Abstract
Forced expression of transcription factors epigenetically reprograms somatic cells harvested from routine skin biopsies into induced pluripotent stem cells (iPSCs). Human iPSCs are key resources for drug discovery, regenerative medicine and tissue engineering. Here we developed a materials approach to explore how culture substrates could impact factor-mediated reprogramming of human fibroblasts. A materials library consisting of nanofibrous substrates with randomly oriented and aligned structures was prepared by electrospinning four polymers [polylactic acid (PLA), polycaprolactone (PCL), thermoplastic polyurethane (TPU) and polypropylene carbonate (PPC)] into nanofiber orientations. Adsorbing protein to each substrate permitted robust attachment of fibroblasts to all substrates. Fibroblasts on aligned substrates had elongated nuclei, but after reprogramming factor expression, nuclei became more circular. Reprogramming factors could override the nuclear shape constraints imposed by nanofibrous substrates, and the majority of substrates supported full reprogramming. Early culture on PCL and TPU substrates promoted reprogramming, and TGF-β repressed substrate effects. Partial least squares modeling of the biochemical and biophysical cues within our reprogramming system identified TGF-β and polymer identity as important cues governing cellular reprogramming responses. We believe that our approach of using a nanofibrous materials library can be used to dissect molecular mechanisms of reprogramming and generate novel substrates that enhance epigenetic reprogramming.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Dramatic changes in cell fate and function can be achieved via forced expression of transcription factors.14 These changes are coordinated by particular combinations of factors (e.g., Oct4, Sox2, Klf4 and c-Myc, or Oct4, Sox2, Nanog and Lin2840,45) and are molecularly encoded at the epigenetic level. Such epigenetic reprogramming processes are typically multi-step and noisy, generating many intermediate, partially reprogrammed cell types during the process.2,17 Improving methods to accelerate reprogramming and generate high quality, fully reprogrammed cells would advance the field and open up new applications in disease modeling,34 tissue engineering41 and drug discovery.15
While extensive studies have been conducted on the roles of transcription factors, miRNA, small molecules and other soluble factors,12,16 very little is known about how biophysical properties of cell culture substrates influence somatic cell reprogramming. Recent work with mouse fibroblasts indicated that altering the biophysical microenvironment through the use of microgroove channels and electrospun fibers induced hyperacetylation of histones that synergized with Oct4, Sox2, Klf4 and c-Myc reprogramming factors.10 Hyperacetylation of histones has been shown to prevent chromatin from compacting into a 30-nm fiber.38 Such hyperacetylation could promote promiscuous transcription by favoring a more open chromatin conformation that permits binding of the reprogramming factors and transcriptional machinery. Indeed, an open chromatin structure induced by two histone variants, TH2A and TH2B, was linked to enhanced reprogramming.39 Further, hyperacetylation of specific sites in the genome or at specific lysines on histone 3 could directly serve as recognition sites for factors that promote transcription.1 For instance, acetylation of lysine 27 on histone 3 (H3K27ac), an enhancer mark, was increased in MBD3-depleted cells that had accelerated reprogramming kinetics.32 Acetylation machinery could complex with other cofactors (e.g., WDR5) and reprogramming factors to hyperacetylate chromatin at specific loci to locally enhance transcription1,20 to enhance reprogramming.10,13 Together, these results suggest that materials that force nuclei to have a more open chromatin environment by increasing acetylation both globally and at specific sites in the genome could synergize with reprogramming factors to drive epigenetic reprogramming.
Because nanofibrous materials were successful in mouse reprogramming10 and may be able to perturb nuclei to adopt a more open conformation amenable to reprogramming, we have taken an empirical approach to identifying nanofibrous materials that could promote human reprogramming. Synthetic polymers—PCL, PLA, PPC, and TPU—are commonly used in manufacturing and polymer processing to produce highly-aligned scaffolds that may be able to alter nuclear shape. Furthermore, these materials can be synthesized to mimic the fibrous nature of native extracellular matrix (ECM), which has been shown to change the degree of cellular adhesion to substrates (e.g., through variations in ligand density, integrin clustering and cytoskeletal organization22).
Here we describe the synthesis and characterization of a material library of nanofibrous substrates generated by electrospinning PCL, PLA, PPC, and TPU. Reprogramming of human fibroblasts by transcription factors was performed on this library to identify substrates that enhanced factor-mediated reprogramming. We found that nuclear shape can be perturbed on these nanofibrous substrates, but reprogramming factors rapidly erase these perturbations and induce a circular shape early in the process. Several nanofibrous substrates promoted reprogramming, and elevated reprogramming did not correlate with materials-induced global hyperacetylation of histone 3. Regression of our experimental dataset indicated a multivariate response of cells to the biochemical and biophysical properties of their microenvironment during reprogramming. We expect this work to inform the development of design rules for materials that can drive the epigenetic reprogramming of patients’ cells.
Materials and Methods
Materials
Polylactic acid (PLA, 2002 D) and poly(propylene carbonate) (PPC, QPAC® 40) were purchased from Natureworks LLC and Empower Materials Inc., respectively. Medical grade thermoplastic polyurethane (TPU, Texin® Rx85A) was generously donated by Bayer Material Science Inc. Polycaprolactone (PCL, Mn = 80000 g/mol), polyacrylic acid (PAA, Mn = 50000 g/mol), chloroform (CF, ACS regent), N,N-dimethyl formamide (DMF, ACS regent) and pentyl acetate were all purchased from Sigma-Aldrich (Milwaukee, WI, USA). Manufacturers for all other reagents are listed below within the text.
Substrate Preparation
Each polymer solution was prepared at various concentrations as described in Supplementary Table 1. Electrospinning was carried out using a custom-built electrospinning device as previously described.29 The prepared solution was loaded in a plastic syringe connected to an 18-gauge blunt-end needle and then mounted on a digital syringe pump (Harvard Bioscience Company). Spinning conditions consisted of a voltage of 18 kV, a 15 cm needle-to-target distance, and a 0.5 mL/h flow rate. Two parallel steel plates separated by a 3 cm distance were used to collect the aligned fibers. Fibers were then applied to sterilized, round stainless steel washers (inner diameter of 8.33 mm, McMaster-Car; Part C0532R) to maintain structural integrity for cell culture applications. Random fibers were spun directly onto the washers.
