Abstract
A novel quasi-three-dimensional (quasi-3D) microscopy technique has been developed to enable visualization of a cell under dynamic loading in two orthogonal planes simultaneously. The three-dimensional (3D) dynamics of the mechanical behavior of a cell under fluid flow can be examined at a high temporal resolution. In this study, a numerical model of a fluorescently dyed cell was created in 3D space, and the cell was subjected to uniaxial deformation or unidirectional fluid shear flow via finite element analysis. Therefore, the intracellular deformation in the simulated cells was exactly prescribed. Two-dimensional fluorescent images simulating the quasi-3D technique were created from the cell and its deformed states in 3D space using a point-spread function (PSF) and a convolution operation. These simulated original and deformed images were processed by a digital image correlation technique to calculate quasi-3D-based intracellular strains. The calculated strains were compared to the prescribed strains, thus providing a theoretical basis for the measurement of the accuracy of quasi-3D and wide-field microscopy-based intracellular strain measurements against the true 3D strains. The signal-to-noise ratio (SNR) of the simulated quasi-3D images was also modulated using additive Gaussian noise, and a minimum SNR of 12 was needed to recover the prescribed strains using digital image correlation. Our computational study demonstrated that quasi-3D strain measurements closely recovered the true 3D strains in uniform and fluid flow cellular strain states to within 5% strain error.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent experimental and theoretical studies have shown that intracellular deformation of cells under fluid flow is inhomogeneous and location and height dependent,1,5,14 highlighting the importance of accurately measuring the three-dimensional (3D) intracellular strains. One typical image-based method to measure intracellular strains under mechanical loading is to use fluorescent dyes or proteins to visualize intracellular structures and then record their displacements using microscopy. Numerical methods are then utilized on these images to calculate the intracellular deformation.6,12
The most common microscopy techniques to visualize intracellular structures are wide-field fluorescent microscopy or confocal/deconvolution microscopy. However, traditional wide-field fluorescent microscopes are limited by their lack of z-direction (depth) information, and confocal/deconvolution techniques, while 3D, lose temporal resolution due to the necessity of raster scanning a z-stack.11 Previously, our laboratory developed a quasi-three-dimensional (quasi-3D) imaging system that can observe a single cell in two orthogonal planes simultaneously by using two wide-field fluorescent microscopes, thereby allowing additional z-direction visualization without a loss in temporal resolution. Details about the quasi-3D image system can be found in a previously published article.2 The quasi-3D technique was used to calculate quasi-3D intracellular stain fields of a single cell under fluid flow.
Strain is defined as the deformation of a point in a 3D space. However, 2D images captured by wide-field microscopy contain information from an in-focus plane of a 3D object (with a thickness related by the depth of field for the imaging system) but also contain information from blurred out-of-focus planes related in Fourier optics by a point spread function (PSF) that may distort the accuracy of image-based strain calculations. Furthermore, the relationship and accuracy between the “quasi-3D” strain field calculated through a “compressed” two dimensional image to the real strain in 3D space is unknown.
In this study, a computational model was developed to investigate prescribed 3D strain states with wide-field quasi-3D microscopy-based strain measurements using the PSF and convolution operation. A “virtual”, simulated 3D fluorescently dyed cell was input into a finite element computational model and mechanically deformed by a fluid-structure-coupled finite element analysis (FEA) model to generate precise levels of intracellular strain. Quasi-3D images were generated from the model cell and its deformed states using a PSF and convolution operation.3,10
A digital image correlation technique used in the actual experimental measurements was employed to calculate the quasi-3D strains for comparison to the finite element prescribed strains. Therefore, the accuracy and precision of quasi-3D intracellular strain measurements were investigated theoretically.
The objectives of this study were to: (1) establish a numerical mathematical model to mimic cell deformation under fluid flow based on FEA; (2) obtain simulated quasi-3D images of the cell states in (1) using PSF and convolution and calculate intracellular strain fields using the same digital image correlation and differentiation technique in previous quasi-3D measurements; and (3) determine the accuracy of quasi-3D image-based strain calculations vs. the prescribed deformations by FEA.
