Abstract
Recently the ATP-binding cassette (ABC) efflux pumps have been proved to be a major component of drug resistance in Mycobacterium tuberculosis. The objective of this study was to investigate the expression profiles of Rv1456c-Rv1457c-Rv1458c efflux system in clinical isolates of M. tuberculosis and its involvement in drug-resistance mechanisms. Significantly increased mRNA expression of Rv1456c, Rv1457c, and Rv1458c appeared among the clinical isolates (P < 0.05), which are resistant to at least one of the four first-line drugs including rifampin, isoniazid, streptomycin, and ethambutol. In addition, overexpression of this efflux system was more frequently found in multidrug-resistant and extensively drug-resistant M. tuberculosis strains. Therefore, Rv1456c-Rv1457c-Rv1458c efflux pumps may play an important role in drug resistance of treatment of M. tuberculosis. Further investigation of this gene may lead to the development of countermeasures against M. tuberculosis drug resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycobacterium tuberculosis (TB) infection causes significant morbidity and mortality throughout the world, particularly in developing countries (Morrison et al. 2008). The infection rate of M. tuberculosis in China was particularly high, with six million infected patients and 250,000 deaths per year (Smith 2003; National Technic Steering Group of the Epidemiological Sampling Survey for Tuberculosis and Duanmu 2002; Jin et al. 2009). One of the major problems in combating this disease is the intrinsic and acquired resistance to therapeutic agents of M. tuberculosis (Zahrt 2003; Haydel 2010). Although acquired drug resistance is mainly due to mutational alterations of the drug target (Shi et al. 2007), it has become clear that multidrug efflux systems also play important roles in drug resistance of M. tuberculosis (Putman et al. 2000).
The ATP-binding cassette (ABC) efflux pump is one of the best known and characterized families of transporter proteins, involved in the protection of organisms against deleterious compounds (Shilling et al. 2006; Ouellette et al. 1994). These transporters belong to the class of primary active transporters, which use energy derived from the hydrolysis of the diphosphate bond of ATP to drive transport of compounds against a concentration gradient. In M. tuberculosis, the genes encoding the predicted ABC transporters occupy about 2.5% of the genome (Braibant et al. 2000). Recent research has shown that multidrug resistance of M. tuberculosis may associate with constitutive or inducible expression of ABC efflux systems, therefore knowledge of these M. tuberculosis ABC efflux systems is critical for understanding their involvement in the development of drug resistance in M. tuberculosis.
To date, several ABC efflux systems have been identified in M. tuberculosis (Choudhuri et al. 2002; Pasca et al. 2004; Molle et al. 2004) and one member of them, Rv2686c-Rv2687c-Rv2688c, has been conferred resistant to fluoroquinolone. The structure of Rv1456c-Rv1457c-Rv1458c operon is similar to Rv2686c-Rv2687c-Rv2688c, so in this paper we investigated the role of Rv1456c-Rv1457c-Rv1458c efflux pump in clinical isolates of drug-resistant M. tuberculosis.
Materials and methods
M. tuberculosis isolates
This study was conducted with the approval of the local ethics committee of Wuxi Hospital of Infectious Disease and Jiangsu Institute of Nuclear Medicine. A total of 35 clinical isolates of M. tuberculosis along with M. tuberculosis reference strain H37Rv were included in the present study. These isolates were collected between July 2009 and July 2010 from patients hospitalized at Wuxi Hospital of Infectious Disease.
Susceptibility testing
The WHO recommended drug susceptibility testing is done by the proportion method on Lowenstein–Jensen medium. It takes 3–6 weeks to obtain the initial positive culture with an additional 3 weeks for susceptibility testing recommendations to determine strain susceptibility. Resistance to four first-line anti-TB drugs including isoniazid (INH), rifampicin (RIF), streptomycin (STR), and ethambutol (EMB) and eight second-line anti-TB drugs including pyrazinamide, ofloxacin, aminosalicylic acid capreomycin, isoniazid aminosalicylate, amikacin, protionamide, and rifapentin capsules of 35 M. tuberculosis clinical isolates has been tested.
