Abstract
Palmitic acid (PA) is the most common saturated long-chain fatty acid in food that causes cell apoptosis. However, little is known about the molecular mechanisms of PA toxicity. In this study, we explore the effects of PA on proliferation and apoptosis in human osteoblast-like Saos-2 cells and uncover the signaling pathways involved in the process. Our study showed that endoplasmic reticulum (ER) stress and autophagy are involved in PA-induced Saos-2 cell apoptosis. We found that PA inhibited the viability of Saos-2 cells in a dose- and time-dependent manner. At the same time, PA induced the expression of ER stress marker genes (glucose-regulated protein 78 (GRP78) and CCAAT/enhancer binding protein homologous protein (CHOP)), altered autophagy-related gene expression (microtubule-associated protein 1 light chain 3 (LC3), ATG5, p62, and Beclin), promoted apoptosis-related gene expression (Caspase 3 and BAX), and affected autophagic flux. Inhibiting ER stress with 4-PBA diminished the PA-induced cell apoptosis, activated autophagy, and increased the expression of Caspase 3 and BAX. Inhibiting autophagy with 3-MA attenuated the PA and ER stress-induced cell apoptosis and the apoptosis-related gene expression (Caspase 3 and BAX), but seemed to have no obvious effects on ER stress, although the CHOP expression was downregulated. Taken together, our results suggest that PA-induced Saos-2 cell apoptosis is activated via ER stress and autophagy, and the activation of autophagy depends on the ER stress during this process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diets rich in high-fat foods, especially saturated fats, cause high plasma lipid levels in blood, leading to obesity and insulin resistance. A connection between obesity and osteoporosis has been suggested (Zhao et al. 2007a; Zhao et al. 2007b). Observational studies have shown that patients with type 2 diabetes mellitus (T2DM) have higher risks of osteoporosis (Zhao et al. 2007b). Palmitic acid (PA) is the most common saturated long-chain fatty acid in food and is present in plant and animal products, such as palm oil, coconut oil, cheese, butter, and lard. Extensive epidemiological data show that PA triggers apoptosis in many cell types, including neuronal cells (Hsiao et al. 2014), granulosa cells (Mu et al. 2001), cardiomyocytes (Leroy et al. 2008), stem cells (Lu et al. 2012), hepatocytes (Mei et al. 2011) and endothelial cells (Jiang et al. 2010). Lipotoxicity can induce cell apoptosis via numerous mechanisms, including endoplasmic reticulum (ER) stress (Cunha et al. 2008) and autophagy (Jaishy and Abel 2016; Las and Shirihai 2010; Li et al. 2014).
The ER is an indispensable and elaborate eukaryotic organelle that controls protein biogenesis and trafficking and maintains cellular homeostasis (Ma et al. 2010). Many perturbations, including protein degradation, calcium imbalance, hypoxia, and cellular redox regulation, can affect ER homeostasis and trigger a protective mechanism known as the unfolded protein response (UPR), which aims to sustain cell survival and helps to reestablish homeostasis (Szpigel et al. 2018). Glucose-regulated protein 78-kDa (GRP78) is an ER chaperone that interacts with activating transcription factor 6 (ATF6), inositol requiring enzyme 1α (IRE1α), and pancreatic ER kinase (PERK) in the ER membrane, which maintains these transmembrane proteins in an inactive configuration (Yang et al. 2016a). The UPR assists protein synthesis by eliminating misfolded or unfolded proteins through ER-associated degradation (ERAD) and lysosome-mediated autophagy (Cawley et al. 2011; Chen et al. 2016). However, when the UPR fails to manage misfolded and unfolded proteins, the cellular apoptosis pathways are triggered (Ben et al. 2015; Walter and Ron 2011). ER stress often induces cell apoptosis via the CCAAT/enhancer-binding protein homologous protein (CHOP).
Autophagy is a complex catabolic process in eukaryotes for degrading and recycling the cytoplasmic components through autophagosome formation and lysosome degradation (Hsu et al. 2017; Liang and Kobayashi 2016). Cellular stresses such as nutrient deprivation, oxidative stress, or lipotoxicity block autophagy, thus causing cellular apoptosis, whereas the activation of autophagy alleviates damage and restores cellular function (Han and Wang 2017), which indicates that autophagy is essential for the maintenance of cellular homeostasis during various stress conditions (Liu and Levine 2015). However, there is increasing evidence that excessive or uncontrolled levels of autophagy may be essential for cell apoptosis in certain settings (Levine and Yuan 2005; Liu and Levine 2015; Matsui et al. 2007). Moreover, a recent study reported inhibiting autophagy against cardiac hypoxia/reoxygenation injury in H9c2 cells (Huang et al. 2017). As a set of evolutionarily conserved proteins, the autophagy-related proteins, mediate the homeostasis functions of autophagy through the formation of a double-membrane bound structure termed the autophagosome (Liu and Levine 2015). In addition, the cytosolic form of the microtubule-associated protein 1 light chain 3 (LC3) I (LC3I) transforms to the lipidated form of LC3II by the cysteine protease Atg5 (Dikic and Elazar 2018).
As mentioned above, both autophagy and ER stress play a fundamental role in the maintenance of cellular homeostasis. Moreover, recent studies also showed that ER stress (Yamada et al. 2015) and autophagy (Xia et al. 2010; Zhang et al. 2016) are involved in diabetic osteoporosis. In addition, ER stress could be a novel autophagy inducer (Zeng et al. 2017).
In this study, we aimed to explore the potential molecular mechanisms of PA-induced Saos-2 cell apoptosis. Herein, we investigated whether ER stress and autophagy are involved in PA-induced Saos-2 cell apoptosis and explored the relationship between ER stress and autophagy in PA-induced Saos-2 cell apoptosis.
