Abstract
Uropathogenic Escherichia coli (UPEC) isolates contain large genomic segments, termed pathogenicity islands (PAIs), that contribute to their virulence. A total of 150 UPEC and 50 commensal E. coli isolates from outpatients were investigated for antimicrobial susceptibility and the presence of eight PAI markers. One hundred ninety (95 %) isolates were resistant to one or more antimicrobial agents. The most frequent resistance found against amoxicillin (68 %), amoxicillin/clavulanic acid (55 %), aztreonam (50 %), trimethoprim/sulfamethoxazole (46 %) and tetracycline (43.5 %). Antimicrobial resistance among UPEC isolates was higher than that of commensals. PAI markers were detected in substantial percentage of commensal (88 %) and UPEC isolates (98.6 %) (P > 0.05). The most prevalent PAI marker among UPEC and commensal isolates was PAI IV536 (98.7 % UPEC vs. 84 % commensal). We found a high number of PAI markers such as PAI ICFT073, PAI IICFT073, PAI I536, PAI II536, PAI III536 and PAI IIJ96 significantly associated with UPEC. PAI III536 (21.3 %) and PAI IIJ96 (8 %) were detected only in the uropathogenic isolates. Several different combinations of PAIs were found among UPEC isolates. Comparison of PAIs among UPEC and commensal isolates showed that many UPEC isolates (79.3 %) carried two or more PAI markers, while 6 % of commensals had two PAI markers (P < 0.05). The most frequent combinations of PAI markers in UPEC isolates were PAI IV536 + PAI IICFT073 (18 %) and PAI IV536 + PAI ICFT073 + PAI IICFT073 (18 %). These results indicate that PAI markers are widespread among commensal and UPEC isolates and these commensal isolates may be reservoirs for transmission of these markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli is the prototypic commensal species of the facultatively anaerobic microbiota in the human large intestine, but certain strains also cause extraintestinal infections (extraintestinal pathogenic Escherichia coli—ExPEC) including urinary tract infections (UTIs), meningitis, pneumonia, skin and soft-tissue infections, and sepsis (Koga et al. 2014; Östblom et al. 2011). Uropathogenic Escherichia coli (UPEC) is one of the primary etiological agents of UTIs, accounting for 75–90 % of community-acquired UTIs and approximately 50 % of nosocomial UTIs (Oliveira et al. 2011; Copur-Cicek et al. 2014).
The emergence of antimicrobial resistant E. coli has become a serious public health threat worldwide (Rice 2009). The intensive use of antimicrobial agents in human and veterinary medicine is associated with an emerging resistance against therapeutic drugs, followed by the selection of virulence and resistance gene cassettes carrying E. coli strains in humans, animals and the environment (Sepp et al. 2009). Horizontal transfer of these gene cassettes seems to be the main cause of the rapid spread of antibiotic-resistance genes across a wide diversity of bacteria. Beyond the horizontal gene transfer, the loss and acquisition of functional modules are important in the process of rapid bacterial development of resistance (Wozniak and Waldor 2010).
Commensal and ExPEC isolates typically differ with respect to phylogenetic groups and virulence attributes (Sabaté et al. 2006). ExPEC pathogenicity is due to the presence of virulence genes located on chromosome or plasmids which are infrequent among commensal E. coli isolates. These virulence genes on the chromosome are typically found in specific regions called pathogenicity islands (PAIs) (Ananias and Yano 2008). Increasing evidence shows that differences in virulence between pathogenic and nonpathogenic bacterial strains can be attributed in part to virulence genes located in PAIs (Wenting et al. 2013). Pathogenicity islands were described for the first time in uropathogenic E. coli strain 536 in the late 1980s by Hacker et al. (1990). The virulence determinants encoded on different PAIs of UPEC strains 536, J96 and CFT073 are shown in Table 1. PAIs are distinct genetic elements of pathogens encoding various virulence factors such as protein secretion systems, host invasion factors, iron uptake systems and toxins (Yoon et al. 2005). PAIs are a subset of genomic islands and can be identified by features such as the large size (>10 kb), frequent association with tRNA encoding genes or other att sites for temperate bacteriophages, and a G + C content different from host bacterial core genome. These elements are frequently flanked by repeated sequences and carry many fragments of other mobile and accessory genetic elements such as bacteriophages, plasmids and insertion sequence (IS) elements. Some PAIs are unstable regions and can spontaneously disappear from the chromosome. Therefore, PAIs are considered to have evolved from mobile genetic elements by horizontal gene transfer. It can also be assumed that these DNA regions underwent and will continue to undergo further evolutionary changes, resulting in an ongoing evolution of bacterial pathogens (Dobrindt et al. 2002). Identification of PAIs is essential in understanding the development of disease and the evolution of bacterial pathogenesis (Yoon et al. 2005). The objectives of the present case–control study were to compare the presence of various PAI markers among UPEC and commensal E. coli isolates from the stools of healthy subjects and to determine the antimicrobial resistance profiles of these isolates.