Total RNA extraction
M. tuberculosis clinical isolates were grown on Lowenstein–Jensen medium at 37°C for 3 to 4 weeks. Fresh bacterial cells were collected and ground in which the lysozyme was used to break down the cell wall. Then total RNA was extracted using MagNA Pure LC RNA Isolation Kit-High Performance (Roche) according to the manufacture’s protocol.
Quantitative RT-PCR
Total RNA was submitted to cDNA synthesis using PrimeScriptTM RT reagents Kit (Takara). Quantitative RT-PCR was performed with the cDNA using SYBR Green PCR Master Mix (Takara) and specific primers for Rv1456c, Rv1457c, and Rv1458c were designed for this study using the Primer Express Software version 2.0 (Applied Biosystems, Table 1). To assure the specific amplification, melting curves of each reaction were assessed and each sample was run in duplicate. A gene encoding heat shock protein 65 (Hsp65) was used as housekeeping gene for normalization. Relative quantification of the target gene expression was analyzed with the Q-Gene software.
Statistical analysis
Analysis of data was performed using SPSS 15.0. Differences between variables were compared by the unpaired Student’s t test, and differences in proportions were compared by chi-square test as appropriate. A P value of <0.05 was considered to be significant.
Results
Antibiotic susceptibility
Thirty-five clinical isolates of M. tuberculosis were collected from sputa of patients with active pulmonary tuberculosis, and their antibiotic susceptibility testing was performed according to the WHO recommended standard conventional proportional method. Among the 35 clinical isolates, 12 were susceptible to all twelve first-line and second-line antibiotics, whereas 23 were resistant to at least one of four first-line anti-TB drugs and at least one of eight second-line anti-TB drugs. Among the 23 antibiotic-resistant clinical isolates, 13 were resistant to RIF, 17 were resistant to INH, 15 were resistant to STR, and 7 were resistant to EMB. Furthermore, 14 of 23 antibiotic-resistant clinical isolates were polydrug-resistant M. tuberculosis strains (PDR-TB), 5 were multidrug-resistant M. tuberculosis strains (MDR-TB), and 4 were extensively drug-resistant M. tuberculosis strains (XDR-TB, Table 2).
Gene expression of Rv1456c-Rv1457c-Rv1458c
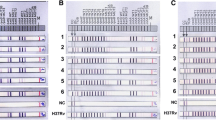
To investigate whether the ABC efflux pump, Rv1456c-Rv1457c-Rv1458c is involved with the drug resistance of all the tested clinical isolates of M. tuberculosis, the relative expression profiles of these three genes in the recruited clinical strains were examined and compared to the reference strain H37Rv. As shown in Fig. 1, significantly increased expression of Rv1456c, Rv1457c, and Rv1458c genes was observed in drug-resistant clinical isolates, while not in the drug-susceptible groups. Furthermore, quantitative analysis revealed that expression level of Rv1456c, Rv1457c, and Rv1458c respectively increased 3.4-, 4.6-, and 5.4-fold in drug-resistant strains when comparing to susceptible group (P < 0.05).
Relative expression profile of Rv1456c, Rv1457c, and Rv1458c in the M. tuberculosis clinical isolates. The results from real-time PCR showed higher mRNA levels of Rv1456c, Rv1457c, and Rv1458c from drug-resistant clinical isolates compared to drug-susceptible groups and the difference was statistically significant. *P < 0.05; **P < 0.01
Then the association between the expression profiles of Rv1456c, Rv1457c, and Rv1458c and drug-resistance mechanism of these clinical isolates was analyzed. The percentage of isolates with increased expression of Rv1456c, Rv1457c, and Rv1458c genes in each drug-resistant group was listed in Table 3.