Materials and methods
Materials
3-methyladenine (3-MA, autophagy inhibitor), PA, rapamycin (also named AY-22989, autophagy agonist), 4-phenylbutyrate (4-PBA, ER stress inhibitor), and thapsigargin (TG, ER stress agonist) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). McCoy’s 5a medium and fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NY, USA). Human osteoblast-like Saos-2 cells were purchased from the cell bank of the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). Total Protein Extraction Kit and BCA Protein Assay Kit were purchased from Nanjing Keygen Biotech Co., Ltd. (Nanjing, Jiangsu, China). The Annexin V-PE/PI Apoptosis Analysis Kit was purchased from Beyotime Institute of Biotechnology (Shanghai, China). All the antibodies were obtained from Abcam Biotechnology (Shanghai, China) or Santa Cruz (Shanghai, China). The details of antibodies are listed in Supplementary Table S1. PVDF membranes were obtained from Millipore (Bedford, MA). Caspase 3 Activity Colorimetric Assay Kit were obtained from Nanjing Biobox Biotech (Nanjing, China). mCherry-GFP-LC3 plasmid was purchased from Fenghui Shengwu Biotech Co., Ltd. (Wuhan, Hubei, China). Lipofectamine 2000™ reagent was obtained from Thermo Fisher Scientific (Shanghai, China).
Cell culture and treatment
Saos-2 cells were cultured in McCoy’s 5a medium supplemented with 10% FBS 50 μg/mL penicillin and 50 μg/mL gentamicin in a humidified incubator at 37 °C and 5% CO2. The growth medium was refreshed every 3 days. When the cells reached 70–80% confluence, they were treated with different concentrations (0–800 μM) of PA, and the morphology of Saos-2 cell was imaged in supplementary Fig. S1. At various times (0–48 h) during the treatment, cells were collected and processed for further experiments. In other experiments, Saos-2 were exposed to 200 μM PA in the presence or absence of 500 nM 4-PBA, 5 mM 3-MA, 5 μM AY-22989, or 500 nM TG for 24 h. After culture, the cells were collected for CCK8, flow cytometry, mCherry-GFP-LC3, Caspase 3 activity measurement, and western blotting analysis.
Measurement of cell viability
Saos-2 cells proliferation was monitored by CCK8 according to the guidelines. Cells were plated at a density of 2 × 104 per well in 96-well plates. After treatment with 0–800 μM PA for 24 h, 10 μl of CCK8 was added to each well, and the cells were incubated for 2 h at 37 °C. The number of viable cells was measured at 450 nm by a Microplate Reader (Bio-Rad 680). The results are presented as a percentage of the values measured for untreated control cells. The experiment was repeated independently three times.
Cell apoptosis measurement
The apoptosis level of Saos-2 cells was quantified with an Annexin V/PI Apoptosis Analysis Kit according to the manufacturer’s instructions. The cells were analyzed using a fluorescence-activated cell sorter (Becton, Dickinson and Company, USA) within 1 h. Early apoptotic cells were determined by counting the percentage of Annexin V+/PI− cells; progressed apoptotic cells were obtained by counting the percentage of Annexin V+/PI+ cells; necrotic cells were detected by counting the percentage of Annexin V-/PI+ cells, and Annexin V−/PI− cells were considered as surviving cells as the previous reported (Lin et al. 2015; Lin et al. 2012; Wang et al. 2016; Wang et al. 2015).
Western blot
After the treatment, Saos-2 cells were collected and washed with ice-cold PBS and lysed with RIPA buffer, and the total protein concentration was measured by the BCA assay. Fifty micrograms of total protein from each sample was loaded into each well of a 15% SDS-PAGE gel, and then proteins were separated by electrophoresis. Proteins were then transferred to PVDF membranes. After blocking in TBST with 10% nonfat milk for 2 h, the samples were incubated with anti-CHOP antibody (diluted 1:1000), anti-Beclin1 antibody (diluted 1:1000), anti-GRP78 antibody (diluted 1:1000), anti-LC3 antibody (diluted 1:1000), anti-ATG5 antibody (diluted 1:1000), anti-BAX antibody (diluted 1:500), anti-p62 antibody (diluted 1:1000), and anti-β-actin antibody (diluted 1:2000) overnight at 4 °C. After washing, the membranes were incubated with secondary antibody conjugated to horseradish peroxidase at 37 °C for 30 min. The immunoreactive bands were visualized using a Super Signal West Pico kit, according to the manufacturer’s instructions, and the density of the protein bands (Supplementary Fig. S1 and S2) were measured by densitometric analysis using ImageJ (Version 1.49).
mCherry-GFP-LC3 analysis
mCherry-GFP-LC3 analysis was used to evaluate autophagic flux. The mCherry-GFP-LC3 plasmid was transfected into Saos-2 cells using the Lipofectamine 2000™ reagent according to the manufacturer’s instructions. Briefly, the cells were seeded at 1 × 106 per well in a 6-well plate. After 24 h, the cells were transfected with 4 μg of plasmids and 10 μl of Lipofectamine. After 6 h of incubation, the medium was removed and replaced with fresh culture medium for 42 h. The red fluorescent proteins (RFP) and green fluorescent proteins (GFP) were observed using a laser scanning microscope system (Olympus Corporation, Tokyo, Japan). GFP degrades in an acidic environment while RFP does not. Thus, yellow spots (formed out of the overlap between red and green) indicate autophagosomes, while red spots indicate autophagic lysosomes. If autophagy is activated, the red signal will dominate over yellow (Amplified figures in Supplementary Fig. S4, S5 and S6). If autophagy is suppressed, there will be more yellow signal than red signal (Feng et al. 2017).