Materials and methods
Bacterial isolation
In this case–control study, between March 2013 and February 2014, a total of 200 E. coli strains were isolated from urine and stool specimens of three major university hospitals in Zanjan, Iran. One hundred fifty strains were isolated from urine samples of adult outpatients with symptomatic urinary tract infections (UTIs) and 50 strains from stool specimens of healthy subjects with no history of diarrhoea and antibiotic therapy for at least 1 month (control group). Criteria for symptomatic UTI (all cases of UTIs were cystitis episodes) included dysuria, urgency and frequency of micturition. Patients with fever, nausea and vomiting symptoms, catheterization and those yielding mixed infection were excluded from the study. Specimens were cultured on MacConkey agar (Merck, Germany), and one isolate from each patient was analysed. Verified isolates of E. coli were preserved at −70 °C in trypticase soy broth (Merck) containing 20 % (v/v) glycerol for further analysis.
Antimicrobial susceptibility testing
Susceptibility of isolates to the following antibiotics was examined using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute 2013): Amoxicilin (25 μg), Aztreonam (30 μg), Amikacin (30 μg), Cefotaxime (30 μg), Cefoxitine (30 μg), Ceftazidime (30 μg), Ciprofloxacin (5 μg), Amoxicillin/clavulanic acid (30 μg), Trimethoprim/sulfamethoxazole (25 μg), Cefepime (10 μg), Gentamicin (10 μg), Imipenem (10 μg) and Tetracycline (30 μg) (MAST, Merseyside, UK). Isolates shown to be resistant to at least three different classes of antimicrobial agents were determined to be multidrug resistant (MDR). E. coli ATCC 25922 was used as control for antibiotic resistance.
Detection of PAI markers in E. coli isolates
The presence of the eight PAIs, PAI I536, PAI II536, PAI III536, PAI IV536, PAI ICFT073, PAI IICFT073, PAI IJ96 and PAI IIJ96 was assessed using the previously described primers listed in Table 2 (Sabaté et al. 2006). Template DNA was extracted from E. coli isolates by boiling. One isolate from each patient was grown in LB (Luria Bertani Broth, Merck) until the exponential phase with 2 McFarland turbidity. Then, 500 μL of bacterial suspension was centrifuged at 8000 rpm for 5 min and pellet suspended in 200 μL sterile deionized water and boiled at 100 °C for 15 min. After centrifugation at 13000 rpm for 3 min, supernatant was used for PCR. Simplex PCR was performed using DreamTaq PCR Master Mix (Thermo Fisher Scientific), which contains Taq polymerase, dNTPs, MgCl2 and the appropriate buffer. Each PCR tube contained 25 μL reaction mixture composed of 12.5 μL of the master mix, 1.5 μL of each forward and reverse primer solution (in a final concentration of 200 nmol/L), 5 μL of DNA with concentration of 400 ng/μL and nuclease-free water to complete the final volume. PCR was performed using the Gene Atlas 322 system (ASTEC). Amplification involved an initial denaturation at 94 °C, 5 min followed by 35 cycles of denaturation (94 °C, 1 min), annealing (55 °C, 1 min) and extension (72 °C, 1 min), with a final extension step (72 °C, 8 min). The amplified DNA was separated by submarine gel electrophoresis on 1.5 % agarose, stained with ethidium bromide and visualized under UV transillumination. UPEC strains 536 and J96 were used as PAI marker controls.
Statistical analysis
The data were analysed with SSPS version 17.0 software (SPSS). A chi-square test was used to determine the statistical significance of the data. A P value of <0.05 was considered significant.
Results
Distribution of PAI markers in commensal and uropathogenic E. coli isolates
Overall, 192 (96 %) isolates were positive for the presence of PAI markers, 44 commensal isolates (88 %) and 148 UPEC isolates (98.7 %) from patients with symptomatic urinary tract infections.