In addition, we summarized the expressional profile of the Rv1456c, Rv1457c, and Rv1458c efflux gene in each drug-resistant category of M. tuberculosis isolates (Table 4). Interestingly, high expression of three genes was observed in most of MDR and XDR strains but comparatively lower in PDR-TB strains. Our results suggested that Rv1456c-Rv1457c-Rv1458c efflux pump play an important role in the drug resistance of M. tuberculosis.
Discussion
Drug-resistance rate among M. tuberculosis clinical isolates has increased over the last few years, single mutation in certain genes has been found to be sufficient for high-level antibiotic resistance in M. tuberculosis (Abbadi et al. 2009; Riska et al. 2000). Increased efflux of the antibiotics has also been reported to be a common mechanism and represents the first step in the acquisition of antibiotic resistance (Poole 2007). ABC transporter is the main efflux pump involved in determining intrinsic levels of resistance in M. tuberculosis (Rodriguez and Smith 2006; Braibant et al. 1996). To date, several ABC efflux pumps including Drr and Rv2686c-Rv2687c-Rv2688c operon, involved in drug resistance of M. tuberculosis has been reported (Choudhuri et al. 2002; Pasca et al. 2004; Molle et al. 2004), but the correlation between the expression profiles of efflux pump genes and drug resistance of M. tuberculosis has not been established.
In this study, we focused on the Rv1456c-Rv1457c-Rv14588c efflux system. The structure of Rv1456c-Rv1457c-Rv14588c operon is similar to Rv2686c-Rv2687c-Rv2688c, which could be composed of two copies of the nucleotide-binding domains (Rv1458c), and one copy of each membrane-spanning domains (Rv1456c and Rv1457c). Rv1458c protein is more likely involved in ATP hydrolysis. Here we first demonstrated that the mRNA expression of Rv1456c, Rv1457c, and Rv1458c efflux genes in M. tuberculosis clinical isolates from China and investigated the association between the above-mentioned gene expression and drug-resistant mechanisms.
Thirty-five clinical isolates of M. tuberculosis were collected from patients hospitalized at Wuxi Hospital of Infectious Disease between July 2009 and July 2010, which have been tested with drug susceptibility as well. Among total 35 clinical isolates, 12 were susceptibility strains and 23 were resistant to at least one of four first-line anti-TB drugs and at least one of eight second-line drugs. The transcriptional expressions of Rv1456c, Rv1457c, and Rv1458c genes were evaluated in these thirty-five clinical isolates compared to the reference strain H37RV. Our results demonstrated that overexpression of the Rv1456c, Rv1457c, and Rv1458c were more frequent among drug-resistant M. tuberculosis clinical isolates and respectively increased 3.4-, 4.6-, and 5.4-fold compared to drug-susceptible group. We also observed the correlation between drug resistance of M. tuberculosis and the overexpression of our target genes. Elevated expression of Rv1456c, Rv1457c, and Rv1458c appeared in all the clinical isolates resistant to at least one of four first-line drugs including rifampin, isoniazid, streptomycin, and ethambutol. Furthermore, overexpression of these three efflux transporter genes was more frequent in MDR-TB and XDR-TB strains.
Our studies suggested that the overexpression of Rv1456c-Rv1457c-Rv1458c efflux genes may play an important role in drug resistance of the studied clinical isolates of M. tuberculosis. Further experiments are required to identify the exact mechanism leading to the increased expression of Rv1456c-Rv1457c-Rv14588c efflux system in drug-resistant M. tuberculosis. Our study will provide the basis for functional characterization of Rv1456c-Rv1457c-Rv1458c efflux pump and for the future development of therapeutic agents against the multidrug-resistant M. tuberculosis.