Because we were unable to make aligned TPU fibers without the addition of PAA, 1 wt% PAA was added to the TPU solution before electrospinning to improve conductivity. However, dense fiber mats of random TPU fibers could be created for water contact angle (WCA) and Brunauner-Emmett-Teller (BET) assay without the addition of PAA. To prepare the PCL nanofibrous substrates with a “shish kebab” structure (PCL-SK), 30 μL of 0.8% PCL in the pentyl acetate was dropped onto the prepared PCL nanofibers and the samples were placed in the fume hood to let solvents evaporate overnight.
Scanning Electron Microscopy Imaging and Analysis
Each nanofibrous substrate was imaged by scanning electron microscopy (SEM, JEOL 6500, Nikon) using an accelerating voltage of 10 kV. The electrospun substrates were cut with a scalpel and sputter-coated with gold for 40 s before imaging. Images were converted to binary format using ImageJ, where the fibers appeared black and the pores white. Fiber alignment, fiber diameter and substrate porosity were determined by ImageJ software analysis. The percentage of the white pixels relative to the total scaffold surface area in each image was defined as the percent porosity.33 For alignment and diameter measurements, 50 fibers were profiled for each sample from 3 images. The fiber diameter was the average value of at least 50 fibers measured using 3 images.
Water Contact Angle
WCA was calculated for each substrate in the random orientation and before protein adsorption. Each polymer sample was electrospun for up to 3 h in order to create the proper thickness to support a water droplet. Each sample was tested using the sessile drop method during atmospheric conditions using a video contact angle instrument (Dataphysics OCA 15) and a droplet size setting of 4 μL. Three samples for each substrate were tested.
Mechanical Properties and Flexibility
Electrospun membranes and TCPS were configured into a rectangular shape, with the dimensions of 50 × 10 mm.40 Tensile tests were performed on an Instron 5565 universal testing machine using a 250 N load cell with a crosshead speed of 5 mm/min. At least four samples were tested for each type of electrospun fibrous membrane. The Young’s modulus and elongation-at-break were obtained from the stress–strain curves.
BET Surface Area
Surface areas of PLA, PCL and TPU substrates were measured by a Brunauner–Emmett–Teller instrument (Micromeritics Gemini VII Version 2.00) using a nitrogen gas physical adsorption method. The instrument accuracy had a lower limit of 0.1 m2 and therefore only randomly oriented substrates, which could be synthesized at large scale, were measured. Prior to the BET measurement, samples were degassed in a vacuum oven at 45 °C for 24 h. A relative pressure range, P/P o, of 0.05–0.25 was used for calculating the BET surface area using the Gemini VII software. The individual mass of each substrate (used in surface area per substrate calculations) was assayed by measuring the mass of each polymer (n = 15) with washer and then subtracting the washer’s mass.
Protein Adsorption
The amount of protein adsorbed to each substrate was calculated using the Pierce BCA Protein Assay Kit and protocol (Thermo Fisher Scientific Inc., 2013) per manufacturer’s instructions. Proteins were allowed to adsorb onto the materials overnight, using the same media conditions that were used when seeding cells. A standard graph was made following the BCA standard kit protocol using bovine serum albumin (BSA) as standards. Absorbance at 560 nm of each sample (n = 3) was measured using a plate reader (Promega GloMax®-Multi Microplate Multimode Reader), and protein content was inferred from the standard graph.
Substrate Processing for Cell Culture
After electrospinning, each fiber substrate was allowed to air dry in a fume hood for at least 24 h, to allow full dissipation of any residual solvents. Samples were sterilized via ultraviolet light from the biosafety cabinet (Baker Company) for 20 min on each side of the scaffold. All cell culture experiments were conducted in 24-well plates that were pre-treated with polyhydroxyethylmethacrylate (poly-HEMA) (Sigma) to prevent cell attachment to culture wells. Poly-HEMA, 2 wt% solution was made with 100% ethanol (Pharmco-AAPER, ACS/USP Grade). Each scaffold was immersed in a protein solution to allow for protein adsorption before cells were seeded. The protein solution contained the following components: DMEM-high glucose (Life Technologies), supplemented with 5% FBS (Life Technologies), 2% Penicillin/Streptomycin (Life Technologies) and growth-factor-reduced Matrigel (BD) coated at a concentration of 0.02 mg per well, for a minimum of 1 h inside an incubator at 37 °C.
Media Conditions
Fibroblast cells were maintained in a “fibroblast medium” consisting of DMEM-high glucose (Life Technologies), supplemented with 10% FBS (Life Technologies), 1 mM l-glutamine (Life Technologies), 1% nonessential amino acids (Life Technologies) and Penicillin/Streptomycin (Life Technologies). Fibroblast medium was used during routine maintenance of the cells as well as during seeding onto the scaffolds.
“E8 medium” was made in house following the previously published protocol.4 E8 medium was then modified by excluding TGF-β, which is then referenced as “E7 medium.” Both E8 and E7 are a defined, xeno-free human pluripotent stem cell culture media and do not contain serum. Both E8 and E7 were used to culture human embyronic stem cell (hESC) lines, WA01 and WA01+H2B-mCherry cells, as well as the C1.2 iPSC line.