The frequency of each PAI marker in the patient and control groups is shown in Table 3. Comparison of PAI marker distribution among UPEC and commensal isolates showed that UPEC isolates harboured markers with higher frequency than in commensal isolates except PAI IJ96 (P < 0.05). The most prevalent PAI marker among UPEC isolates was PAI IV536 (98.7 %), followed by PAI IICFT073 (61.3 %), PAI ICFT073 (43.3 %), PAI III536 (21.3 %), PAI I536 (16.7 %), PAI II536 (12 %), PAI IIJ96 (8 %) and PAI IJ96 (0.7 %). The frequency of PAI IV536, PAI ICFT073 and PAI IICFT073 in commensal isolates was 84, 6 and 4 %, respectively. PAI III536 and PAI IIJ96 were not detected in commensal isolates.
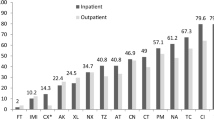
The presence of multiple PAIs with different combinations was found among UPEC isolates. Figure 1 and Table 4 show that many UPEC isolates (79.3 %) carry two or more PAIs compared with commensal isolates (6 %) (P < 0.05). The number of PAIs per isolate and their specific combinations are shown in Table 4. The mean number of PAIs per isolate was higher in UPEC isolates than commensals (P < 0.05). The most frequent combinations of PAI markers in UPEC isolates were PAI IV536 + PAI IICFT073 (18 %) and PAI IV536 + PAI ICFT073 + PAI IICFT073 (18 %), followed by PAI IV536 + PAI ICFT073 (7.3 %). The most commensal isolates (82 %) had only one PAI marker.
Susceptibility to antimicrobial agents
Antimicrobial resistance patterns of isolates are presented in Table 5. In all, 190 (95 %) isolates were resistant to one or more of the 13 tested antimicrobial agents, with the most frequent resistance found against amoxicillin (68 %), amoxicillin/clavulanic acid (55 %), aztreonam (50 %), trimethoprim/sulfamethoxazole (46 %) and tetracycline (43.5 %). Imipenem showed the highest activity against isolates and only 2 % of isolates were imipenem resistant. Prevalence of antibiotic resistance varied among UPEC and commensal isolates, and the frequency of resistance to amikacine, tetracycline, cefepime and ciprofloxacin in UPEC isolates was significantly higher than commensals (P < 0.05). Antimicrobial drug resistance among UPEC isolates carrying different PAI combinations is presented in Table 6. A total of 132 (66 %) isolates were resistant to at least three different classes of antimicrobial agents and considered as multidrug resistance (MDR): 110 of the UPEC isolates (73.3 %) and 22 of the commensal isolates (44 %) (P < 0.05). The most prevalent MDR pattern was resistance to β-lactams, tetracycline, gentamicin and trimethoprim/sulfamethoxazole. All MDR isolates of UPEC and commensal were positive for the presence at least one PAI marker. One hundred two (92.7 %) multidrug-resistant UPEC isolates carry two or more PAI markers (Table 7).
Discussion
Horizontal gene transfer (HGT) seems to be an important mechanism for bacterial evolution, let alone genome complexity and plasticity. PAIs, which are large genomic segments and most likely transferred by HGT, contribute to the virulence and survival of the hosting bacterial strain in a particular environment (Middendorf et al. 2004; Gal-Mor and Finlay 2006; Che et al. 2014). PAIs have been studied widely in the genomes of pathogenic bacteria, but little attention has been given to PAIs in the genomes of commensal members of a species (Sabaté et al. 2006). In our study, the presence of eight PAI markers was detected among UPEC and commensal E. coli isolates from the stools of healthy subjects. PAI markers were detected in substantial percentage of commensal (88 %) and UPEC isolates (98.6 %) (P > 0.05). According to our results and previous studies, some isolates of E. coli from the intestinal tract of healthy people can be considered potentially virulent, as some isolates showed two PAI markers. Already, it has been reported that ExPEC can asymptomatically colonize the intestinal tract (Koga et al. 2014). Furthermore, there was evidence that the intestinal niche may harbour E. coli isolates, with a large number of accumulated PAIs (Sabaté et al. 2006). However, in our study, commensal isolates contained fewer PAI combinations than UPEC isolates. Previous studies showed that UPEC isolates harboured PAI markers with significantly higher frequency than commensals (Sabaté et al. 2006; Navidinia et al. 2013a). Distribution of various PAIs in our study showed the same pattern with other studies (Sabaté et al. 2006; Li et al. 2010; Navidinia et al. 2013b). PAI IV536, also termed high-pathogenicity island (HPI), was found most frequently in both commensal and UPEC isolates and is reported to be the most ubiquitous PAI found in Enterobacteriaceae. In previous studies (Sabaté et al. 2006; Navidinia et al. 2013b), PAI IV536 was detected in 38 and 18 % of faecal E. coli isolates respectively, compared with 84 % of isolates in the present study. The high frequency of PAI IV536 in commensal isolates has led to the suggestion that HPI may be a fitness island rather than a pathogenicity island (Sabaté et al. 2006). Previous studies (Middendorf et al. 2004) demonstrated that PAI IV is stable in E. coli 536, a fact that could explain its high frequency. The remaining PAI markers except PAI IJ96 had a similar distribution in commensal and UPEC isolates but with significantly higher frequency in UPEC isolates. Similar to our results, it was suggested that PAI IJ96 might not be important in the pathogenesis of urosepsis (Sabaté et al. 2006). Several different combinations of PAI markers were found among 79.3 % of UPEC isolates, while 6 % of commensals had two PAI markers. Our results showed that the mean number of PAIs per isolate was higher among UPEC in comparison with commensals (P < 0.05).