Methods
The details of quasi-3D microscopy and related intracellular strain calculations can be found elsewhere.2 Briefly, a custom-designed dual-microscope system consisted of an inverted microscope and an upright microscope. A custom-built holder aligned a 45° mirror in the light path of the upright microscope to obtain “side-view” images of a single cell, while the inverted microscope obtained regular “bottom-view” images of the same cell simultaneously (Fig. 1a). The cell was imaged in a glass tube with a rectangular cross-section, which provided perpendicular glass planes for high-resolution imaging and also served as a fluid flow chamber. Both microscopes used 60× LUCPLNFLN long working distance objective lenses. Epi-fluorescent images (0.215 μm/pixel resolution) captured from both microscopes were processed by digital image correlation for determination of displacement fields. The resulting x–y (for bottom-view) and x–z (for side-view) displacement fields (see axis definitions in Fig. 1a) were smoothed and differentiated using a 2D Savitsky-Golay bilinear least-squares filter.9 Finite Lagrangian strain fields were calculated from the differentiation fields (three bottom-view strains: E yy , bottom-view E xx , and E xy ; and three side-view strains: E zz , side-view E xx , and E xz ). Subcellular regions in a cell were also defined for analyzing heterogeneous strain distributions in both bottom-view and side-view (Fig. 1b).
(a) Schematic of the quasi-3D microscopy setup. (b) Axes for x–y plane bottom-view and x–z plane side-view images in the quasi-3D system defined. Subcellular location divisions are depicted. Subcellular thirds for bottom and side views are in blue lines, and subcellular heights for side-view are in red lines. The axes of the cube model were the same as in the cell shape model
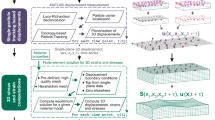
The computational approach involved generation of fluorescent images of cell slices using a digital fluorescence algorithm (Fig. 2). The original and deformed objects represented by their resultant fluorescent images have a one-to-one correspondence with their respective models in FEA. Therefore, the deformation strain states of these objects were precisely quantified. These original and deformed image slices of an object were convolved to create a “simulated” quasi-3D image of the object using the PSF of our microscopy system. A digital image correlation technique was used to calculate the quasi-3D strains for comparison to the finite element prescribed strains.
Flow chart of the procedure to compare 3D FEA-based strains with quasi-3D-based strain measurements. Fluorescent images of a single cell were simulated and their deformed counterparts were created via FEA. The intracellular deformation in the simulated cells was precisely determined. Quasi-3D images of cells were simulated by convolving these 3D cells with a PSF. These simulated original and deformed images were processed by digital image correlation to calculate quasi-3D strains. These calculated strains were compared to the FEA predicted strains, thus providing a theoretical basis for the measurement of the accuracy of quasi-3D intracellular strain measurements
Digital Fluorescence Algorithm
To mimic the light properties of randomly distributed intracellular fluorescent dye particles, randomly distributed points in 3D space in an object were treated as particle point sources of light with an assigned value of light intensity. The mean fluorescent light intensity for a cell measured in our quasi-3D images was used to mimic that of a simulated cell. However, the light intensity of each individual particle was assigned a randomly generated value from a Gaussian distribution. The light intensity values generated by the Gaussian distributor were in the same range as those typically seen in experimentally obtained quasi-3D images in our system. A distance decay function was adopted to assign the light intensity from each particle point to a surrounding 5 × 5 × 5 image pixel grid to create a nonuniform light intensity pattern similar to fluorescent images of cells with punctate structures ~1 μm in size. The distance between two adjacent grid points, i.e., the image pixel resolution, was the same as the quasi-3D system, 0.215 μm. The distance decay function was:
where I 0(0) is the initial light intensity and the parameter α is the bandwidth. We set α = 10 μm. z is the distance from the points (i, j) to the adjacent point (i + Δi, j + Δj). From this procedure, a “virtual”, simulated 3D fluorescently dyed cube or a hemi-spherical cell-like object was created. Figure 3 shows representative 2D fluorescent sections of a cube and a realistic cell. By running the Gaussian random light intensity value generator several times, five sets of virtual cubes or hemi-spherical cells (n = 5 each) were created for subsequent processing and analyses. The edge length of the cube was 15 μm, and the diameter of a typical rounded, realistic cell was about 15 μm as well. The realistic cell 3D shape was reconstructed from previous quasi-3D cell membrane images.2 It should be noted that the spatial positions of the fluorescent dye particles were used as nodes of tetrahedral elements in the finite element model of the same cell to precisely prescribe cellular deformations.