Caspase 3 activity measurement
Caspase 3 activity was measured using a Caspase 3 Activity Colorimetric Assay Kit as previous reported (Lin et al. 2015; Wang et al. 2015). Simply, after PA exposure, the cells were harvested by centrifugation and incubated in lysis buffer on ice for 15 min. The lysates were then centrifuged at 15,000 rpm and 4 °C for 15 min, and the protein content was determined using the BCA Protein Assay Kit according to the manufacturer’s instructions. Then, each sample was incubated with the Caspase 3 substrate at 37 °C in a microplate for 4 h. The samples were measured at 405 nm using a microplate reader (Bio-Rad 680).
Statistical analyses
All experiments were repeated at least three times for each group, and the data are presented as the mean ± SEM. The data were analyzed by ANOVA, followed by Fisher’s least significant difference test and independent samples Student’s t test, with SPSS software, version 13.0 (SPSS, Chicago, IL, USA).
Results
Effect of PA on the proliferation and apoptosis in Saos-2 cells
To detect the toxic effect of PA on Saos-2 cells, the cells were treated with 0–800 μM PA for 24 h. CCK8 results showed that PA treatment reduced the cell viability in a dose-dependent manner and the minimum effective dose was 100 μM PA (Fig. 1a). Flow cytometry analysis revealed that PA treatment increased the percentage of apoptotic Saos-2 cells in a dose-dependent manner compared with the control (Fig. 1b). In addition, the IC50 value was approximately 200 μM. These results showed that PA reduced cell viability and induced cell apoptosis in a dose-dependent manner.
Effect of PA on the growth and apoptosis of Saos-2 cells. a Cells were treatment with different concentrations (0–800 μM) of PA for 24 h and then processed for the cell activity analysis. b Cells were treatment with different concentrations (0–800 μM) of PA for 24 h and then processed for apoptosis assay. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Effect of PA on Caspase 3 activity and BAX expression in Saos-2 cells
To further explore the mechanism of PA on cell apoptosis of Saos-2 cells, apoptosis-related gene expression (Caspase 3 and BAX) was measured by colorimetric assay and western blot analysis, respectively. The results showed that Caspase 3 activity was similar to BAX expression during the culture at different times or with different doses. PA enhanced the levels of Caspase 3 activity and BAX protein in a dose-dependent manner at 24 h (Fig. 2a, b). At the same time, PA-induced Caspase 3 activation and BAX expression started from 12 to 48 h, and the highest Caspase 3 activation and BAX expression were observed at 48 h (Fig. 2c, d). These results showed that PA-induced cell apoptosis was related to the Caspase 3 activation and BAX expression.
PA induces the apoptosis-related gene expression in Saos-2 cells. a The Caspase 3 activity in Saos-2 cells after treatment with different doses of PA (0–800 μM) for 24 h. b The relative BAX expression in Saos-2 cells after treated with different doses of PA (0–800 μM) for 24 h. c The Caspase 3 activity in Saos-2 cells after treatment with 200 μM PA for different times (0–48 h). d The relative BAX expression in Saos-2 cells after treatment with 200 μM PA for different times (0–48 h). BAX expression was normalized to β-actin. Statistical analysis is shown in the bar graphs. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Effect of PA on ER stress in Saos-2 cells
To determine the role of ER stress in PA-induced in Saos-2 cell apoptosis, ER stress marker genes (GRP78 and CHOP) expression was detected by western blot (Fig. 3). PA treatment significantly induced the expression of GRP78 in a dose-dependent manner, although treatment with 400 and 800 μM PA did not produce differences at 24 h (Fig. 3a). Moreover, GRP78 protein levels gradually increased at 3 h and remained a high level at 24 h but decreased at 48 h after treatment with 200 μM PA (Fig. 3b). The expression levels of the CHOP protein were increased similar with GRP78 expression after treatment with different doses of PA at 24 h (Fig. 3a). However, CHOP was weakly detected after treatment with 200 μM PA for 3 h and 6 h but began to increase at 12 h, and the highest expression levels of CHOP were found at 24 h (Fig. 3b). These results indicated that ER stress is related to PA-induced Saos-2 cell apoptosis.
PA induces the expression of the ER stress-related proteins expression in Saos-2 cells. a The relative expression of GRP78 and CHOP in Saos-2 cells after treatment with different doses of PA (0–800 μM) for 24 h. b The relative expression of GRP78 and CHOP after treatment with 200 μM PA for different times (0–48 h). Protein expression was normalized to β-actin. Statistical analysis is shown in the bar graphs. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Effect of PA on autophagy in Saos-2 cells
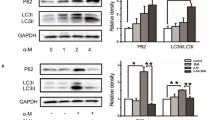
To determine the effects of PA on autophagy of Saos-2 cells, the expression levels of autophagy-related genes (Beclin1, ATG5, p62, and LC3) were studied via western blot. The results showed that the expression pattern of Beclin1, ATG5, and LC3II was similar during the culture at different times or with different doses of PA, but not p62. The levels of Beclin1, ATG5, and LC3II protein was increased in a dose-dependent manner, and the p62 was decreased after PA treatment (Fig. 4a). Moreover, activated autophagy (the increased expression levels of Beclin1, as shown in Fig. 4b) occurred as early as at 3 h and remained at a high level for 48 h. However, the expression of p62 was increased between 3 and 6 h and decreased again from 12 to 48 h (Fig. 4b). These results indicated that autophagy is related to PA-induced Saos-2 cell apoptosis.
