Abstract
The molecular chaperone DnaK binds to exposed hydrophobic segments in proteins, protecting them from aggregation. DnaK interacts with protein substrates via its substrate-binding domain, and the affinity of this interaction is allosterically regulated by its nucleotide-binding domain. In addition to regulating interdomain allostery, the nucleotide state has been found to influence homo-oligomerization of DnaK. However, the architecture of oligomeric DnaK and its potential functional relevance in the chaperone cycle remain undefined. Towards that goal, we examined the structures of DnaK by negative stain electron microscopy. We found that DnaK samples contain an ensemble of monomers, dimers, and other small, defined multimers. To better understand the function of these oligomers, we stabilized them by cross-linking and found that they retained ATPase activity and protected a model substrate from denaturation. However, these oligomers had a greatly reduced ability to refold substrate and did not respond to stimulation by DnaJ. Finally, we observed oligomeric DnaK in Escherichia coli cellular lysates by native gel electrophoresis and found that these structures became noticeably more prevalent in cells exposed to heat shock. Together, these studies suggest that DnaK oligomers are composed of ordered multimers that are functionally distinct from monomeric DnaK. Thus, oligomerization of DnaK might be an important step in chaperone cycling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat shock protein 70 (Hsp70) belongs to a family of highly conserved molecular chaperones that play central roles in protein homeostasis via their involvement in protein folding, degradation, transport, and complex remodeling (Hesterkamp and Bukau 1998; Parsell and Lindquist 1993; Young et al. 2003). Hsp70s have been found to be essential in the cellular response to a variety of stressors, including heat shock and oxidation (Delaney 1990; Paek and Walker 1987; Winter et al. 2005). In addition, they have been proposed as a promising drug target in a variety of diseases including cancer and neurodegeneration (Evans et al. 2010; Patury et al. 2009). Thus, understanding the structure and function of Hsp70 family members is expected to enhance our understanding of cellular proteostasis, while also advancing our ability to design new disease treatments.
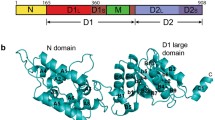
Towards those goals, extensive studies have focused on the prokaryotic Hsp70, DnaK. Like other Hsp70s, Escherichia coli DnaK is a 70-kDa chaperone composed of a nucleotide binding domain (NBD) and a substrate binding domain (SBD) (Bertelsen et al. 2009). The SBD can be further subdivided into a beta sandwich domain, responsible for binding substrates, and an alpha helical lid that closes over bound substrates (Bertelsen et al. 2009). The NBD and SBD are connected by a flexible linker and communicate allosterically to link substrate binding to nucleotide hydrolysis (Slepenkov and Witt 1998). In the ATP bound state, DnaK (DnaK-ATP) has relatively weak affinity for substrates, but, upon ATP hydrolysis, structural changes lead to an increase in substrate affinity by lowering the off-rate (Palleros et al. 1993; 1994). Thus, DnaK is believed to carry out its molecular chaperone activities by repeatedly binding and releasing client proteins, a process made possible by coupling substrate affinity to ATP hydrolysis. While DnaK alone has a very slow intrinsic ATPase rate, interactions with co-chaperones, DnaJ and GrpE, stimulate ATP turnover. Briefly, DnaJ interacts directly with DnaK via a conserved J-domain, and this interaction stimulates the hydrolysis of ATP (Wittung-Stafshede et al. 2003). Furthermore, DnaJ is known to bind exposed hydrophobic regions of unfolded proteins (Liberek et al. 1991; Szabo et al. 1996). Thus, DnaJ both localizes DnaK to client proteins and induces ATP hydrolysis. Meanwhile, GrpE serves as a nucleotide exchange factor, leading to the release of ADP and the re-binding of ATP by DnaK, a process that releases bound client proteins and completes the ATPase cycle (Liberek et al. 1991).
Defining the structural changes that occur in response to nucleotide cycling is central to understanding the molecular mechanisms by which DnaK functions as a chaperone. Consistent with this idea, a wide variety of biochemical and biophysical methods have been utilized to characterize the structural changes that occur in response to nucleotides. Together, these studies support a model in which the motions of the NBD and SBD are coupled in the ATP bound state (DnaK-ATP), which destabilizes the substrate binding pocket and favors opening of the alpha helical lid (Buchberger et al. 1995; Rist et al. 2006; Shi et al. 1996; Swain et al. 2007). Conversely, in the ADP-bound state (DnaK-ADP), the NBD and the SBD are observed to move independently, ordering the substrate binding pocket and closing the lid. These insights have benefitted from structural analyses of both isolated domains (Mayer and Bukau 2005) and two-domain structures. For example, the ATP-bound form of DnaK has been modeled based on the X-ray crystal structure of Hsp110, a distant homolog of Hsp70 (Liu and Hendrickson 2007), while the two-domain solution structure of DnaK-ADP in complex with a peptide substrate has been determined by NMR (Bertelsen et al. 2009). Together, these structures and others provide insight into the key interfaces involved in nucleotide state-driven changes in structure.