Deformed Cube Images
In order to precisely create deformation of the cube, we created a 15 μm cube finite element model using commercial finite element software ADINA 8.6.2 (ADINA R&D, Waterman, MA). The fluorescent particle points were used as nodes of the tetrahedral elements in the model. The size of each element was about 1 μm. Three uniaxial tensile (E xx , E yy , and E zz ) and two uniaxial shear strain conditions (E xy and E xz ) were applied to the model. The uniaxial strains in the cube were homogeneous in the applied direction. A linear elastic solid model was adopted as the constitutive cube material model. The Young’s modulus was set as 1 kPa, and the Poisson’s ratio was 0.45.
Appropriate uniaxial displacement boundary conditions were imposed to ensure uniaxial tension or shear strains. The cube was deformed in 20 uniform incremental deformation steps. All the element nodes were deformed within the whole cube. Therefore, the deformed positions of fluorescent particle points were tracked and used to generate the corresponding simulated deformed images of the cell. Uniaxial strain and simple shear cases were applied to the cube model in directions corresponding to the measurable strains in the quasi-3D technique (i.e., E xx bottom-view, E xy , E zz , E yy , E xx side-view, and E xz ).
Realistic Cell Images under Fluid Flow Deformation
The fluorescent particle positions of a 3D realistic cell were used as nodes and input into a fluid–structure interaction finite element model using ADINA. The fluid and solid solution variables were fully coupled. The flow boundary conditions were set to give a wall shear stress of 7 dynes/cm2 in a tube of the same rectangular cross-section as in our previous experimental study. Flow was applied for 10 s in the FEA model followed by a 10 s rest period. The cell was deformed in 27 steps with the same incremental deformation amplitude over 20 s. The deformed positions of fluorescent particle points were tracked and used to generate the corresponding simulated images of the deformed cell.
Quasi-3D Image Simulation and Strain Calculation
The quasi-3D technique consists of two wide-field fluorescent microscopes that can simultaneously and independently obtain 2D images of a cell in the orthogonal “bottom-view” x–y and “side-view” x–z planes. To mimic this image technique, the 3D simulated fluorescent images (n = 5 for either cube or cell images) were convolved with a PSF onto x–y or x–z plane slices to create simulated bottom or side view fluorescent wide-field microscopy images for quasi-3D analysis. The PSF describes the response of an imaging system to a point source or point object of light. With linear optics, the image of a complex object can then be seen as a convolution of the true object and the PSF. In this case, the “true” object was the simulated image of a cell. With information of the specific PSF for our microscopy system, we generated 2D bottom or side views of the cell. We adopted a PSF that used an analytical derivation using Fraunhofer diffraction along with a 0.7 Numerical Aperture, 60× magnification, 510 nm wavelength of emission light, and CCD pixel size (12.9 μm), to generate a 3D PSF.4,7,8 The convolution of the PSF and the light intensity matrix containing the cell and all related calculations were performed in MATLAB 2009b (The Mathworks, Natick, MA). Furthermore, to test the effect of general types of image noise on the accuracy of the PSF and digital image correlation, additive steps of Gaussian noise were uniformly applied to the quasi-3D image slices of the cell model under fluid shear to decrease the signal-to-noise ratio (SNR). SNR was defined as the mean intensity of the inside of the cell subtracted by the mean of the background non-cell region, divided by the standard deviation of the background. Convolved sets of quasi-3D images of the object and its deformed states were processed to obtain strain fields according to a previously established technique.2 Briefly, each deformed image was compared against the original using a digital image correlation technique to obtain displacement fields inside the object. The displacement fields were smoothed and differentiated using a bilinear least squares filter and then Lagrangian strains were determined.9 The E xx , E yy , and E xy were calculated for the bottom-view images, and E zz , E xx , and E xz were calculated for the side-view images. These average quasi-3D strains were compared with the finite element prescribed strain values. To analyze the effect of focal plane choice in the accuracy of the quasi-3D strain calculations, 5 different planes from the PSF-convolved cube were chosen (Fig. 4), and these planes were tracked through the steps of applied deformation. 3 planes were inside the cube, and 2 planes were outside the cube.