Generation of H2B-mCherry Transgenic Cells
hESC line WA01 (WiCell Research Institute) underwent genome modification to create an H2B-mCherry positive stem cell line via CRISPR/Cas9 gene editing. The H2B-mCherry plasmid was generated by cloning the H2B-mCherry sequence (Addgene # 20972) into the GFP sequence of the AAV-CAGGS-EGFP plasmid (Addgene #22212). This plasmid was electroporated with Cas9 plasmids as described previously.26,28 Isolated cell lines retained constitutive expression during culture and differentiation into HEF1 (fibroblast) cells (see next section). The hESC WA01+H2B-mCherry cell line was used for cell attachment and proliferation data. The cells were maintained on 6-well plates coated with Matrigel at a concentration of 8.6 µg/cm2. All stem cell lines were maintained with E8 medium, fed on a daily basis and typically passaged every 4 days using EDTA (Versene, Life Technologies).4 Before seeding the hESCs onto the corresponding substrates the cells were pretreated with a ROCK inhibitor (Y-27632, Selleck Chemical) at a 10 µM concentration for a minimum of 2 h. ROCK inhibition prevents apoptosis of single cells during passaging of human pluripotent stem cells.5,6,30,44 The passaged cells were seeded at a density of 4 × 104 cells per well with E8 medium for attachment and proliferation studies. Cells were fed on a daily basis and cultured at incubator settings of 5% CO2 and 37 °C.
Fibroblast Differentiation
WA09-H2B-mCherry hESCsand C1.2 iPSCs were made into embryoid bodies via the Aggrewell method (Stem Cell Technologies) per manufacturer’s instructions and cultured in non-adherent cell culture plates (Corning Inc.) to form aggregates in “differentiation medium” comprised of 80% knockout Dulbecco’s modified Eagle’s medium (KO-DMEM) (Invitrogen), 1 mM l-glutamine, 20% FBS, and 1% nonessential amino acids. After 4 days in suspension, aggregates were transferred onto matrigel-coated plates and cultured for an additional 9 days. After three passages in differentiation medium, cells were transitioned to fibroblast media. Most cells in culture appeared fibroblast-like after three to five passages. To confirm a fibroblast phenotype after differentiation, cells were stained with anti-prolyl 4-hydroxylase beta (mouse monoclonal clone #3-2B12A, Acris Antibodies) as described previously.19
Fibroblast Reprogramming
C1.2 fibroblasts contain the doxycycline (DXC)-inducible transcription factors Oct4, Sox2, Klf4 and c-Myc.19 The C1.2 cell line was prepared by infecting C1 fibroblasts with a inducible c-Myc virus19 to generate a C1.2 iPSC line that could be differentiated into “secondary” fibroblasts as described previously.19 C1.2 fibroblasts were cultured in fibroblast medium. Cells were maintained between 5 and 15 passages with media changes every 2 days and passaged every 6–7 days with TrypLE (Invitrogen) on TCPS plates or flasks.
Cells were seeded at a density of 4 × 104 per well of a 24-well plate (BD) for reprogramming experiments. DXC-treated cells were cultured with doxycycline hyclate (Sigma Cat. D9891) at a final concentration of 2 µg/mL. During reprogramming, cells were fed daily with fibroblast medium with DXC and 1 µM hydrocortisone4 (Sigma). The DXC stock concentration of 2 mg/mL solution was typically vortexed prior to addition.
Immunofluorescence
Samples were washed with PBS, fixed with 4% paraformaldehyde in PBS for 15 min, and permeabilized with 0.5% Triton X-100 in PBS for 30 min. Primary antibodies were allowed to bind overnight at 4 °C in a 5% donkey serum blocking buffer. Secondary antibodies were obtained from Life Technologies and used at a 1:400 dilution in 5% donkey serum for an incubation period of 1 h at 4 °C. Primary antibody dilutions are listed in (Supplemental Table 2). Imaging was performed using a Nikon Eclipse Ti.
Flow Cytometry
Data was acquired with an Accuri C6 (BD Biosciences) and analyzed using FlowJo (TreeStar). Samples that were antibody-stained were detached from substrates by two incubations of 10 min each in Accutase (Life Technologies) at 37 °C followed by the immunofluorescence staining protocol as described above. For attachment and proliferation studies, flow cytometry data was collected 24 h post-seeding for initial cell attachment data (n = 3), and again 72 h post seeding for proliferation data (n = 3). Each well containing the substrate was initially washed twice with PBS to ensure that the cells collected were not growing in suspension within the poly-HEMA coated well. For cell counting, TrypLE (Life Technologies) was used to detach the cells for 5 min at 37 °C. After incubation the cells were collected and centrifuged at 200×g for 5 min. The supernatant was then aspirated, and the cells were resuspended in 600 μL of PBS and filtered through a 100 µm mesh (BD) prior to analysis.
Nuclear Shape and Size Analysis
Nuclear size and shape index was calculated by manually tracing ellipses around individual Hoescht-stained nuclei. Nuclear shape index is given by the formula
where A and P represent the area and perimeter of a nucleus, respectively. Analysis was performed using Fiji software.37
Statistics
Statistical analysis was performed using Microsoft Excel via an unpaired, 2-tailed Student’s t test for single-variable analysis and a two-factor ANOVA test with replicates for two-variable analysis.
Partial Least Squares Regression
Analysis was performed in MATLAB using the SIMPLS method.8 The model was analyzed for goodness of fit (R 2 Y) as well as goodness of prediction (Q 2). Q 2 was validated by 14-fold cross validation so that each observation was left out exactly once.11 Inputs to the model consisted of an explanatory matrix. Columns in the matrix represented biochemical and biophysical cues. Each row represented a different combination of cues. A response matrix was generated from experimental data with columns representing different cellular responses and rows corresponding to the above cue combinations. Raw explanatory and response values were mean centered and variance scaled prior to analysis.