PA induces the expression of the autophagy-related proteins in Saos-2 cells. a The relative expression of Beclin1, LC3, ATG5, and p62 after treatment with different doses of PA (0–800 μM) for 24 h. b The relative expression of Beclin1, LC3, ATG5, and p62 after treatment with 200 μM PA for different times (0–48 h). The expression of Beclin1, p62, ATG5, and the ratio LC3II/LC3I was normalized to β-actin. Statistical analysis is shown in the bar graphs. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Role of ER stress on PA-induced apoptosis in Saos-2 cells
To confirm the role of ER stress and its relation with autophagy in PA-induced cell apoptosis, we used 4-PBA to inhibit ER stress or TG to activate ER stress during the treatment. Similar results were observed using the CCK8 assay and flow cytometry (Fig. 5a, b, c). Further, both PA and TG obviously reduced cell viability, induced apoptosis, promoted Caspase 3 activity, and enhanced BAX expression. Treatment with 4-PBA promoted cell viability, prevented cell apoptosis, and inhibited the apoptotic regulatory genes, Caspase 3 and BAX (Fig. 5a, b, c, f, g). Meanwhile, 4-PBA successfully reduced the protein levels of GRP78 and CHOP (Fig. 5g). Western blot analyses also revealed that the protein levels of Beclin1, ATG5, and LC3II in the PA-induced Saos-2 cell were decreased, but not p62, after treatment of 4-PBA (Fig. 5g). In addition, PA and TG increased the percentage of red and yellow puncta, and 4-PBA treatment inhibited PA-induced the increased percentage of red puncta and yellow puncta, suggesting that 4-PBA treatment significantly alleviated PA-induced autophagy (Fig. 5d, e). These results indicated that ER stress is involved in PA-induced Saos-2 cell apoptosis and can activate autophagy in PA-induced cell apoptosis.
Effect of 4-PBA on the growth and apoptosis of PA-treated Saos-2 cells. a Cell viability was measured by CCK8 assay. b, c Apoptosis analysis was detected via flow cytometry. d, e Autophagic flux was evaluated by mCherry-GFP-LC3 analysis. f The Caspase 3 activity in Saos-2 cells. g The relative expression of related proteins. The ratio LC3II/LC3I and other protein expression levels was normalized to β-actin. PA, 200 μM palmitic acid; 4P, 500 nM 4-PBA; TG, 500 nM thapsigargin. Statistical analysis is shown in the bar graphs. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Role of autophagy in PA-induced cell apoptosis in Saos-2 cells
To confirm the role of autophagy and its relation with ER stress in PA-induced cell apoptosis, we used 3-MA to inhibit autophagy or AY-22989 to activate autophagy during the treatment. The results showed that 3-MA effectively increased the cell viability, suppressed cell apoptosis, decreased Caspase 3 activity, and inhibited BAX expression caused by PA at 24 h compared with the control (Fig. 6a, b, c, f, g). Western blot showed that 3-MA successfully inhibited the expression of Beclin1, ATG5, and LC3 induced by PA but seemed to increase the p62 accumulation (Fig. 6g). At the same time, 3-MA suppressed CHOP expression caused by PA, but not GRP78 (Fig. 6g). However, autophagy agonist AY-22989 had no obvious effects on the cell viability and cell apoptosis compared with the control (Fig. 6a, b). In addition, 3-MA treatment decreased PA-induced the formation of autophagosome and autolysosomes (Fig. 6d, e). These results demonstrated that autophagy is involved in PA-induced Saos-2 cell apoptosis but may have no obvious effect on ER stress.
Effect of 3-MA on the growth and apoptosis of PA-treated Saos-2 cells. a Cell viability was measured by CCK8 assay. b, c Apoptosis analysis was detected via flow cytometry. d, e Autophagic flux was evaluated by mCherry-GFP-LC3 analysis. f The Caspase 3 activity in Saos-2 cells. g The relative expression of related proteins. The ratio LC3II/LC3I and other protein expression levels was normalized to β-actin. PA, 200 μM palmitic acid; 3MA, 5 mM 3-methyladenine; AY: 5 μM AY-22989. Statistical analysis is shown in the bar graphs. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Role of autophagy in ER stress-induced apoptosis in Saos-2 cells
The above data indicated that ER stress was the upstream event leading to autophagy during PA-induced Saos-2 cell apoptosis. To further explore the role of autophagy in ER stress-induced apoptosis in Saos-2 cells, we added 3-MA or AY-22989 to the culture medium with or without TG. The results showed that 3-MA successfully rescued the TG-induced cell apoptosis and the cell viability decrease caused by PA (Fig. 7a, b, and c). At the same time, 3-MA treatment altered PA-induced expression of the apoptotic regulatory genes (Caspase 3 and BAX, Fig. 7f, g). In addition, 3-MA altered PA-induced autophagy-related gene (Beclin1, ATG5, p62, and LC3II) expression (Fig. 7g). Consistently, 3-MA treatment partly decreased TG-induced autophagic flux (Fig. 7d, e). These results demonstrated that autophagy is involved in ER stress-mediated Saos-2 cell apoptosis.
Effect of 3-MA on the growth and apoptosis of TG-treated Saos-2 cells. a Cell viability was measured by CCK8 assay. b, c Apoptosis analysis was detected via flow cytometry. d, e Autophagic flux was evaluated by mCherry-GFP-LC3 analysis. f The Caspase 3 activity in Saos-2 cells. g The relative expression of related proteins. The ratio LC3II/ LC3I and other proteins expression levels was normalized to β-actin. 3MA, 5 mM 3-methyladenine; TG, 500 nM thapsigargin; AY, 5 μM AY-22989. Statistical analysis is shown in the bar graphs. Data are presented as the mean ± SEM of three independent experiments. Bars with different letters are significantly different (p < 0.05)
Discussion
Numerous studies have demonstrated the relationship between obesity and osteoporosis, and epidemiological evidence has shown that obesity is correlated with increased bone mass. However, the detailed mechanisms of diabetic osteoporosis have not been fully elucidated. In the present study, we use PA to study the mechanisms of lipotoxicity on Saos-2 cells. We demonstrate that PA-induced Saos-2 cell apoptosis is activated via ER stress-mediated autophagy.
