Abstract
The purpose of this study was to evaluate whether disodium gadoxetate (Gd-EOB-DTPA) affects proton density fat fractions (PDFFs) during use of the multiecho Dixon (meDixon) method in phantom and simulation magnetic resonance imaging (MRI) studies at 3 T. Fat–water phantoms comprising vegetable fat–water emulsions with varying fat volume percentages (0, 5, 10, 15, 20, 30, 40, and 50) and Gd-EOB-DTPA concentrations (0 and 0.4 mM) were prepared. Phantoms without Gd-EOB-DTPA were defined as precontrast, and those with Gd-EOB-DTPA were defined as postcontrast. All phantoms were scanned with a 3 T MRI system using the meDixon method, and precontrast and postcontrast PDFFs were calculated. Simulated pre and postcontrast PDFFs in the liver were calculated using a theoretical formula. The relationship between PDFFs measured in the pre and postcontrast phantoms was evaluated using linear regression and Bland–Altman analysis. The regression analysis comparing the pre and postcontrast PDFFs yielded a slope of 0.77 (P < 0.001). The PDFFs on the postcontrast phantom were smaller than those on the precontrast phantom. The mean difference between the PDFFs on the pre and postcontrast phantoms was 6.12% (95% confidence interval 3.13 to 9.10%; limits of agreement −0.88 to 13.11%). The simulated PDFF on the postcontrast phantom was smaller than that on the precontrast phantom. We demonstrated that the PDFF measured using the meDixon was smaller on postcontrast than on precontrast at 3 T, if a low flip angle was used. This tendency was also seen in the simulation study results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nonalcoholic fatty liver disease, including nonalcoholic steatohepatitis, is a focus of considerable research interest [1]. Nonalcoholic steatohepatitis differs from simple steatosis and can progress to fibrosis, cirrhosis, and hepatocellular carcinoma [2,3,4]. Therefore, managing the amount of fat stored in the liver is important. Liver biopsy is the gold standard for the quantification of fat content in the liver. However, this method is invasive and its evaluation is subjective [5, 6]. Furthermore, the fat distribution within the liver is not uniform [7], which can lead to sampling errors [8]. Therefore, an alternative quantification method is needed. Magnetic resonance spectroscopy (MRS) is considered a reference standard for the quantification of liver fat content [9]. However, it is difficult to assess the spatial distribution of fat throughout the entire liver using MRS alone. Another image-based method, i.e., the Dixon technique [10], allows calculation of the proton density fat fraction (PDFF) for the entire liver. The conventional Dixon method uses in-phase and out-of-phase images to account for the 3.5-ppm chemical shift between water and fat [7]. However, this method cannot correct the T2* effect and becomes problematic during evaluation of fat content in the liver, which includes both fat and iron [11]. Iron markedly decreases the T2* value in the liver, which changes the signal intensity in the liver and confounds the estimation of fat on magnetic resonance imaging (MRI). The novel multiecho Dixon (meDixon) method [12] allows a flexible echo time setting (rather than in-phase or out-of-phase) and T2* correction. A recent study showed that meDixon and MRS provide robust and congruent results when used to quantify liver fat [12].