Results
Materials Library Characteristics
A biomimetic substrate library with ECM-like nanofibrous structure was successfully generated through electrospinning.42 Synthetic polymers PCL, PLA, PPC and TPU were individually electrospun as a membrane onto electrically conductive parallel plates to align fibers. Electrospinning without an organized electric field at the collector plate created substrates that were not aligned in a particular direction, termed “random” substrates (Fig. 1 and Supplementary Fig. 1). Scanning electron microscope (SEM) analysis confirmed a nanofibrous structure with fiber diameters of 500–1000 nm within the range of native ECM3 (Table 1). For the aligned substrates, the distribution of fiber diameter of TPU, PPC and PCL fibers were fairly concentrated within 10° of the desired direction of alignment (Table 1 and Supplementary Fig. 2). Aligned substrates also differed from randomly spun substrates in porosity (Table 1). Finally, a “shish kebab” structure was prepared with PCL nanofibrous substrates (PCL-SK) to produce a variant topology compared to standard PCL substrates43 (Supplemental Fig. 1 and Supplemental Table 3).
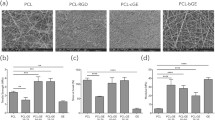
Nanofibrous substrate library with attached fibroblasts. Electron microscopy images of each polymer substrate [polylactic acid (PLA), polycaprolactone (PCL), thermoplastic polyurethane (TPU)]. Below each image is an immunocytochemistry image of HEF1 fibroblasts grown on each substrate. Cells were fixed after 5 days in culture. Nuclei and F-actin were stained by Hoechst dye and phalloidin, respectively. Arrows indicate fiber orientation. TCPS tissue culture polystyrene
Each substrate in the library was characterized for its surface area, surface hydrophobicity, elastic modulus, flexibility and porosity. A nitrogen adsorption isotherm on a large electrospun sample was utilized for surface area calculations. Areas per substrate reached three orders of magnitude higher than tissue culture polystyrene (TCPS, Table 1). The surface hydrophobicity of the substrates were characterized via WCAs and were higher than 90°, indicating that all the prepared substrates were hydrophobic (Table 1). TCPS had the smallest WCA, which is consistent with prior reports.23 The mechanical properties of substrates were significantly softer than TCPS but stiffer than native ECM,9 with a Young’s Modulus range between 7 and 40 MPa (Table 1). Flexibility of substrates was assessed by elongation at break. Nanofibrous substrates were more flexible than TCPS (Table 1).
Prior to cell seeding, nanofibrous substrates were pre-coated with a thin adsorbed layer of Matrigel, because pre-coating synthetic polymeric substrates with proteins can be important for cell-surface interaction with human pluripotent stem cells.23 Matrigel adsorbed well to nanofibrous substrates as measured through a bicinchoninic acid assay (Supplementary Fig. S3; Supplementary Table 3). Shortly after seeding, fibroblasts on randomly oriented fibers exhibited a morphology similar to fibroblasts grown on TCPS (Fig. 1). On all of the aligned substrates, fibroblasts were more elongated in comparison to cells on random substrates and TCPS (Fig. 1, white arrows indicate fiber orientation). Despite these differences in cell morphology, fibroblasts on all substrates were able to attach and proliferate at similar levels (Supplemental Fig. 4). Human pluripotent stem cells were also able to attach and proliferate at similar levels on all substrates (Supplemental Fig. 5). These results demonstrate that all of our matrigel-coated nanofibrous substrates can support the attachment and proliferation of both the starting and ending cell types—fibroblasts and pluripotent stem cells, respectively—of reprogramming experiments.
Reprogramming on Nanofibrous Substrates
Fibroblasts were next seeded on the material library and induced to express Oct3/4, Sox2, Klf4 and c-Myc reprogramming factors. We utilized a “secondary” reprogramming fibroblast system19 that harbored doxycycline (DXC)-inducible transgenes for the reprogramming factors to achieve transgene expression during reprogramming without viral infection. For subsequent reprogramming experiments that required culture over three to 4 weeks, we focused on TPU, PLA and PCL substrates due to their ease of synthesis and handling. A large fraction of PPC substrates tore or broke during routine handling in the initial cell attachment and proliferation studies. These substrates were deemed to be too fragile to be immersed in media and transported to and from standard incubators for multiple weeks.
Fibroblasts express a cell-surface marker, CD44, and gradually lose expression of CD44 as cells progress in reprogramming to intermediate states31 (Fig. 2a). In contrast, fully reprogrammed cells express the pluripotency marker, TRA-1-60, and are negative for CD44 (Fig. 2a). At 7, 12, 17 and 22 days after factor induction, cells were isolated from each substrate, co-stained for CD44 and TRA-1-60 via immunocytochemistry and analyzed for levels of these markers by flow cytometry. By following the percentage of cells in each gate (Figs. 2a and 2b), we were able to follow the progression and kinetics of cellular reprogramming on the materials library.
Nanofibrous substrates support the reprogramming of human fibroblasts. (a) Schematic of reprogramming. Mature fibroblast cells are CD44 positive (red) and, upon factor expression, progress stochastically to an intermediate state between fibroblasts and pluripotent stem cells (PSCs), which are positive for TRA-1-60 (green). Phase-contrast images of fibroblasts and PSCs were taken on TCPS. (b) Time course of marker levels during reprogramming on TPU-R as measured by flow cytometry. Numbers within plot indicate percent of total cells in each gate (shaded in red, grey or green). A.U., arbitrary fluorescent units. (c) Pluripotency marker staining of reprogrammed cells after removal of reprogramming factors. Central colonies are positive for all the pluripotency markers, Nanog, Oct3/4, SSEA-4, Sox2 and TRA-1-60. (d) Quantification of the kinetics of reprogramming on nanofibrous substrates. Amount of time required to lose fibroblast identity (50% of cells are CD44−) correlated with the number of reprogrammed cells at day 22. Dashed line represents linear regression best-fit line. All experiments were conducted in E7 media with hydrocortisone and doxycycline
On all nanofibrous substrates, cells initially were CD44+ and then ~40–60% of these cells lost CD44 expression by Day 7, thereby losing fibroblast identity and gaining an intermediate phenotype (Fig. 2b and Supplemental Fig. 6). Over the subsequent 15 days, a small percentage of intermediate cells gained hallmarks of pluripotent cells, leading to 1–7% of the overall population being TRA-1-60+/CD44− by day 22 (Fig. 2b and Supplemental Fig. 6). After reprogramming culture on nanofibrous substrates, DXC was removed from the media to shutdown reprogramming factor expression, and cells were grown and passaged for several weeks to see whether the observed TRA-1-60+/CD44− cells would retain their pluripotent markers. For all substrates, reprogrammed colonies could be identified that stained positive simultaneously for pluripotency markers: Nanog, Oct4, Sox2, SSEA-4 and TRA-1-60 (Fig. 2c). These reprogrammed cells were factor independent and acquired all marks of pluripotency, indicating robust reprogramming of fibroblast state.16 The nanofibrous substrates had similar kinetics of reprogramming overall (Supplemental Fig. 6), but important differences in the progression to intermediate states could be identified (Fig. 2d). The average time that it took cells to transit out of the fibroblast CD44+ state correlated strongly with the number of TRA-1-60+/CD44− pluripotent cells seen on day 22 (Fig. 2d). Top performing substrates were TPU-R, TCPS, TPU-A and PCL-R.