Results
Cube Cell Model under Unixial Strains
A homogeneous uniaxial strain model of a cube and a heterogeneous shear flow model of a cell coupled with a digital fluorescence algorithm were successfully created. These 3D models were convolved with a PSF function specific for the experimental quasi-3D setup.2 Sample 2D PSF-convolved images of the fluorescent cube and cell before and after deformation are shown in Fig. 5. It should be noted that the precise deformation of these simulated images was known and was able to be compared with quasi-3D calculations.
Cube and cell image simulation. The fluorescently dyed 3D cube or cell was convolved with a PSF, and individual planes were extracted to obtain 2D images. Sample 2D images of the (a) cube and (b) cell are provided before and after deformation by uniaxial tension for the cube and fluid shear for the cell. Arrows represent the direction of deformation
In the uniaxial deformation cube model, 5 different PSF-convolved planes in and around the cube were analyzed to determine the effect of focal plane position and the blur induced by a PSF on strain accuracy. Positions of the different focal planes are defined in Fig. 4. Planes outside the cube had a blurry appearance with degradation of cube features. Planes inside the cube had more distinct and sharp features. Strains calculated from planes inside the cube matched closely with the finite element-prescribed strain values, but strains from planes outside the cube were consistently lower than the actual value (Fig. 6). This demonstrates that uniaxial homogeneous strains calculated by digital image correlation are relatively unaffected by the added blur from neighboring planes by PSF convolution as long as the focal plane is within the area being deformed and not outside.
Six different uniaxial strain or pure shear cases were applied to the PSF-convolved simple cube model. The applied strain was then calculated in the middle plane of the cube in the view that the applied strain was visible (e.g., E zz was measured using the side-view x–z plane). The results are presented in Table 1. The quasi-3D calculated strains closely matched the prescribed strains (from FEA) in the simple cube model in both uniaxial and shear strain cases. The relative error for these measurements was less than 5%.
Realistic Cell Model under Fluid Flow
The 3D cell fluid–structure interaction finite element model was used to investigate the whole-cell strains of a simulated cell under fluid shear loading. In the FEA simulation, average strains of the whole cell were calculated for each time point. Whole-cell E xz showed viscoelastic creep and creep recovery strain effects from the fluid flow, but near-zero strains were observed in all other 3D FEA strain components (Fig. 7).
Time course of whole-cell strains for (a) E zz , (b) E xx , and (c) E xz in the cell shear flow model (n = 5). FEA strains are in dashed lines and quasi-3D strains are in half-shaded squares. Viscoelasticity is visible in shear E xz under fluid shear flow. Quasi-3D strains matched well with the FEA strains
The cell model was then convolved with a PSF, and the quasi-3D strains were calculated from selected planes in the bottom-view x–y and side-view x–z. Near-zero strains were observed in the bottom-view x–y plane, but significant strains were seen in side-view x–z plane (Fig. 7). Overall, the calculated quasi-3D strains were similar to the FEA strains inside the cell. Whole-cell E xz showed large viscoelastic behavior from the fluid flow, while the E zz and E xx side-view strain components hovered near zero throughout the flow pattern. However, E zz was slightly overestimated during the flow period (Fig. 7a).