Yokoo et al. [13] evaluated the effect of gadobenate dimeglumine (Gd-BOPTA; MultiHance, Bracco Diagnostics, Princeton, NJ, USA) on PDFF using a conventional two-point Dixon method. Gd-BOPTA is an extracellular intravenous contrast agent that can also be used as a hepatocyte-specific contrast medium, because 3–5% of the agent is taken up by hepatocytes. Yokoo et al. demonstrated that a gadolinium (Gd)-based contrast agent caused no significant change in PDFF, because the T 1 bias was minimized using a low flip angle. T 1 bias means that the T 1 of water in the liver is longer than the T 1 of fat in the liver. However, T 2* correction could not be performed in that study because of an insufficient number of echoes. T 2* correction is important in the liver, because iron deposition confounds PDFF measurements [11]. Disodium gadoxetate (Gd-EOB-DTPA; Primovist, Bayer HealthCare, Osaka, Japan) is a hepatocyte-specific contrast agent that can acquire hepatobiliary phase images, as well as dynamic images. More recently, Hernand et al. [14] showed that PDFF using T 2* correction was unaffected by Gd-EOB-DTPA at a low flip angle (slope = 1.02 ± 0.02; P = 0.32). Both the above-mentioned studies were performed at only 1.5 T MRI [13, 14], and it is well known that the T 1 value at 3 T is longer than that at 1.5 T, although the difference depends on the tissue involved [15]. For example, the T 1 values for the liver were significantly different (586 ms at 1.5 T and 809 ms at 3 T), while those of fat were not (343 ms at 1.5 T and 382 ms at 3 T) [15]. Therefore, we hypothesized that the greater difference in T 1 values between the liver and fat would affect the PDFF at 3 T MRI. To the best of our knowledge, there has been no research addressing the influence of Gd-based contrast agents on PDFF using the Dixon method at 3 T. Here, we evaluated whether Gd-EOB-DTPA affects the PDFF in use of the meDixon method in phantom and simulation studies using 3 T MRI.
2 Materials and methods
2.1 Phantoms
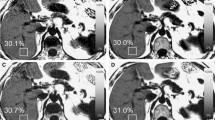
Glass bottles with a diameter of 2 cm containing vegetable fat–water emulsions in varying fat volume percentages (0, 5, 10, 15, 20, 30, 40, and 50) and Gd-EOB-DTPA concentrations (0 and 0.4 mM) were prepared for the fat–water phantoms (Fig. 1). The fat structures with a chemical shift from water and the abundance ratio in the phantom are similar to those in the living liver [16]. Purified water containing soy lecithin (15 g soy lecithin/2000 mL of purified water) was used to prepare homogenous fat–water emulsions. When Gd-EOB-DTPA was used, it was diluted with purified water to create a 10 mM Gd-EOB-DTPA solution. The required volume of the solution was then used to generate 0.4 mM concentrations. All the phantoms contained agar powder for stability. Each phantom with fat and water was slowly stirred using a hand mixer in hot water to homogenously dissolve the fat and water. The phantoms were then cooled to normal temperature. The volumes of soy lecithin and agar powder were ignored, because they were considerably smaller than those of water and fat. The glass bottles were inserted into a plastic container with a diameter of 18 cm for each Gd-EOB-DTPA concentration. The fat volume percentage was the ratio of the volume of vegetable oil to that of vegetable oil and water. Phantoms without and with Gd-EOB-DTPA were defined as pre and postcontrast phantoms, respectively. The T 1 and T 2 values for the phantoms are shown in Table 1. The T 1 and T 2 values of the phantoms were measured using the inversion-recovery and spin-echo methods, respectively. The magnetic resonance (MR) parameters used for measurement of T 1 were as follows: fixed repetition time (TR), 10,000 ms; fixed echo time (TE), 12 ms; and inversion times, 100, 150, 200, 400, 600, 800, 1200, 2000, and 3500 ms. The MR parameters used for measurement of T 2 were as follows: fixed TR, 3000 ms; and TE, 20, 40, 60, 80, 100, 130, 160, 190, 220, 250, 280, 310, and 350 ms. The reference hepatic T 1 values for the pre- and postcontrast phantoms at 3 T were 851 and 385 ms, respectively [17].
a Photographic image of phantoms containing different percentages of fat (0, 5, 10, 15, 20, 30, 40, and 50). b Image showing the proton density fat fraction using the multiecho Dixon technique. The fat volumes in each phantom were determined for a Gd-EOB-DTPA concentration of 0 mM and for a Gd-EOB-DTPA concentration of 0.4 mM. The tags show the average proton density fat fraction for five scans of each of the phantoms
2.2 MRI protocols
All phantoms were scanned with a 3 T MRI scanner (Skyra; Siemens Healthcare, Erlangen, Germany) using the meDixon method. In the sequence, the optical T 2* and the signal intensities of water (S w) and fat (S f) at a TE of 0 ms were calculated from the signal intensities of multiecho images [18]. The PDFF map was generated automatically from the S w and the S f. The scan imaging parameters were as follows: field of view, 300 × 300 mm; matrix size, 128 × 128; TR, 9.0 ms; TE, 1.05, 2.46, 3.69, 4.92, 6.15, 7.38 ms; slice thickness, 5 mm; flip angle, 4°; scan time, 15 s; and bandwidth, 1300 Hz/pixel. A low flip angle is recommended for fat quantification using the Dixon method to avoid T 1 independence. The average of five scans was used for the analysis.