Early Effects of Nanofibrous Substrates
To narrow the time window over which nanofibrous substrates affect reprogramming, we switched substrates in the middle of reprogramming, dividing the process into an “early” (day 0–17) and “late” phase (day 18–24). For the “early” conditions, reprogramming factor-expressing fibroblasts were grown on a nanofibrous substrate for the first 17 days and passaged to TCPS for the last 7 days (Fig. 3). For the “late” conditions, reprogramming factor-expressing fibroblasts were cultured on TCPS for the first 17 days and passaged to a nanofibrous substrate for the last 7 days (Fig. 3). Flow cytometric analysis of cells at the end of these experiments indicated that early culture on nanofibrous substrates could boost reprogramming, with up to 30% of cells being reprogrammed, while late culture did not boost reprogramming over TCPS levels (Fig. 3). The top performing nanofibrous substrates that had faster kinetics—TPU-Random (TPU-R), TPU-Aligned (TPU-A) and PCL-Random (PCL-R) (Fig. 2d)—performed well in the early conditions only (Fig. 3).
Nanofibrous substrates enhance early events during reprogramming. Top: Schematic of experimental design. Reprogramming was carried out for 24 days under two conditions: early vs. late culture on nanofibrous scaffolds. “Early Nanofibers” labels culture on indicated nanofibrous substrate during days 0–17 of factor expression, while “Late Nanofibers” labels culture on indicated nanofibrous substrate during days 18–24 of factor expression. Cells were cultured on TCPS during the other periods of reprogramming. Bottom: Percent of TRA-1-60+ cells after 24 days of reprogramming in both conditions. Error bars denote 95% confidence intervals. Asterisks indicate p values <0.001 from an unpaired Student’s t test
Reprogramming Factors Quickly Induce Circular Nuclei
We reasoned that early effects of nanofibrous substrates could be mediated by early perturbations of nuclear shape, area and structure. Immunocytochemical analysis of cells on scaffolds permits the tracking of nuclear size and nuclear shape index (NSI), which ranges from 1 for circular nuclei to near zero for a more elongated shape (Fig. 4a). Fibroblasts grown in basal media conditions adopted a more elongated morphology on aligned substrates compared to TCPS and random substrates, consistent with results seen in the fibroblast attachment and proliferation experiments (Figs. 1 and 4b, Supplemental Fig. 4). Reprogramming factor expression abolished nuclear shape differences (Fig. 4b), indicating all nuclei on average have the same shape on all of the substrates. Similarly, nuclear area was uniform after 7 days of factor expression (Supplemental Fig. 7A), and reduced slightly in reprogramming conditions compared to basal media conditions, as previously reported.27 We probed the global histone 3 acetylation (AcH3) levels of these nuclei via immunocytochemistry followed by flow cytometry analysis (for representative data, see Supplemental Fig. 7B). For all conditions, global histone acetylation levels decreased modestly within the nuclei of cells after 7 days of induction of reprogramming factors (Fig. 4c). Only a weak correlation between AcH3 levels and NSI (R 2 = 0.51) was observed (Supplemental Fig. 7C). To summarize, aligned nanofibrous substrates altered nuclear shape during routine culture, but expression of reprogramming factors abolished these changes within 7 days, leading to smaller nuclei without any elevation in global AcH3 levels.
Reprogramming factors induce circular nuclear shape for all conditions. (a) Diagram of the nuclear shape index (NSI) metric. Grey-scale image for nuclei stained with Hoechst. The NSI of a perfect circle is equal to one, while the NSI of a line approaches zero. (b) Effects of nanofibrous substrates on NSI of C1.2 fibroblast cells during normal growth (Basal, E7 media, light blue) and during reprogramming (E7 media with hydrocortisone and doxycycline, dark blue). After 7 days of treatment, cells were fixed and stained nuclei were analyzed. Error bars denote 95% confidence intervals. Asterisks indicate p values from an unpaired Student’s t test (*p < 0.01, **p < 0.001, ***p < 10−5). (c) Mean histone 3 acetylation (AcH3) levels in cells with and without reprogramming factors for 7 days, as measured by flow cytometry
TGF-β Inhibits Effects of Nanofibrous Substrates
Given that transforming growth factor beta (TGF-β) can perturb early phases of reprogramming by inhibiting the mesenchymal to epithelial transition (MET),24,25,36 this growth factor was added to our reprogramming media to see if it would affect reprogramming on nanofibrous substrates. TGF-β slowed reprogramming in all conditions, as measured by the lower percentage of CD44− cells that have lost fibroblast identity throughout the time course (Fig. 5a). Analysis at the end of reprogramming also indicated that TGF-β decreases the number of intermediate and reprogrammed cells across all conditions (Fig. 5b, Supplementary Fig. 8). Notably, TGF-β abolished the previously observed differences among the different nanofibrous substrates (Fig. 5c), and no nanofibrous substrates in media conditions with TGF-β outperformed TCPS.