A minimum SNR of 12 was necessary to maintain accurate strains with a mean error of less than .004 (Fig. 8). At an SNR of 9 and lower, the mean strain error jumped to greater than 0.01. Interestingly, while normal strains were inaccurate at a SNR of 9, shear strain accuracy was relatively unaffected at an SNR 9.
Means of the absolute error of calculated quasi-3D whole-cell strains in (a) bottom-view and (b) side-view of the simulated cell shear flow model (n = 5). Gaussian noise was added to the quasi-3D images prior to the calculation of strains to decrease the SNR. A critical SNR of 9 was found to cause a sharp increase in the error of quasi-3D strains vs. FEA strains, especially in the normal strains
Subcellular Strains
Strains at the end of the flow loading period were compared between FEA and side-view quasi-3D strains to determine the accuracy of subcellular average strain measurements. The results are presented in Fig. 9. All quasi-3D strains matched the 3D strain in subcellular regions with high accuracy. The absolute errors of the strains were around 0.005. The trailing edge of the cell had a slightly larger absolute error than the other regions. The FEA nodal displacement patterns in this region were the most complex and non-uniform, possibly contributing to the higher error in the quasi-3D calculation.
Discussion
A previous study in our laboratory developed a quasi-3D imaging technique to view a cell in two orthogonal planes. Strains were determined in both planes and were dubbed “quasi-3D strains” to represent, in some sense, the 3D deformation of the cell. This study focused on validating the accuracy of quasi-3D strains against the real 3D deformation within a cell from a theoretical perspective. To achieve this, a computational model was developed to be able to directly compare prescribed 3D strains in a cell vs. their quasi-3D counterparts. A finite element model of a cube and cell was deformed with simple and complex deformation patterns. Each deformation state was then convolved with a PSF and then quasi-3D images were extracted and processed to obtain quasi-3D strains.
The quasi-3D strains correlated well with the prescribed 3D strain values in both uniform uniaxial and shear cases and also in realistic fluid flow-induced deformation. The computational model was able to determine how the PSF affects wide-field microscopy-based measurements of strains. The additional error from the PSF was less than 5%, showing that the quasi-3D technique can accurately capture the real 3D deformation of a cell. A lower SNR limit of 9 was determined in this model system when calculating normal strains. This SNR limit was lower than other simulated image studies involving digital image correlation.15 However, most studies focus on displacement error as an output of SNR. The output in this study was strain, a differentiated value of the displacement field, and may be less sensitive to error due to the additional smoothing and differentiation operations.9,13 Normal strain errors at an SNR of 9 in E zz and E xx were higher than in E yy due to the difficulty of tracking larger displacements in noisy images of the model cell under fluid shear in the z and x directions than in the y direction. Interestingly, shear strain error was relatively unaffected at this SNR value, demonstrating that the accuracy of calculated normal strains may be more sensitive to Gaussian noise than shear strain in fluid shear experiments. This should be considered when determining the acceptable level of noise of wide-field microscopy images used in mechanical analyses of strain.
Several parameters that could potentially influence the accuracy of the quasi-3D calculations were absent from this idealized model. Factors such as photon noise (of a Poisson distribution) and complex inhomogeneous deformation patterns could decrease the ability of quasi-3D strains to accurately represent the real 3D strains. However, this study focused on the main source of error inherent to the quasi-3D system itself, i.e., the blur caused by out-of-focus planes.
The quasi-3D technique represents a novel technique to overcome limitations in temporal and spatial resolution of current microscopy techniques to measure cellular events in 3D. This study provides additional theoretical validation of this novel quasi-3D technique and of wide-field microscopy in calculating strains and deformations in 3D.