2.3 Simulation
Simulated pre and postcontrast PDFFs in the liver were calculated using theoretical formulae [19]. In the present study, the T 2* effects were ignored, because T 2* correction was performed using multiecho. Thus, the terms related to T 2* were eliminated from the formula. The PDFF was defined as follows:
where S w and S f are water and fat signals, respectively. The S w and S f are given by the following equations:
and
where M w and M f are the water and fat proton densities, respectively. T 1w and T 1f are water and fat T 1 values, respectively, and α is a flip angle. The simulated PDFFs were calculated using T 1w = 933 ms and T 1f = 361 ms for precontrast and T 1w = 358 ms and T 1f = 359 ms for postcontrast, as reported in a previous study [20]. The TR and α were set at 9.0 ms and 4°, respectively—the same as the values in the phantom study.
2.4 Statistical analysis
The statistical analysis was performed using BellCurve for Excel version 2.11 (Social Survey Research Information Co., Ltd., Tokyo, Japan). The relationship between the PDFFs measured in the pre and postcontrast phantoms was evaluated using linear regression (the intercept was defined to be zero) and Bland–Altman analysis. In the Bland–Altman analysis, 95% limits of agreement were defined as the mean difference ±1.96 × the standard deviation. Values of P < 0.05 were considered statistically significant.
3 Results
The PDFFs measured using the meDixon method are summarized in Table 2. Figure 2 shows the results of the regression analyses comparing the pre and postcontrast PDFF in the phantoms. The data had a slope of 0.77 (P < 0.001). The PDFFs on the postcontrast phantom were smaller than those on the precontrast phantom. The mean difference between the PDFFs on the pre and postcontrast phantoms was 6.12% (95% confidence interval 3.13 to 9.10%; limits of agreement −0.88 to 13.11%; Fig. 3). The simulated pre versus postcontrast PDFF is shown in Fig. 2. The simulated postcontrast PDFF was smaller than the precontrast PDFF. This tendency was consistent with the phantom study.
Pre versus postcontrast PDFF in the phantom (a) and simulation (b) studies. The solid lines in the phantom (a) and simulation (b) studies indicate the lines of best fit for the data and simulation data, respectively. In both studies, the postcontrast PDFF was larger than the precontrast PDFF. PDFF proton density fat fraction
4 Discussion
In the phantom study, we demonstrated that the PDFF measured using the meDixon method was reduced post versus precontrast on 3 T MRI. This tendency was consistent with the simulated results. The previous authors have shown that Gd does not affect PDFF using the Dixon method at 1.5 T MRI [13, 14]. Hence, our findings are important, given that the influence of Gd on PDFF is dependent on the strength of the magnetic field.
In a previous study, Hamilton et al. [20] evaluated the T 1 values of water and fat in 59 patients who underwent a Gd-EOB-DTPA MRI examination of the liver. The authors showed that Gd-EOB-DTPA significantly reduced T 1w (precontrast, 933 ms; postcontrast, 358 ms), but did not change T 1f (precontrast, 361 ms; postcontrast, 359 ms), meaning that the longitudinal magnetization of water in postcontrast recovered more rapidly than that in precontrast. Thus, in the postcontrast state, the signal intensity of water is increased, which leads to a relatively decreased PDFF. Another study demonstrated that Gd increased the water signal relative to the fat signal [21]. In the present study, the PDFF in the postcontrast phantom was smaller than that in the precontrast phantom. Furthermore, a decreased PDFF was demonstrated in our simulation using the changes in T 1 between pre and postcontrast conditions. The findings of previous studies [20, 21] and our simulation results suggest that the values recorded in the present study are correct.