We first detected the effect of PA on the proliferation and apoptosis of Saos-2 cells. The data from our study showed that PA inhibited cell viability and induced cell apoptosis in a dose-dependent manner. Moreover, the IC50 value was approximately 200 μM PA, therefore in the next study, we used 200 μM PA as the induction concentration. These results further support the previous studies that PA promotes apoptosis in many cell types and cell lines, such as neuronal cells (Hsiao et al. 2014), cardiomyocytes (Leroy et al. 2008), stem cells (Lu et al. 2012), and hepatocytes (Mei et al. 2011).
To further clarify PA’s role in cell apoptosis, we measured the apoptosis-related genes expression (Caspase 3 and BAX). Caspase 3 acts as an executioner in Caspase-mediated apoptosis, and the expression of Caspase 3 positively correlates with the rate of apoptosis in cells (Lakhani et al. 2006; Porter and Jänicke 1999; Wen et al. 2017). Moreover, in the BCL 2 family, BAX has a proapoptotic effect (Chen et al. 2010; He et al. 2018; Mu et al. 2001). In the present study, the Caspase 3 activation (Zhu et al. 2016) and BAX expression (He et al. 2018) was upregulated after PA treatment, as previously shown in other studies. Based on these results, we concluded that PA can regulate Caspase 3 activation and BAX expression to affect cell apoptosis in Saos-2 cells.
Many studies have provided evidence that ER stress is involved in the process of osteoporosis (Hino et al. 2010; Li et al. 2017; Lisse et al. 2008). An increasing number of studies also reported that ER stress was involved in cell apoptosis (Chamolstad et al. 2016; Gunn 2007; Gwak et al. 2016; Qaisiya et al. 2016; Szegezdi et al. 2006). To detect effect of PA on ER stress in Saos-2 cells, the cells were cultured with different dose of PA for different times. As expected, PA treatment induced the expression of GRP78 and CHOP in a dose-dependent manner. Moreover, PA induced the activation of ER stress from 3 to 48 h. These results revealed that ER stress was activated in PA-treated cells in a marked dose- and time-dependent manner. This is similar to the results of our previous study demonstrating that zearalenone induced RAW 264.7 macrophage apoptosis via ER stress (Chen et al. 2015).
In addition, PA induced upregulation of Beclin1, ATG5, and LC3 protein expression in dose- and time-dependent manner, which indicated that PA also activated autophagy in Saos-2 cells. This study confirmed the previous study that PA can induce cell apoptosis and autophagy activation in endothelial cells (Tan et al. 2012), hepatocytes (Tu et al. 2014), podocytes (Jiang et al. 2017), and intestinal epithelial cells (Maeyashiki et al. 2017). However, the expression of p62 was different than expected. The levels of p62 expression were decreased from 12 to 48 h, and the downregulation occurred in dose-dependent manner. The continued decrease in expression levels of p62 in the PA-treated cells may be due to excessive induction of autophagy in Saos-2 cells treated with PA. In addition, the excessive autophagy induced cell apoptosis (Huang et al. 2017).
To decipher the role of ER stress in PA-mediated Saos-2 cell apoptosis, we suppressed ER stress by 4-PBA during the culture. Our data indicated that 4-PBA obviously inhibited the related genes, GRP78 and CHOP that were induced by PA in Saos-2 cells. In addition, 4-PBA partially reversed PA-induced cell apoptosis and viability decrease. The results were consistent with previous studies that PA induced ER stress and apoptosis in hepatoma cells (Yong et al. 2012; Zhang et al. 2011), renal cells (Katsoulieris et al. 2009), and pancreatic islet β-cells (Martinez et al. 2008). Our study revealed for the first time that ER stress is involved in PA-induced Saos-2 cell apoptosis.
To test the role of autophagy in PA-mediated Saos-2 cell apoptosis, Saos-2 cells were treated with PA in the presence or absence of the autophagy inhibitor 3-MA. Our results indicated that blocking of autophagy with 3-MA partially inhibited PA-induced cell apoptosis, and decreased vitality, the expression of Beclin1, ATG5, and LC3 II, and autophagic flux, while increasing p62 expression (p62 protein is an important substrate for autophagy degradation). This is the first report that autophagy is involved in PA-induced Saos-2 cell apoptosis. However, active autophagy by AY-22989 appeared to have no obvious effects on cell viability and apoptosis. The possible explanation may be that a certain level of autophagy has no obvious effect on cell function or is helpful for cells to maintain cell survival. However, excessively autophagy activation induced by ER stress or PA can lead to cell apoptosis (Zeng et al. 2017).
Previous studies demonstrated that ER stress is an autophagy inducer (Ma et al. 2014; Momoi 2006; Zeng et al. 2017). Some studies also indicated that autophagy is involved in PA-induced cell apoptosis (Jiang et al. 2017; Tan et al. 2012). Furthermore, our study found that both ER stress and autophagy are involved in PA-induced Saos-2 cell apoptosis. Therefore, we further tested the potential relations of ER stress and autophagy in PA-induced cell apoptosis. Our research found that inhibition of ER stress by 4-PBA partially blocks the expression of Beclin1, ATG5, and LC3, and the PA-induced autophagic flux, indicating that ER stress is an inducer of autophagy (Zeng et al. 2017).