Antimicrobial resistance and the spread of resistance genes among pathogenic E. coli isolates have become a major public health problem in developing countries (Su et al. 2006). Treatment of infections associated with multidrug-resistant E. coli is further complicated in Asian countries such as Taiwan, India and Iran (Rice 2009; Phongpaichit et al. 2011). In our study, 95 % of E. coli isolates were resistant to one or more antimicrobial agents and 66 % were multidrug resistant. High frequency of antibiotic resistance among UPEC isolates was reported in previous studies in Iran (Farshad et al. 2012; Rezaee et al. 2011; Neamati et al. 2015). The high incidence of amoxicillin (68 %) and amoxicillin/clavulanic acid (55 %) resistance in the present study is most probably due to the widespread use of these antimicrobial agents in our country. Furthermore, the loss and gain of resistance genes by mobile genetic elements are an important mechanism in the development of multidrug-resistant isolates (Wozniak and Waldor 2010). Similar to our study, Navidinia et al. (2013a) and Neamati et al. (2015) were reported 78 and 51.4 % resistance to aztreonam. High-level resistance to aztreonam was reported by Pobiega et al. (2013) among the extended-spectrum β-lactamases (ESBL)-producing E. coli isolates. In a study carried out in Tamilnadu, India, the majority of UPEC isolates from UTI patients exhibited MDR phenotype including amoxicillin (96.2 %), tetracycline (72.7 %), cefotaxime (74.8 %) and ciprofloxacin (70.4 %) resistance (Murugan et al. 2012). According to our results, imipenem (98 %) showed the lowest resistance against UPEC and commensal isolates, and only 2 % of isolates were imipenem resistant. In previous studies (Rezaee et al. 2011; Neamati et al. 2015), resistance to imipenem was detected in 1.4 and 0.7 % of UPEC isolates, respectively. According to results, antimicrobial resistance among UPEC isolates was higher than commensal isolates. The high prevalence of antibiotic resistance in UPEC isolates may be due to acquisition of the resistance genes from intestinal microbiota as reservoirs for transmission of these genes (Ravi et al. 2014).
Ethical considerations
This study was performed using routine samples obtained from patients admitted to hospitals, and thus, ethical approval was not required.
References
Ananias M, Yano T (2008) Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Braz J Med Biol L Res 41(10):877–883
Che D, Hasan MS, Chen B (2014) Identifying pathogenicity islands in bacterial pathogenomics using computational approaches. Pathogens 3:36–56
Clinical and Laboratory Standards Institute (2013) Performance standards for antimicrobial susceptibility testing; 23th informational supplement. CLSI document M100-S23. Vol. 31 No.1
Copur-Cicek A, Ozgumus OB, Saral A, Sandalli C (2014) Antimicrobial resistance patterns and integron carriage of Escherichia coli isolates causing community-acquired infections in Turkey. Ann Lab Med 34:139–144
Dobrindt U, Blum-Oehle G, Nagy G, Schneider G, Johann A, Gottschalk G (2002) Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect Immun 70(11):6365–6372
Farshad S, Ranjbar R, Japoni A, Hosseini M, Anvarinejad M, Mohammadzadegan R (2012) Microbial susceptibility, virulence factors, and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom, Iran. Arch Iran Med 15(5):312–316
Gal-Mor O, Finlay BB (2006) Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol 8(11):1707–1719
Hacker J, Bender L, Ott M, Wingender J, Lund B, Marre R, Goebel W (1990) Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microbial pathogenesis 8(3):213–225
Koga VL, Tomazetto G, Cyoia P S, Neves M S, Vidotto M C, Nakazato G, Kobayashi R K T (2014) Molecular screening of virulence genes in extraintestinal pathogenic Escherichia coli isolated from human blood culture in Brazil. Biomed Res Int Article, ID 465054; http://dx.doi.org/10.1155/2014/465054.
