Preslia 74:27–44
Ruiz E, Crawford DJ, Stuessy TF, Gonzalez F, Samuel R, Becerra J, Silva M (2004) Phylogenetic relationships and genetic divergence among endemic species of Berberis, Gunnera, Myrceugenia and Sophora of the Juan Fernandez Islands (Chile) and their continental progenitors based on isozymes and nrITS sequences. Taxon 53:321–332
Samuel R, Pinsker W, Ehrendorfer F (1990) Allozyme polymorphism in diploid and polyploid populations of Galium. Heredity 65:369–378
Schlaepfer DR, Edwards PJ, Semple JC, Billeter R (2008) Cytogeography of Solidago gigantea (Asteraceae) and its invasive ploidy level. J Biogeogr 35: 2119–2127
Schmitt T (2009) Biogeographical and evolutionary importance of the European high mountain systems. Frontiers Zool 6:9
Schuhwerk F (1996) Published chromosome-counts in Hieracium. Available at: http://www.botanischestaatssammlung.de/projects/chrzlit.html
Šingliarová B, Hodálová I, Mráz P (2011) A biosystematic study of the diploid-polyploid Pilosella alpicola group with variation in breeding system: patterns and processes. Taxon 60:450–470
Šingliarová B, Mráz P (2009) A taxonomic revision of the Pilosella alpicola group (Asteraceae, Lactuceae) in the Carpathians. Preslia 81:23–41
Šingliarová B, Chrtek J Jr, Mráz P (2008) Loss of genetic diversity in isolated populations of an alpine endemic Pilosella alpicola subsp. ullepitschii: effect of long-term vicariance or long-distance dispersal? Pl Syst Evol 275:181–191
Sipes SD, Wolf PG (1997) Clonal structure and patterns of allozyme diversity in the rare endemic Cycladenia humilis var. jonesii (Apocynaceae). Amer J Bot 84:401–409
Soltis DE, Rieseberg LH (1986) Autopolyploidy in Tolmiea menziesii (Saxifragaceae): genetic insights from enzyme electrophoresis. Amer J Bot 73:310–318
Soltis DE, Soltis PS (1989) Genetic consequences of autopolyploidy in Tolmiea (Saxifragaceae). Evolution 43:586–594
Soltis DE, Soltis PS (1990) Chloroplast DNA and nuclear rDNA variation: insights into autopolyploid and allopolyploid evolution. In Kawano S (ed) Biological approaches and evolutionary trends in plants. Academic Press, San Diego, pp 97–117
Soltis DE, Soltis PS (1993) Molecular data and the dynamic nature of polyploidy. Crit Rev Pl Sci 12:243–273
Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the succes of polyploids. Proc Natl Acad Sci USA 97:7051–7057
Stebbins GL (1980) Polyploidy in plants: unsolved problems and prospects. In Lewis WH (ed) Polyploidy, biological relevance. Plenum Press, New York and London, pp 495–520
Štorchová H, Chrtek J Jr, Bartish IV, Tetera M, Kirschner J, Štěpánek J (2002) Genetic variation in agamospermous taxa of Hieracium sect. Alpina (Compositae) in the Tatry Mts (Slovakia). Pl Syst Evol 235:1–17
Stuessy TF, Weiss-Schneeweiss H, Keil DJ (2004) Diploid and polyploid cytotype distribution in Melampodium cinereum and M. leucanthum (Asteraceae, Heliantheae) Amer J Bot 91:889–898
Sun M, Wong KC (2001) Genetic structure of three orchid species with contrasting breeding systems using RAPD and allozyme markers. Amer J Bot 88:2180–2189
Sydes MA, Peakall R (1998) Extensive clonality in the endangered shrub Haloragodendron lucasii (Haloragaceae) revealed by allozymes and RAPDs. Molec Ecol 7:87–93
Těšitel J, Malinová T, Štech M, Herbstová M (2009) Variation in the Melampyrum sylvaticum group in the Carpathian and Hercynian region: two lineages with different evolutionary histories. Preslia 81:1–22
Thompson JD, Lumaret R (1992) The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends Ecol Evol 7:302–307
Trávníček P, Dočkalová Z, Rosenbaumová R, Kubátová D, Szeląg Z, Chrtek J (2011) Bridging global and microregional scales: ploidy distribution in Pilosella echioides (Asteraceae) in central Europe. Ann Bot (Oxford) 107:443–454
Tyler T (2001) Forslag till ny taxonomisk indelning av stangfibblorna (Pilosella) i Norden (The genus Pilosella in the Nordic countries). Svensk Bot Tidskr 95:39–67
Tyler T (2005) Patterns of allozyme variation in Nordic Pilosella. Pl Syst Evol 250:133–145
Varga ZS, Schmitt T (2008) Types of oreal and oreotundral disjunctions in the western Palearctic. Biol J Linn Soc 93:415–430
Weeden NF, Wendel JF (1989) Genetics of plant isozymes. In Soltis P, Soltis D (eds) Isozymes in plant biology. Dioscorides Press, Portland, pp 46–72
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Weiss H, Dobeš C, Schneeweiss GM, Greimler J (2002) Occurrence of tetraploid and hexaploid cytotypes between and within populations in Dianthus sect. Plumaria (Caryophyllaceae). New Phytol 156:85–94
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Yahara T (1990) Evolution of agamospermous races in Boehmeria and Eupatorium. Pl Spec Biol 5:183–196
Yeh FC, Yang R-C, Boyle T (1999) POPGENE version 1.32. Microsoft Window-based freeware for population genetic analysis. Available at: http://www.ualberta.ca/~fyeh/
Zahn KH (1922–1930) Hieracium. In Ascherson P, Graebner P (eds) Synopsis der mitteleuropäischen Flora 12 (1), Gebrüder Borntraeger, Leipzig
Acknowledgements
We would like to thank Róbert Lakoštík (Bratislava) for his valuable assistance in the field, Róbert Šuvada (Brzotín) for helping with the map preparation, Frederic Rooks (Praha) for language correction and two anonymous reviewers for helpful suggestions. This study was financially supported by the Grant Agency of the Ministry of Education of the Slovak Republic and the Slovak Academy of Sciences (VEGA 1/0028/08 to PM), the Academy of Sciences of the Czech Republic (AV0Z 60050516, to JC) and the Ministry of Education, Youth and Sports of the Czech Republic (0021620828, to JC).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic Supplementary Material 1
Allelic frequencies in eight polymorphic loci in all analyzed populations of the Pilosella alpicola group. Species-unique alleles are indicated in bold (DOC 132 kb)
Rights and permissions
About this article
Cite this article
Šingliarová, B., Chrtek, J., Plačková, I. et al. Allozyme Variation in Diploid, Polyploid and Mixed-Ploidy Populations of the Pilosella alpicola Group (Asteraceae): Relation to Morphology, Origin of Polyploids and Breeding System. Folia Geobot 46, 387–410 (2011). https://doi.org/10.1007/s12224-011-9102-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12224-011-9102-0