We detected the expression of GRP78, CHOP, BAX, and Caspase3 activation in Saos-2 cells when 3-MA was added to the culture medium. Our results showed that inhibition of autophagy by 3-MA had no obvious effects on the expression of GRP78, but significantly decreased the expression of CHOP, BAX, and Caspase 3 activation. Our results suggest that that GRP78 is antiapoptotic during the ER stress (Zheng et al. 2014), but CHOP, BAX, and Caspase3 (Yang et al. 2016b; Zheng et al. 2014) are proapoptotic in cell apoptosis. Moreover, 3-MA lowered PA-induced cell apoptosis, which indirectly inhibit CHOP, BAX, and Caspase 3 expression.
We further study the role of autophagy in ER stress-mediated Saos-2 cell apoptosis. We also found that inhibition of autophagy by 3-MA attenuated TG-induced apoptosis. This was further supported by the decrease in expression of CHOP, BAX, and Caspase 3 activation. In addition, 3-MA treatment suppressed autophagy-related genes expression and the number of autophagosomes and autolysosomes. This result confirms that autophagy is involved in ER stress-mediated cell apoptosis, and ER stress-induced autophagy may be due to excessive autophagy, which leads to cell apoptosis (Feng et al. 2017).
Conclusions
In summary, our study demonstrates that both ER stress and autophagy are involved in PA-induced Saos-2 cell apoptosis, and the activation of autophagy depends on the ER stress during this process. This study offers new insights into the molecular mechanisms of lipotoxicity in diabetic osteoporosis.
References
Ben SI, Prola A, Boussabbeh M, Guilbert A, Bacha H, Abid-Essefi S, Lemaire C (2015) Crocin and quercetin protect HCT116 and HEK293 cells from Zearalenone-induced apoptosis by reducing endoplasmic reticulum stress. Cell Stress Chaperones 20:927–938
Cawley K, Deegan S, Samali A, Gupta S (2011) Assays for detecting the unfolded protein response. Methods Enzymol 490:31–51
Chamolstad H, Yu JE, Lee SH, Kim JG, Sung KS, Hwang J, Yoo YD, Lee YJ, Kim ST, Lee DH (2016) Modulation of SQSTM1/p62 activity by N-terminal arginylation of the endoplasmic reticulum chaperone HSPA5/GRP78/BiP. Autophagy 12:426–428
Chen YJ, Liu WH, Kao PH, Wang JJ, Chang LS (2010) Involvement of p38 MAPK- and JNK-modulated expression of Bcl-2 and Bax in Naja nigricollis CMS-9-induced apoptosis of human leukemia K562 cells. Toxicon 55:1306–1316
Chen F, Li Q, Zhang Z, Lin P, Lei L, Wang A, Jin Y (2015) Endoplasmic reticulum stress cooperates in zearalenone-induced cell death of RAW 264.7 macrophages. Int J Mol Sci 16:19780–19795
Chen F, Lin P, Wang N, Yang D, Wen X, Zhou D, Wang A, Jin Y (2016) Herp depletion inhibits zearalenone-induced cell death in RAW 264.7 macrophages. Toxicol in Vitro 32:115–122
Cunha DAD, Hekerman P, Ladriere L, Bazarracastro A, Ortis F, Wakeham MC, Moore F, Rasschaert J, Cardozo AK, Bellomo EA (2008) Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci 121:2308–2318
Dikic I, Elazar Z (2018) Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19:349–364
Feng D, Wang B, Wang L, Abraham N, Tao K, Huang L, Shi W, Dong Y, Qu Y (2017) Pre-ischemia melatonin treatment alleviated acute neuronal injury after ischemic stroke by inhibiting ER stress-dependent autophagy via PERK and IRE1 signalings. J Pineal Res 62:e12395
Gunn P (2007) ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129:1337–1349
Gwak H, Kim S, Dhanasekaran DN, Song YS (2016) Resveratrol triggers ER stress-mediated apoptosis by disrupting N-linked glycosylation of proteins in ovarian cancer cells. Cancer Lett 371:347–353
Han J, Wang Y (2017) mTORC1 signaling in hepatic lipid metabolism. Protein Cell 1–7
He Y, Zhou L, Fan Z, Liu S, Fang W (2018) Palmitic acid, but not high-glucose, induced myocardial apoptosis is alleviated by N-acetylcysteine due to attenuated mitochondrial-derived ROS accumulation-induced endoplasmic reticulum stress. Cell Death Dis 9:568
Hino S, Kondo S, Yoshinaga K, Saito A, Murakami T, Kanemoto S, Sekiya H, Chihara K, Aikawa Y, Hara H (2010) Regulation of ER molecular chaperone prevents bone loss in a murine model for osteoporosis. J Bone Miner Metab 28:131–138
Hsiao YH, Lin CI, Liao H, Chen YH, Lin SH (2014) Palmitic acid-induced neuron cell cycle G2/M arrest and endoplasmic reticular stress through protein palmitoylation in SH-SY5Y human neuroblastoma cells. Int J Mol Sci 15:20876–20899
Hsu HC, Li SJ, Chen CY, Chen MF (2017) Eicosapentaenoic acid protects cardiomyoblasts from lipotoxicity in an autophagy-dependent manner. Cell Biol Toxicol 1–13
Huang Z, Wu S, Kong F, Cai X, Ye B, Shan P, Huang W (2017) MicroRNA-21 protects against cardiac hypoxia/reoxygenation injury by inhibiting excessive autophagy in H9c2 cells via the Akt/mTOR pathway. J Cell Mol Med 21:467–474
Jaishy B, Abel ED (2016) Lipids, lysosomes and autophagy. J Lipid Res 57:1619–1635