TGF-β inhibits effects of nanofibrous substrates. (a) Kinetics of reprogramming on nanofibrous substrates when media is supplemented with TGF-β. Non-fibroblast population (Intermediate+iPSC fraction) was analyzed by flow cytometry (described in Fig. 2b). (b) Change in the Intermediate+iPSC fraction on various substrates when media was supplemented with TGF-β. (c) When TGF-β was added to the media, amount of time required to lose fibroblast identity did not correlate with number of reprogrammed cells at day 22. Red dashed line represents linear regression best-fit line. Blue dashed line represents linear regression best-fit line in control media (as shown in Fig. 2d). Error bars denote 95% confidence intervals. Asterisks indicate p values <0.001 from an unpaired Student’s t test
Cues Controlling Reprogramming
To gain deeper insight into biophysical and biochemical cues that correlate with observed reprogramming responses, we performed partial least squares regression (PLSR) between cues and responses. A PLSR model was built using datasets with materials properties and media conditions as the multivariate cues and the number and timing of reprogramming as the response inputs. When the experimental values of the numbers and times of reprogramming were plotted against those predicted by the PLSR model (Fig. 6a), a linear relationship with a relatively low root mean square error of prediction (RMSPE) was observed, suggesting a good predictive ability for the cues within the training set (RMSPE ~ 1). Cues with high weights in a two-component model were TGF-β and polymer identity, while fiber diameter, WCA, protein adsorption and alignment had low weights (Fig. 6b, Supplemental Fig. 9A). PLSR modeling of all materials properties and cues with all cellular responses yielded a predictive model (Supplemental Fig. 9B) with low error (Supplemental Fig. 9C) while using five components (Supplemental Fig. 9D). In this expanded model, TGF-β again was heavily weighted.
Partial least squares regression of biophysical and biochemical cues and cellular responses. (a) Diagram of the predictability of PLSR model. A perfectly predicted model will follow the line y = x. R 2 values are 0.76 and 0.82 for time to 50% fibroblast cells and number or iPSCs per substrate respectively. (b) Biophysical cues with the largest contribution to principal component transformation of number of iPSCs in a two-component model. Negative regression coefficients indicates inhibition of cellular reprogramming responses, while positive indicates promotion. (c) Root mean square error in predictive calculation. Increased number of components leads to greater error in predictions due to overfitting. (d) Cumulative sum of variance explained by increasing number of components
Discussion
Our results demonstrate robust reprogramming of human cells on nanofibrous substrates. Reprogrammed cells were similar to those generated on standard polystyrene substrates, were factor-independent, and had all markers of pluripotent cells as reported in prior work.16,35 Top-performing substrates accelerated early stages of reprogramming that intersect with the timing of MET during reprogramming.24,25,36 These substrates may be able to activate pathways involving actin filament organization that have been implicated in mesenchymal–epithelial transitions.18
Because top-performing substrates varied along many different properties, it is likely that a combination of material characteristics led to enhanced reprogramming. PLSR modeling indicated that polymer identity, following TGF-β, was the next most important cue governing reprogramming response. Surface area and porosity also had a modest effect. Further molecular characterization of the substrates may be necessary to understand the different rates of reprogramming on our library. It is possible that the top performing nanofibrous substrates permit an advantageous conformation of Matrigel23 that encourages integrin engagement, focal adhesion formation or growth factor stabilization21 to promote reprogramming. Because the distribution of cytoskeletal stress fibers can be sensitive to geometry of underlying substrates,20 nanofibrous substrates could also change the degree of actin polymerization and actomyosin contractility. Further characterization of these molecular differences involving cell–material interactions among the substrates in our library could provide more insight into why observed differences were seen in the kinetics and efficiencies of reprogramming. Finally, expanding the library to sample material property space evenly may be able to provide a more comprehensive picture of how reprogramming occurs on nanofibrous materials. In our library, the total amounts of protein adsorbed to these substrates did not vary more than an order of magnitude. Likewise, mechanical properties and WCAs did not vary greatly across the library.
A surprising result was that aligned substrates did not retain an elongated nuclear shape during reprogramming. Reprogramming factors within 7 days could bypass biophysical constraints imposed by our nanofibrous substrates. The mechanisms by which this rapid change occurred was suppressed by TGF-β and likely intersects with MET pathways.7,24,25,36
In a mouse reprogramming system, aligned substrates significantly enhanced reprogramming of mouse fibroblasts,10 but markedly different responses were observed here with human fibroblasts among randomly spun and aligned nanofibrous substrates. In contrast to mouse reprogramming results on aligned nanofibrous substrates, nuclei were not hyper acetylated on histone 3 in our experiments. No correlation of hyperacetylation with enhanced reprogramming was observed, and AcH3 regression coefficients in PLSR models were near zero (Supplemental Fig. 9B–D). This disparity may be attributed to known differences in the signaling requirements between mouse and human pluripotent cells.16 Profiling local acetylation of particular genomic loci, rather than averaging acetylation globally over the whole genome, may be required to understand the differences between the mouse and human results on nanofibrous substrates.
Conclusions
This work represents the first report of reprogramming of human cells on nanofibrous polymeric substrates. All of the polymers tested—PLA, TPU and PCL—supported fibroblast and pluripotent cell attachment and full reprogramming to factor-independent pluripotent stem cells. TPU-R, TPU-A and PCL-R were the best substrates in our library, outperforming standardly used polystyrene substrates. The enhanced reprogramming on nanofibrous substrates occurred during the first 17 days of reprogramming and was inhibited by TGF-β. These results suggest a possible role of nanofibrous substrates to enhance MET of fibroblasts during early stages of reprogramming. Notably, reprogramming factors were able to change nuclear shape of cells on all substrates within 7 days regardless of the initial nuclear shape constrained by the substrate. PLSR modeling of several materials properties revealed a complex relationship between polymer identity and reprogramming with less pronounced roles for fiber diameter, fiber alignment, protein adsorbtion and WCA. We believe further work with the identified top-performing substrates will lead to the design of substrates that promote epigenetic reprogramming.