Li B, Sun JY, Han LZ, Huang XH, Fu Q, Ni YX (2010) Phylogenetic groups and pathogenicity island markers in fecal Escherichia coli isolates from asymptomatic humans in China. Appl Environ Microb 76(19):6698–6700
Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J (2004) Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol 186:3086–3096
Murugan K, Savitha T, Vasanthi S (2012) Retrospective study of antibiotic resistance among uropathogens from rural teaching hospital, Tamilnadu, India. Asian Pacific J Tropical Dis 2(5):375–380
Navidinia M, Najar Peerayeh S, Fallah F, Bakhshi B (2013a) Phylogenetic groups and pathogenicity island markers in Escherichia coli isolated from children. Jundishapur J Microbiol 6(10):e8362
Navidinia M, Najar Peerayeh S, Fallah F, Bakhshi B, Adabian S, Alimehr S, Gholinejad Z (2013b) Distribution of the pathogenicity islands markers (PAIs) in uropathogenic E.coli isolated from children in Mofid children hospital, Pediatric Infections Research Center. Arch Pediatr Infect Dis 1(2):75–79
Neamati F, Firoozeh F, Saffari M, Zibaei M (2015) Virulence genes and antimicrobials resistance pattern in uropathogenic Escherichia coli isolated from hospitalized patients in Kashan, Iran. Jundishapur J Microbiol 8(2):e17514
Oliveira FA, Paludo KS, Arend LN, Farah SM, Pedrosa FO, Souza EM, Surek M, Picheth G, Fadel-Picheth CM (2011) Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res 10(4):4114–4125
Östblom A, Adlerberth I, Wold AE, Nowrouzian FL (2011) Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants’ commensal microbiotas. Appl Environ Microb 77(7):2303–2308
Phongpaichit S, Tunyapanit W, Pruekprasert P (2011) Antimicrobial resistance, class I integrons and extended spectrum betalactamases in E. coli clinical isolates from patients in south Thailand. J Health Sci 57:281–288
Pobiega M, Wojkowska-Mach J, Chmielarczyk A, Romaniszyn D, Adamski P, Heczko PB et al (2013) Molecular characterization and drug resistance of Escherichia coli strains isolated from urine from long-term care facility residents in Cracow, Poland. Med Sci Monit 19:317–326
Ravi A, Avershina E, Ludvigsen J, L’Abée-Lund TM, Rudi K (2014) Integrons in the intestinal microbiota as reservoirs for transmission of antibiotic resistance genes. Pathogens 3:238–248
Rezaee MA, Sheikhalizadeh V, Hasani A (2011) Detection of integrons among multi-drug resistant (MDR) Escherichia coli strains isolated from clinical specimens in northern west of Iran. Braz J Microbiol 42(4):1308–1313
Rice LB (2009) The clinical consequences of antimicrobial resistance. Curr Opin Microbiol 12:476–481
Sabaté M, Moreno E, Pérez T, Andreu A, Prats G (2006) Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect 12:880–886
Schmidt H, Hensel M (2004) Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17(1):14–56
Sepp E, Stsepetova J, Lõivukene K, Truusalu K, Kõljalg S, Naaber P, Mikelsaar M (2009) The occurrence of antimicrobial resistance and class 1 integrons among commensal Escherichia coli isolates from infants and elderly persons. Annals of Clinical Microbiology and Antimicrobials 8:34
Su J, Shi L, Yang L, Xiao Z, Li X, Yamasaki S (2006) Analysis of integrons in clinical isolates of Escherichia coli in China during the last six years. FEMS Microbiol Lett 254:75–80
Wenting J, Jinling S, Magaly T, Shaohua Z, Jianghong M (2013) Distribution of pathogenicity islands OI-122, OI-43/48, and OI-57 and a high-pathogenicity island in shiga toxin-producing Escherichia coli. Appl Environ Microb 79(11):3406–3412
Wozniak RAF, Waldor MK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8(8):552–563
Yoon SH, Hur CG, Kang HY, Kim YH, Oh TK, Kim JF (2005) A computational approach for identifying pathogenicity islands in prokaryotic genomes. BMC Bioinformatics 6:184
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Samei, A., Haghi, F. & Zeighami, H. Distribution of pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Folia Microbiol 61, 261–268 (2016). https://doi.org/10.1007/s12223-015-0433-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-015-0433-8