Jiang H, Liang C, Liu X, Jiang Q, He Z, Wu J, Pan X, Ren Y, Fan M, Li M (2010) Palmitic acid promotes endothelial progenitor cell apoptosis via p38 and JNK mitogen-activated protein kinase pathways. Atherosclerosis 210:71–77
Jiang X, Chen X, Wan J, Gui H, Ruan X, Du X (2017) Autophagy protects against palmitic acid-induced apoptosis in podocytes in vitro. Sci Rep 7:42764
Katsoulieris E, Mabley JG, Samai M, Green IC, Chatterjee PK (2009) Alpha-linolenic acid protects renal cells against palmitic acid lipotoxicity via inhibition of endoplasmic reticulum stress. Eur J Pharmacol 623:107–112
Lakhani SA, Masud A, Kuida K, Jr PG, Booth CJ, Mehal WZ, Inayat I, Flavell RA (2006) Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311:847–851
Las G, Shirihai OS (2010) The role of autophagy in β-cell lipotoxicity and type 2 diabetes. Diabetes Obes Metab 12:15–19
Leroy C, Tricot S, Lacour B, Grynberg A (2008) Protective effect of eicosapentaenoic acid on palmitate-induced apoptosis in neonatal cardiomyocytes. BBA-Biomembranes 1781:685–693
Levine B, Yuan J (2005) Autophagy in cell death: an innocent convict? J Clin Invest 115:2679–2688
Li S, Li J, Shen C, Zhang X, Sun S, Cho M, Sun C, Song Z (2014) Tert-butylhydroquinone (tBHQ) protects hepatocytes against lipotoxicity via inducing autophagy independently of Nrf2 activation. BBA-Biomembranes 1841:22–33
Li J, Yang S, Li X, Liu D, Wang Z, Guo J, Tan N, Gao Z, Zhao X, Zhang J (2017) Role of endoplasmic reticulum stress in disuse osteoporosis. Bone 97:2–14
Liang Q, Kobayashi S (2016) Mitochondrial quality control in the diabetic heart. J Mol Cell Cardiol 95:57–69
Lin P, Yang Y, Li X, Chen F, Cui C, Hu L, Li Q, Liu W, Jin Y (2012) Endoplasmic reticulum stress is involved in granulosa cell apoptosis during follicular atresia in goat ovaries. Mol Reprod Dev 79:423–432
Lin P, Chen F, Sun J, Zhou J, Wang X, Wang N, Li X, Zhang Z, Wang A, Jin Y (2015) Mycotoxin zearalenone induces apoptosis in mouse Leydig cells via an endoplasmic reticulum stress-dependent signalling pathway. Reprod Toxicol 52:71–77
Lisse TS, Thiele F, Fuchs H, Hans W, Przemeck GKH, Abe K, Rathkolb B, Quintanillamartinez L, Hoelzlwimmer G, Helfrich M (2008) ER stress-mediated apoptosis in a new mouse model of osteogenesis imperfecta. PLoS Genet 4:e7
Liu Y, Levine B (2015) Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ 22:367–376
Lu J, Wang Q, Huang L, Dong H, Lin L, Lin N, Zheng F, Tan J (2012) Palmitate causes endoplasmic reticulum stress and apoptosis in human mesenchymal stem cells: prevention by AMPK activator. Endocrinology 153:5275–5284
Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM (2010) Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones 15:281–293
Ma XH, Piao SF, Dey S, Mcafee Q, Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G (2014) Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 124:1406–1417
Maeyashiki C, Oshima S, Otsubo K, Kobayashi M, Nibe Y, Matsuzawa Y, Onizawa M, Nemoto Y, Nagaishi T, Okamoto R (2017) HADHA, the alpha subunit of the mitochondrial trifunctional protein, is involved in long-chain fatty acid-induced autophagy in intestinal epithelial cells. Biochem Bioph Res Commun 484:636–641
Martinez SC, Tanabe K, Cras-Méneur C, Abumrad NA, Bernal-Mizrachi E, Permutt MA (2008) Inhibition of Foxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes 57:846–859
Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J (2007) Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 100:914–922
Mei S, Ni H, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding W (2011) Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther 339:487–498
Momoi T (2006) ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 14:230–239
Mu YM, Yanase T, Nishi Y, Tanaka A, Saito M, Jin CH, Mukasa C, Okabe T, Nomura M, Goto K (2001) Saturated FFAs, palmitic acid and stearic acid, induce apoptosis in human granulosa cells. Endocrinology 142:3590–3597
Porter AG, Jänicke RU (1999) Emerging roles of caspase-3 in apoptosis. Cell Death Differ 6:99–104
Qaisiya M, Brischetto C, Jašprová J, Vitek L, Tiribelli C, Bellarosa C (2016) Bilirubin-induced ER stress contributes to the inflammatory response and apoptosis in neuronal cells. Arch Toxicol 91:1–12
Szegezdi E, Logue SE, Gorman AM, Samali A (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7:880–885
Szpigel A, Hainault I, Carlier A, Venteclef N, Batto AF, Hajduch E, Bernard C, Ktorza A, Gautier JF, Ferré P (2018) Lipid environment induces ER stress, TXNIP expression and inflammation in immune cells of individuals with type 2 diabetes. Diabetologia 61:399–412
Tan SH, Shui G, Zhou J, Li JJ, Bay BH, Wenk MR, Shen HM (2012) Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin). J Biol Chem 287:14364–14376