References
Apostolou, E., and K. Hochedlinger. Chromatin dynamics during cellular reprogramming. Nature 502:462–471, 2013.
Armond, J. W., K. Saha, A. A. Rana, C. J. Oates, R. Jaenisch, M. Nicodemi, and S. Mukherjee. A stochastic model dissects cell states in biological transition processes. Sci. Rep. 4:3692, 2014.
Barnes, C. P., S. A. Sell, E. D. Boland, D. G. Simpson, and G. L. Bowlin. Nanofiber technology: designing the next generation of tissue engineering scaffolds. Adv. Drug Deliv. Rev. 59:1413–1433, 2007.
Beers, J., D. R. Gulbranson, N. George, L. I. Siniscalchi, J. Jones, J. A. Thomson, and G. Chen. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 7:2029–2040, 2012.
Chen, G., Z. Hou, D. R. Gulbranson, and J. A. Thomson. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell 7:240–248, 2010.
Chen, G., D. R. Gulbranson, Z. Hou, J. M. Bolin, V. Ruotti, M. D. Probasco, K. Smuga-Otto, S. E. Howden, N. R. Diol, N. E. Propson, R. Wagner, G. O. Lee, J. Antosiewicz-Bourget, J. M. Teng, and J. A. Thomson. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8:424–429, 2011.
Craene, B. D., and G. Berx. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13:97–110, 2013.
de Jong, S. SIMPLS: an alternative approach to partial least squares regression. Chemometr. Intell. Lab. Syst. 18:251–263, 1993.
Discher, D. E., D. J. Mooney, and P. W. Zandstra. Growth factors, matrices, and forces combine and control stem cells. Science 324:1673–1677, 2009.
Downing, T. L., J. Soto, C. Morez, T. Houssin, A. Fritz, F. Yuan, J. Chu, S. Patel, D. V. Schaffer, and S. Li. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 12:1154–1162, 2013.
Eriksson, L. Multi- and Megavariate Data Analysis, MKS Umetrics AB, 2006.
Feng, B., J.-H. Ng, J.-C. D. Heng, and H.-H. Ng. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4:301–312, 2009.
Gaspar-Maia, A., A. Alajem, E. Meshorer, and M. Ramalho-Santos. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12:36–47, 2011.
Graf, T., and T. Enver. Forcing cells to change lineages. Nature 462:587–594, 2009.
Grskovic, M., A. Javaherian, B. Strulovici, and G. Q. Daley. Induced pluripotent stem cells—opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 10:915–929, 2011.
Hanna, J. H., K. Saha, and R. Jaenisch. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143:508–525, 2010.
Hanna, J., K. Saha, B. Pando, J. van Zon, C. J. Lengner, M. P. Creyghton, A. van Oudenaarden, and R. Jaenisch. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462:595–601, 2009.
Haynes, J., J. Srivastava, N. Madson, T. Wittmann, and D. L. Barber. Dynamic actin remodeling during epithelial–mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell 22:4750–4764, 2011.
Hockemeyer, D., F. Soldner, E. G. Cook, Q. Gao, M. Mitalipova, and R. Jaenisch. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3:346–353, 2008.
Jain, N., K. V. Iyer, A. Kumar, and G. V. Shivashankar. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. PNAS 110:11349–11354, 2013.
Jiao, J., Y. Dang, Y. Yang, R. Gao, Y. Zhang, Z. Kou, X. F. Sun, and S. Gao. Promoting reprogramming by FGF2 reveals that the extracellular matrix is a barrier for reprogramming fibroblasts to pluripotency. Stem Cells 31:729–740, 2012.
Kim, I. L., S. Khetan, B. M. Baker, C. S. Chen, and J. A. Burdick. Fibrous hyaluronic acid hydrogels that direct MSC chondrogenesis through mechanical and adhesive cues. Biomaterials 34:5571–5580, 2013.
Kohen, N. T., L. E. Little, and K. E. Healy. Characterization of Matrigel interfaces during defined human embryonic stem cell culture. Biointerphases 4:69–79, 2009.
Li, R., J. Liang, S. Ni, T. Zhou, X. Qing, H. Li, W. He, J. Chen, F. Li, Q. Zhuang, B. Qin, J. Xu, W. Li, J. Yang, Y. Gan, D. Qin, S. Feng, H. Song, D. Yang, B. Zhang, L. Zeng, L. Lai, M. A. Esteban, and D. Pei. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7:51–63, 2010.
Liu, X., H. Sun, J. Qi, L. Wang, S. He, J. Liu, C. Feng, C. Chen, W. Li, Y. Guo, D. Qin, G. Pan, J. Chen, D. Pei, and H. Zheng. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT–MET mechanism for optimal reprogramming. Nat. Cell Biol. 15:829–838, 2013.
Mali, P., L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville, and G. M. Church. RNA-guided human genome engineering via Cas9. Science 339:823–826, 2013.
Mattout, A., A. Biran, and E. Meshorer. Global epigenetic changes during somatic cell reprogramming to iPS cells. J. Mol. Cell Biol. 3:341–350, 2011.
McNulty, J. D., T. Klann, J. Sha, M. Salick, G. T. Knight, L. S. Turng, and R. S. Ashton. High-precision robotic microcontact printing (R-μCP) utilizing a vision guided selectively compliant articulated robotic arm. Lab Chip 14:1923–1930, 2014.
Mi, H.-Y., X. Jing, B. R. Jacques, L.-S. Turng, and X.-F. Peng. Characterization and properties of electrospun thermoplastic polyurethane blend fibers: effect of solution rheological properties on fiber formation. J. Mater. Res. 28:2339–2350, 2013.