References
Adachi, T., Y. Aonuma, K. Taira, M. Hojo, and H. Kamioka. Asymmetric intercellular communication between bone cells: propagation of the calcium signaling. Biochem. Biophys. Res. Commun. 389:495–500, 2009.
Baik, A. D., X. L. Lu, J. Qiu, B. Huo, E. Hillman, C. Dong, and X. E. Guo. Quasi-3D cytoskeletal dynamics of osteocytes under fluid flow. Biophys. J. 99:2812–2820, 2010.
Gardner, M. K., D. J. Odde, and K. Bloom. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods 41:232–237, 2007.
Gardner, M. K., B. L. Sprague, C. G. Pearson, B. D. Cosgrove, A. D. Bicek, K. Bloom, E. D. Salmon, and D. J. Odde. Model convolution: a computational approach to digital image interpretation. Cell. Mol. Bioeng. 3:163–170, 2010.
Helmke, B. P., A. B. Rosen, and P. F. Davies. Mapping mechanical strain of an endogenous cytoskeletal network in living endothelial cells. Biophys. J. 84:2691–2699, 2003.
Knight, M. M., Z. Bomzon, E. Kimmel, A. M. Sharma, D. A. Lee, and D. L. Bader. Chondrocyte deformation induces mitochondrial distortion and heterogeneous intracellular strain fields. Biomech. Model. Mechanobiol. 5:180–191, 2006.
Marian, A., F. Charriere, T. Colomb, F. Montfort, J. Kuhn, P. Marquet, and C. Depeursinge. On the complex three-dimensional amplitude point spread function of lenses and microscope objectives: theoretical aspects, simulations and measurements by digital holography. J. Microsc.-Oxf. 225:156–169, 2007.
Ohkubo, M., S. Wada, T. Matsumoto, and K. Nishizawa. An effective method to verify line and point spread functions measured in computed tomography. Med. Phys. 33:2757–2764, 2006.
Pan, B., H. Xie, Z. Guo, and T. Hua. Full-field strain measurement using a two-dimensional savitzky-golay digital differentiator in digital image correlation. Opt. Eng. 46:033601, 2007.
Preza, C., M. I. Miller, L. J. Thomas, and J. G. McNally. Regularized linear method for reconstruction of 3-dimensional microscopic objects from optical sections. J. Opt. Soc. Am. A-Opt. Image Sci. Vis. 9:219–228, 1992.
Semwogerere, D., and E. R. Weeks. Confocal microscopy. In: Encyclopedia of Biomaterials and Biomedical Engineering. London: Taylor and Francis, 2005.
Ueki, Y., N. Sakamoto, and M. Sato. Direct measurement of shear strain in adherent vascular endothelial cells exposed to fluid shear stress. Biochem. Biophys. Res. Commun. 394:94–99, 2010.
Wang, C. C., J. M. Deng, G. A. Ateshian, and C. T. Hung. An automated approach for direct measurement of two-dimensional strain distributions within articular cartilage under unconfined compression. J. Biomech. Eng. 124:557–567, 2002.
Wang, Y., L. M. McNamara, M. B. Schaffler, and S. Weinbaum. A model for the role of integrins in flow induced mechanotransduction in osteocytes. Proc. Natl Acad. Sci. USA 104:15941, 2007.
Zhou, P., and K. E. Goodson. Subpixel displacement and deformation gradient measurement using digital image/speckle correlation (disc). Opt. Eng. 40:1613–1620, 2001.
Acknowledgments
We would like to thank Mr. Timothy K. Lee for helpful discussions. This work was supported by the National Institutes of Health (AR058453 & AR052417).
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor David J. Odde oversaw the review of this article.
Jun Qiu and Andrew D. Baik contributed equally to this work.
Rights and permissions
About this article
Cite this article
Qiu, J., Baik, A.D., Lu, X.L. et al. Theoretical Analysis of Novel Quasi-3D Microscopy of Cell Deformation. Cel. Mol. Bioeng. 5, 165–172 (2012). https://doi.org/10.1007/s12195-011-0218-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-011-0218-3