Yokoo et al. [13] and Hernand et al. [14] reported that Gd caused no significant change in PDFF in the liver at 1.5 T MRI (pre versus postcontrast PDFF: slope, 0.94; intercept, −0.01; slope, 1.02; and intercept, −0.19; respectively) and believed that the T 1 bias was minimized using a low flip angle. In contrast, the precontrast PDFF was larger than the postcontrast PDFF when using a high flip angle (pre versus postcontrast PDFF: slope, 0.68; intercept, −0.02; slope, 0.61; and intercept, 0.10; respectively). Therefore, if the T 1 bias is considered, the precontrast PDFF must be larger than the postcontrast PDFF. Our present findings indicate that the T 1 bias on 3 T MRI cannot be eliminated because of the large difference in T 1w between the pre and postcontrast liver, even if a low flip angle is used. T 1 values for most tissues (including water) are longer [15] and the T 1-shortening effects of Gd are relatively greater at 3 T than at 1.5 T, whereas there is a little difference in T 1f between 3 T and 1.5 T [15]. Therefore, the change in the difference between T 1w and T 1f on pre and postcontrast should be more prominent at 3 T than at 1.5 T. This phenomenon could account for the difference in findings of the present study at 3 T and the previous studies at 1.5 T.
The simulated PDFF on postcontrast was smaller than that on precontrast; however, the difference was smaller than that in the phantom study. There are possible reasons for these results. First, the T 1 value of the precontrast phantom (906.7–1330.9 ms) was slightly larger than that of the reported precontrast liver (mean, 851 ms; range 702–1058 ms) [17]. Thus, the difference in PDFF in the phantom study might be slightly larger than that in an actual liver. Second, the postcontrast T 1w and T 1f were treated as approximately the same in the simulation study as in an earlier study by Hamilton et al. [20]. However, another study demonstrated that the T 1w must be shorter than the T 1f after Gd-EOB-DTPA uptake into hepatocytes [14]. In that study, a PDFF with a low flip angle was compared with a PDFF with a high flip angle. The precontrast PDFF was overestimated at a high flip angle relative to a low flip angle (slope 1.33; P < 0.001); conversely, the postcontrast PDFF was underestimated (slope 0.81; P < 0.001). This result suggests that T 1w is shorter than T 1f in the postcontrast state, although T 1w is longer than T 1f in the precontrast state. Hence, if the T 1w used for calculating the postcontrast PDFF is smaller than the T 1f, the simulated difference between the pre and postcontrast PDFF will approximate the result for a phantom. A further clinical study is needed to determine the actual difference in PDFF of the liver between the pre and postcontrast images on 3 T MRI.
This study has a limitation in that we could not directly compare the influence of Gd on the PDFF between 1.5 T and 3 T, because we could not use a meDixon sequence at 1.5 T in our facility.
5 Conclusion
We demonstrated in this study that the PDFF measured using the meDixon technique was smaller on postcontrast relative to that on precontrast at 3 T if a low flip angle and T 2* correction were used. This result differs from that in the studies performed using 1.5 T MRI. Therefore, we should consider that the influence of Gd-EOB-DTPA on PDFF differs according to whether MRI is performed at 1.5 T or 3 T.
Change history
14 March 2018
A diameter of glass bottles in phantoms in the above article (2 cm) was incorrect. The correct diameter is 4.5 cm.
References
Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–31.
Adams LA, Lymp JF, St Sauver J, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–21.
Ratziu V, Bonyhay L, Di Martino V, et al. Survival, liver failure, and hepatocellular carcinoma in obesity-related cryptogenic cirrhosis. Hepatology. 2002;35:1485–93.
Kawamura Y, Arase Y, Ikeda K, et al. Large-scale long-term follow-up study of Japanese patients with non-alcoholic fatty liver disease for the onset of hepatocellular carcinoma. Am J Gastroenterol. 2012;107:253–61.