Despite advances in our understanding of the interdomain allostery in DnaK, other nucleotide-dependent structural transitions have been less explored. For example, multiple groups have noted that, while DnaK-ATP is predominantly monomeric, DnaK-ADP is prone to self-oligomerization (Osipiuk et al. 1993; Schonfeld et al. 1995). Furthermore, oligomerization has also been observed in human isoforms of Hsp70, suggesting that it is a conserved structural transition (Angelidis et al. 1999; Benaroudj et al. 1995; Carlino et al. 1992; Toledo et al. 1993). Specifically, these studies suggest that ADP leads to Hsp70 self-oligomerization in a concentration- and temperature-dependent manner in vitro and that this process is reversed with the addition of ATP, excess peptide substrate, and some co-chaperones, such as GrpE (Benaroudj et al. 1996; Schonfeld et al. 1995; Schroder et al. 1993). Finally, oligomeric forms of human Hsp70s may be present in cells (Angelidis et al. 1999; Carlino et al. 1992), and in the case of the ER-resident isoform of Hsp70 called BiP, in vivo oligomerization has been shown to be a regulated process that is promoted by post-translational modifications (Freiden et al. 1992).
Based on these observations, we hypothesized that oligomerization of DnaK may result in the formation of distinct, higher ordered structures that could have functional relevance in the chaperone cycle. To evaluate this idea, we used electron microscopy (EM), cross-linking, and biochemical measurements to explore the architecture and function of DnaK oligomers. We also examined whether they could be found in cells and if thermal stress could impact their formation. Together, these studies suggest that DnaK is preferentially assembled into dimers, trimers, and other small oligomers in response to ADP and that these structures are functionally distinct from monomeric DnaK.
Results
Self-oligomerization of DnaK is influenced by the nucleotide state
As mentioned above, several studies have noted that DnaK and other Hsp70 family members form multimers (Angelidis et al. 1999; Benaroudj et al. 1995; Carlino et al. 1992; Osipiuk et al. 1993; Schonfeld et al. 1995). As a preface for studies into the architecture and function of these assemblies, we expressed and purified DnaK (Online Resource 1) and performed gel filtration studies on the ATP- and ADP-bound states. Consistent with previous reports (Schonfeld et al. 1995), we found that DnaK-ATP was primarily monomeric, while DnaK-ADP samples contained higher order oligomers (Fig. 1a, Online Resource 1). The apparent molecular weights of these peaks (monomers and oligomers ranging from 110 to 670 kDa) were consistent with previously reported values (Schonfeld et al. 1995). Next, to control for any effects of intrinsic ATP hydrolysis, we explored the oligomerization of the T199A mutant of DnaK, which is unable to hydrolyze ATP (see Online Resource 1) (Buchberger et al. 1995). As expected, DnaK-T199A in the ATP-bound state was primarily monomeric by gel filtration, supporting the idea that the ATP-bound form is largely a monomer.
DnaK self-oligomerization occurs in vitro and in vivo. a DnaK in the presence of ATP (DnaK-ATP) is primarily monomeric by gel filtration (black, solid line). The addition of ADP (DnaK-ADP) leads to an increase in higher molecular weight species (red, solid line). A mutant of DnaK that is unable to hydrolyze ATP (T199A-ATP) is also primarily monomeric (black, dotted line). b Fractions collected from gel filtration were subject to both denaturing and native gel electrophoresis. Bands consistent with the molecular weight of trimeric (tri), dimeric (di), and monomeric (m) DnaK were observed. These species are increased in the presence of ADP. c E. coli cells were grown at either 30°C or 43°C, and the lysates were separated by gel filtration and then subject to native and denaturing gel electrophoresis. By Western blot, trimeric (tri), dimeric (di), and monomeric (m) DnaK were observed. The experiment was performed in independent duplicates and both histograms of the resulting band intensities for fractions 12 and 13 mL are shown in the right panel
To further clarify the size distribution of multimers we could isolate by gel filtration, we evaluated each of the gel filtration fractions by both denaturing and native gel electrophoresis. As expected, ADP-favored oligomerization based on the abundance of DnaK bands in earlier fractions (Fig. 1b). Furthermore, we observed monomer and dimer bands even in early fractions of our DnaK-ADP sample, suggesting that these structures are in a dynamic equilibrium, as previously described (Schonfeld et al. 1995) (Fig. 1b). Notably, a small amount of dimeric DnaK was also observed in the presence of saturating concentrations of ATP (Fig. 1b). Together, these results suggest that DnaK reversibly forms oligomers, including dimers, and that this balance is influenced by nucleotide.