Abbreviations
- ABC:
-
ATP-binding cassette
- TB:
-
Mycobacterium tuberculosis
- INH:
-
Isoniazid
- RIF:
-
Rifampicin
- STR:
-
Streptomycin
- EMB:
-
Ethambutol
- PDR:
-
Polydrug resistant
- MDR:
-
Multidrug resistant
- XDR:
-
Extensively drug resistant
References
Abbadi SH, Sameaa GA, Morlock G, Cooksey RC (2009) Molecular identification of mutations associated with anti-tuberculosis drug resistance among strains of Mycobacterium tuberculosis. Int J Infect Dis 13:673–678
Braibant M, Gilot P, Content J (2000) The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol Rev 24:449–467
Braibant M, Lefèvre P, de Wit L, Ooms J, Peirs P, Huygen K, Wattiez R, Content J (1996) Identification of a second Mycobacterium tuberculosis gene cluster encoding proteins of an ABC phosphate transporter. FEBS Lett 394:206–212
Choudhuri BS, Bhakta S, Barik R, Basu J, Kundu M, Chakrabarti P (2002) Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem J 367:279–285
Haydel SE (2010) Extensively drug-resistant tuberculosis: a sign of the times and an impetus for antimicrobial discovery. Pharmaceuticals (Basel) 3:2268–2290
Jin J, Sun L, Jiao W, Zhao S, Li H, Guan X, Jiao A, Jiang Z, Shen A (2009) SLC11A1 (formerly NRAMP1) gene polymorphisms associated with pediatric tuberculosis in China. Clin Infect Dis 48:733–738
Molle V, Soulat D, Jault JM, Grangeasse C, Cozzone AJ, Prost JF (2004) Two FHA domains on an ABC transporter, Rv1747, mediate its phosphorylation by PknF, a Ser/Thr protein kinase from Mycobacterium tuberculosis. FEMS Microbiol Lett 234:215–223
Morrison J, Pai M, Hopewell PC (2008) Tuberculosis and latent tuberculosis infection in close contacts of people with pulmonary tuberculosis in low-income and middle-income countries: a systematic review and meta-analysis. Lancet Infect Dis 8:359–368
National Technic Steering Group of the Epidemiological Sampling Survey for Tuberculosis, Duanmu H (2002) Report on fourth national epidemiological sampling survey of tuberculosis. Zhonghua Jie He He Hu Xi Za Zhi 25:3–7
Ouellette M, Légaré D, Papadopoulou B (1994) Microbial multidrug-resistance ABC transporters. Trends Microbiol 2:407–411
Pasca MR, Guglierame P, Arcesi F, Bellinzoni M, De Rossi E, Riccardi G (2004) Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob Agents Chemother 48:3175–3178
Poole K (2007) Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39:162–176
Putman M, van Veen HW, Konings WN (2000) Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev 64:672–693
Riska PF, Jacobs WR Jr, Alland D (2000) Molecular determinants of drug resistance in tuberculosis. Int J Tuberc Lung Dis 4:S4–10
Rodriguez GM, Smith I (2006) Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J Bacteriol 188:424–430
Shi R, Itagaki N, Sugawara I (2007) Overview of anti-tuberculosis (TB) drugs and their resistance mechanisms. Mini Rev Med Chem 7:1177–1185
Shilling RA, Venter H, Velamakanni S, Bapna A, Woebking B, Shahi S, van Veen HW (2006) New light on multidrug binding by an ATP-binding-cassette transporter. Trends Pharmacol Sci 27:195–203
Smith I (2003) Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev 16:463–496
Zahrt TC (2003) Molecular mechanisms regulating persistent Mycobacterium tuberculosis infection. Microbes Infect 5:159–167
Acknowledgments
This work was financially supported by grant from the Foundation of Department of Public Health of Jiangsu Province (Grant No. H200954). We would like to thank Miao Chen and Wei Liu for the critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hao, P., Shi-Liang, Z., Ju, L. et al. The role of ABC efflux pump, Rv1456c-Rv1457c-Rv1458c, from Mycobacterium tuberculosis clinical isolates in China. Folia Microbiol 56, 549–553 (2011). https://doi.org/10.1007/s12223-011-0080-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-011-0080-7