Piccinino F, Sagenelli E, Pasquale G, Giusti G. Complications following percutaneous liver biopsy. A multicenter retrospective study on 68,276 biopsies. J Hepatol. 1986;2:165–73.
Regev A, Berho M, Jeffers LJ, et al. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am J Gastroenterol. 2002;97:2614–8.
Hayashi T, Saitoh S, Fukuzawa K, et al. Non-invasive assessment of advanced fibrosis using hepatic volume in patients with non-alcoholic fatty liver disease. Hepatol Res. 2017;47:455–64.
Ratziu V, Charlotte F, Heurtier A, et al. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128:1898–906.
Yokoo T, Shiehmorteza M, Hamilton G, et al. Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology. 2011;258:749–59.
Dixon W. Simple proton spectroscopic imaging. Radiology. 1984;153:189–94.
Hines CD, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB. T1 independent, T 2* corrected MRI with accurate spectral modeling for quantification of fat: validation in a fat-water-SPIO phantom. J Magn Reson Imaging. 2009;30:1215–22.
Kukuk GM, Hittatiya K, Sprinkart AM, et al. Comparison between modified Dixon MRI techniques, MR spectroscopic relaxometry, and different histologic quantification methods in the assessment of hepatic steatosis. Eur Radiol. 2015;25:2869–79.
Yokoo T, Collins JM, Hanna RF, Bydder M, Middleton MS, Sirlin CB. Effects of intravenous gadolinium administration and flip angle on the assessment of liver fat signal fraction with opposed-phase and in-phase imaging. J Magn Reson Imaging. 2008;28:246–51.
Hernando D, Wells SA, Vigen KK, Reeder SB. Effect of hepatocyte-specific gadolinium-based contrast agents on hepatic fat-fraction and R2(*). Magn Reson Imaging. 2015;33:43–50.
de Bazelaire CM, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology. 2004;230:652–9.
Fukuzawa K, Hayashi T, Takahashi J, et al. Evaluation of six-point modified dixon and magnetic resonance spectroscopy for fat quantification: a fat-water-iron phantom study. Radiol Phys Technol. 2017;10:349–58.
Onoda M, Hyodo T, Murakami T, et al. Optimizing signal intensity correction during evaluation of hepatic parenchymal enhancement on gadoxetate disodium-enhanced MRI: comparison of three methods. Eur J Radiol. 2015;84:339–45.
Zhong X, Nickel MD, Kannengiesser SA, Dale BM, Kiefer B, Bashir MR. Liver fat quantification using a multi-step adaptive fitting approach with multi-echo GRE imaging. Magn Reson Med. 2014;72:1353–65.
Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med. 2007;58:354–64.
Hamilton G, Middleton MS, Cunha GM, Sirlin CB. Effect of gadolinium-based contrast agent on the relaxation properties of water and fat in human liver as measured in vivo by 1H MRS. In: Book of abstracts: 21th Annual meeting of the International Society for Magnetic Resonance in Medicine; 2013. p. 1516.
Mitchell DG, Stolpen AH, Siegelman ES, Bolinger L, Outwater EK. Fatty tissue on opposed-phase MR images: paradoxical suppression of signal intensity by paramagnetic contrast agents. Radiology. 1996;198:351–7.
Acknowledgements
We thank Editage (www.editage.jp) for English language editing services. This manuscript was partly supported by Akiyoshi Ohtsuka Fellowship of the Japanese Society of Radiological Technology for improvement in English expression of a draft version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human participants and animals
This article does not contain any studies with human participants performed by any of the authors. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent for this study was not required, because no research involving human participants was undertaken by any of the authors.
Additional information
A correction to this article is available online at https://doi.org/10.1007/s12194-018-0450-9.
About this article
Cite this article
Hayashi, T., Fukuzawa, K., Kondo, H. et al. Influence of Gd-EOB-DTPA on proton density fat fraction using the six-echo Dixon method in 3 Tesla magnetic resonance imaging. Radiol Phys Technol 10, 483–488 (2017). https://doi.org/10.1007/s12194-017-0420-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12194-017-0420-7