Tu QQ, Zheng RY, Li J, Hu L, Chang YX, Li L, Li MH, Wang RY, Huang DD, Wu MC (2014) Palmitic acid induces autophagy in hepatocytes via JNK2 activation. Acta Pharmacol Sin 35:504–512
Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086
Wang X, Lin P, Yin Y, Zhou J, Lei L, Zhou X, Jin Y, Wang A (2015) Brucella suis vaccine strain S2-infected immortalized caprine endometrial epithelial cell lines induce non-apoptotic ER-stress. Cell Stress Chaperones 20:399–409
Wang X, Lin P, Li Y, Xiang C, Yin Y, Chen Z, Du Y, Zhou D, Jin Y, Wang A (2016) Brucella suis vaccine strain 2 induces endoplasmic reticulum stress that affects intracellular replication in goat trophoblast cells in vitro. Front Cell Infect Microbiol 6:19
Wen H, Wu Z, Hu H, Wu Y, Yang G, Lu J, Yang G, Guo G, Dong Q (2017) The anti-tumor effect of pachymic acid on osteosarcoma cells by inducing PTEN and Caspase 3/7-dependent apoptosis. J Nat Med 72:1–7
Xia X, Kar R, Gluhakheinrich J, Yao W, Lane NE, Bonewald LF, Biswas SK, Lo W, Jiang JX (2010) Glucocorticoid-induced autophagy in osteocytes. J Bone Miner Res 25:2479–2488
Yamada H, Nakajima T, Domon H, Honda T, Yamazaki K (2015) Endoplasmic reticulum stress response and bone loss in experimental periodontitis in mice. J Periodontal Res 50:500–508
Yang Y, Cheung HH, Tu J, Miu KK, Chan W (2016a) New insights into the unfolded protein response in stem cells. Oncotarget 7:54010–54027
Yang Y, Pei X, Jin Y, Wang Y, Zhang C (2016b) The roles of endoplasmic reticulum stress response in female mammalian reproduction. Cell Tissue Res 363:589–597
Yong Z, Xue R, Zhang Z, Xia Y, Shi H (2012) Palmitic and linoleic acids induce ER stress and apoptosis in hepatoma cells. Lipids Health Dis 11:1
Zeng M, Sang W, Chen S, Chen R, Zhang H, Xue F, Li Z, Liu Y, Gong Y, Zhang H (2017) 4-PBA inhibits LPS-induced inflammation through regulating ER stress and autophagy in acute lung injury models. Toxicol Lett 271:26–37
Zhang Y, Yang X, Shi H, Dong L, Bai J (2011) Effect of α-linolenic acid on endoplasmic reticulum stress-mediated apoptosis of palmitic acid lipotoxicity in primary rat hepatocytes. Lipids Health Dis 10:122
Zhang W, Meng H, Yang R, Yang M, Sun G, Liu J, Shi P, Liu F, Yang B (2016) Melatonin suppresses autophagy in type 2 diabetic osteoporosis. Oncotarget 7:52179–52194
Zhao L, Jiang H, Papasian CJ, Maulik D, Drees BM, Hamilton JJ, Deng H (2007a) Correlation of obesity and osteoporosis: effect of fat mass on the determination of osteoporosis. J Bone Miner Res 23:17–29
Zhao L, Liu Y, Liu P, Hamilton JJ, Recker RR, Deng H (2007b) Relationship of obesity with osteoporosis. J Clin Endocrinol Metab 92:1640–1646
Zheng YZ, Cao ZG, Hu X, Shao ZM (2014) The endoplasmic reticulum stress markers GRP78 and CHOP predict disease-free survival and responsiveness to chemotherapy in breast cancer. Breast Cancer Res Treat 145:349–358
Zhu Q, Yang J, Zhu R, Jiang X, Li W, He S, Jin J (2016) Dihydroceramide-desaturase-1-mediated caspase 9 activation through ceramide plays a pivotal role in palmitic acid-induced HepG2 cell apoptosis. Apoptosis 21:1033–1044
Funding
This research study was funded by the National Natural Science Foundation of China (No. 81660152) and Doctoral Research Start-Up Foundation of Jiujiang University (No. 8879522).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Table 1
(DOCX 16 kb)
Fig. S1
The morphology of Saos-2 cells after treatment with different doses of PA (0–800 μM) for 24 h. (PNG 18533 kb)
Fig. S2
Effects of PA on ER Stress, autophagy, and apoptosis related protein expression. (A) The expression of GRP78 and CHOP in Saos-2 cells after treatment with different doses of PA (0–800 μM) for 24 h; (B) The expression of GRP78 and CHOP after treatment with 200 μM PA for different times (0–48 h); (C) The expression of Beclin1, LC3, ATG5 and p62 in Saos-2 cells after treatment with different doses of PA (0–800 μM) for 24 h; (D) The expression of Beclin1, LC3, ATG5 and p62 after treatment with 200 μM PA for different times (0–48 h); (E) The expression of BAX in Saos-2 cells after treatment with different dose of PA (0–800 μM) for 24 h; (F) The expression of BAX after treatment with 200 μM PA for different times (0–48 h). (PNG 395 kb)
Fig. S3
The related protein expression of ER Stress, autophagy, and apoptosis in different treatment groups. (A) Effect of 4-PBA in PA-treated Saos-2 cells; (B) Effect of 3-MA in PA-treated Saos-2 cells; (C) Effect of 3-MA in TG-treated Saos-2 cells. (PNG 1311 kb)
Rights and permissions
About this article
Cite this article
Yang, L., Guan, G., Lei, L. et al. Palmitic acid induces human osteoblast-like Saos-2 cell apoptosis via endoplasmic reticulum stress and autophagy. Cell Stress and Chaperones 23, 1283–1294 (2018). https://doi.org/10.1007/s12192-018-0936-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-018-0936-8