Ohgushi, M., M. Matsumura, M. Eiraku, K. Murakami, T. Aramaki, A. Nishiyama, K. Muguruma, T. Nakano, H. Suga, M. Ueno, T. Ishizaki, H. Suemori, S. Narumiya, H. Niwa, and Y. Sasai. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7:225–239, 2010.
Quintanilla, R. H., Jr., J. S. T. Asprer, C. Vaz, V. Tanavde, and U. Lakshmipathy. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS ONE 9:e85419, 2014.
Rais, Y., A. Zviran, S. Geula, O. Gafni, E. Chomsky, S. Viukov, A. A. Mansour, I. Caspi, V. Krupalnik, M. Zerbib, I. Maza, N. Mor, D. Baran, L. Weinberger, D. A. Jaitin, D. Lara-Astiaso, R. Blecher-Gonen, Z. Shipony, Z. Mukamel, T. Hagai, S. Gilad, D. Amann-Zalcenstein, A. Tanay, I. Amit, N. Novershtern, and J. H. Hanna. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502:65–70, 2013.
Rnjak-Kovacina, J., S. G. Wise, Z. Li, P. K. Maitz, C. J. Young, Y. Wang, and A. S. Weiss. Tailoring the porosity and pore size of electrospun synthetic human elastin scaffolds for dermal tissue engineering. Biomaterials 32:6729–6736, 2011.
Saha, K., and R. Jaenisch. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell 5:584–595, 2009.
Saha, K., Y. Mei, C. M. Reisterer, N. K. Pyzocha, J. Yang, J. Muffat, M. C. Davies, M. R. Alexander, R. Langer, D. G. Anderson, and R. Jaenisch. Surface-engineered substrates for improved human pluripotent stem cell culture under fully defined conditions. PNAS 108:18714–18719, 2011.
Samavarchi-Tehrani, P., A. Golipour, L. David, H. K. Sung, T. A. Beyer, A. Datti, K. Woltjen, A. Nagy, J. L. Wrana. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7:64–77, 2010.
Schindelin, J., I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, and A. Cardona. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9:676–682, 2012.
Shahbazian, M. D., and M. Grunstein. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100, 2007.
Shinagawa, T., T. Takagi, D. Tsukamoto, C. Tomaru, L. M. Huynh, P. Sivaraman, T. Kumarevel, K. Inoue, R. Nakato, Y. Katou, T. Sado, S. Takahashi, A. Ogura, K. Shirahige, and S. Ishii. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 14:217–227, 2014.
Takahashi, K., and S. Yamanaka. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676, 2006.
Takahashi, K., and S. Yamanaka. Induced pluripotent stem cells in medicine and biology. Development 140:2457–2461, 2013.
Teo, W., and S. Ramakrishna. A review on electrospinning design and nanofibre assemblies. Nanotechnology 17:R89, 2006.
Wang, X., M. R. Salick, X. Wang, T. Cordie, W. Han, Y. Peng, Q. Li, and L. S. Turng. Poly (ε-caprolactone) nanofibers with a self-induced nanohybrid Shish–Kebab structure mimicking collagen fibrils. Biomacromolecules 14:3557–3569, 2013.
Watanabe, K., M. Ueno, D. Kamiya, A. Nishiyama, M. Matsumura, T. Wataya, J. B. Takahashi, S. Nishikawa, S. Nishikawa, K. Muguruma, and Y. Sasai. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25:681–686, 2007.
Yu, J., M. A. Vodyanik, K. Smuga-Otto, J. Antosiewicz-Bourget, J. L. Frane, S. Tian, J. Nie, G. A. Jonsdottir, V. Ruotti, R. Stewart, I. I. Slukvin, and J. A. Thomson. Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920, 2007.
Acknowledgments
We acknowledge generous financial support from the Wisconsin Institute for Discovery (T.C. and K.S.), a Grainger Fellowship (J.C-S.) and the Society in Science Foundation (K.S.). We also would like to thank all members of the Saha lab and BIONATES theme for advice and support throughout this project. We acknowledge Dr. Rob McClain and the University of Wisconsin-Madison Biochemistry department for the use and expertise in collecting surface area data via the BET Micromeritics Gemini VII instrument.
Conflict of interest
Travis Cordie, Ty Harkness, Xin Jing, Jared Carlson-Stevermer, Hao-Yang Mi, Lih-Sheng Turng and Krishanu Saha declare that they have no conflicts of interest. Dr. Saha reports grants from Society in Science Foundation during the conduct of the study.
Ethical Standards
No human and animal studies were carried out by the authors for this article. All work with human embryonic stem cell lines was carried out in accordance with institutional, national, and international guidelines and approved by the Stem Cell Research Oversight Committee at the University of Wisconsin-Madison.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor David Mooney oversaw the review of this article.
This paper is part of the 2014 Young Innovators Issue.
Travis Cordie and Ty Harkness have contributed equally to this work.
Krishanu Saha is an Assistant Professor in the Department of Biomedical Engineering at the University of Wisconsin-Madison. He is also a member of the Wisconsin Institute for Discovery in the bionanocomposite tissue engineering scaffolds (BIONATES) theme. Prior to his arrival in Madison, Dr. Saha studied Chemical Engineering at Cornell University and at the University of California in Berkeley. In his dissertation with Professors David Schaffer and Kevin Healy, he worked on experimental and computational analyses of neural stem cell development, as well as the design of new materials for adult stem cell culture. In 2009 he became a Society in Science: Branco-Weiss fellow in the laboratory of Professor Rudolf Jaenisch at the Whitehead Institute for Biomedical Research at MIT and in the Science and Technology Studies program at Harvard University with Professor Sheila Jasanoff in Cambridge, Massachusetts. Since then, he has performed research on human pluripotent stem cells, disease modeling and synthetic biology.

Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cordie, T., Harkness, T., Jing, X. et al. Nanofibrous Electrospun Polymers for Reprogramming Human Cells. Cel. Mol. Bioeng. 7, 379–393 (2014). https://doi.org/10.1007/s12195-014-0341-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-014-0341-z