DnaK oligomerization occurs within cells and is increased in response to heat and elevated expression
Having confirmed that DnaK forms oligomers in vitro, we were interested in understanding whether this process could occur in E. coli cells. Towards this goal, we separated E. coli cellular lysates by gel filtration and evaluated fractions by denaturing and native gel electrophoresis. In cells grown to mid-log phase at 30°C, we observed bands consistent with the molecular mass of monomeric and dimeric forms of DnaK (Fig. 1c). These results suggest that DnaK homo-oligomerizes in vivo. Next, we wanted to assess if cellular stresses might impact the relative abundance of oligomeric DnaK. Consistent with previous findings, the total amount of DnaK was significantly elevated in response to heat shock (Online Resource 2) (Cowing et al. 1985; Paek and Walker 1987; Tilly et al. 1983). In addition, we observed that DnaK eluted in earlier fractions under these conditions (Fig. 1c), suggesting that it may also be forming higher ordered structures. When these fractions were analyzed by native gel electrophoresis, we found that DnaK was assembled into a discrete series of bands with molecular weights consistent with the trimeric, dimeric, and monomeric forms (Fig. 1c). Although it is not yet possible to ascribe these changes in oligomerization to a specific mechanism, these findings suggest that DnaK forms oligomers in cells under normal growth conditions and that these structures are favored in response to heat stress. Next, we evaluated if this response also occurred in cells facing hydrogen peroxide-induced oxidative stress or stress arising from the stationary phase of cellular growth. Consistent with previous reports (Delaney 1990; Dukan and Nystrom 1998; Spence et al. 1990; Winter et al. 2005), these stresses elevated total DnaK levels, but, interestingly, they did not appreciably alter the extent of oligomerization (Online Resource 2). This result suggests that an increase in DnaK oligomerization is not a general response to cellular stressors. Finally, we wondered whether increasing the levels of DnaK, in the absence of heat stress, would be sufficient to induce oligomerization in cells. To test this idea, we overexpressed DnaK under an inducible promoter in a Δdnak E. coli strain and found that high DnaK levels were sufficient to increase the oligomerization of DnaK (Online Resource 2), suggesting that the amount of oligomeric DnaK within cells is sensitive to its expression level.
DnaK oligomerization is dependent on its concentration, substrate, and co-chaperones
Given the presence of DnaK oligomers within cells, we wanted to better understand the requirements for forming the complex. Previous work has demonstrated that DnaK self-oligomerization in the ADP-bound state occurs in a concentration dependent manner and that ATP, excess peptide substrate, or GrpE lead to monomerization (Benaroudj et al. 1996; Schonfeld et al. 1995, 1993). In our hands, we found very similar results (Online Resource 3A–C). Furthermore, we demonstrated that the J-domain of DnaJ also led to the monomerization of DnaK-ADP (Online Resource 3D (King et al. 1995)). Thus, all the components of the active DnaK molecular chaperone cycle (i.e., substrate, J-domain, and GrpE) promoted monomerization.
Oligomers of DnaK retain ATPase and holdase activities but have reduced foldase activity
These findings lead us to wonder if oligomeric DnaK may represent an enzymatically or functionally inactive pool that might be rapidly monomerized by interaction with other components of the DnaK chaperone system. However, this model seems to be in conflict with the observation of increased oligomerization in response to heat stress (see Fig. 1c), a condition that actively requires DnaK for viability (Paek and Walker 1987). Therefore, we wanted to directly test whether DnaK oligomers have normal or attenuated chaperone activities in vitro. The technical challenge in addressing this issue is the dynamic equilibrium between monomeric and oligomeric DnaK, which makes it difficult to specifically evaluate their individual biochemical properties. To circumvent this problem, we used chemical cross-linking to stabilize oligomeric DnaK. Because the covalent modification of a protein using a chemical cross-linker can sometimes result in deleterious effects, it was essential to control for changes attributable to the cross-linking reaction. First, we treated DnaK-ATP and DnaK-ADP samples with the quenching reagents prior to adding the cross-linker (0.025% gluteraldehyde; see “Materials and methods” for details). As expected, this treatment left DnaK largely monomeric in both the ATP- and ADP-bound states (Fig. 2a). Thus, these samples served as controls for the buffer conditions and handling steps that occur during the reactions. Next, we performed cross-linking on both DnaK-ADP and DnaK-ATP samples using 0.025% gluteraldehyde for 20 min. We expected that higher order structures might be stabilized in both samples but that the ADP sample may be enriched for oligomers and, importantly, that it might contain more oligomers trapped in an ADP-bound conformation. When we visualized the resulting solutions by gel electrophoresis, bands consistent with oligomers and monomer were indeed observed in both ATP and ADP samples, and there was a significant increase in oligomers (57 ± 5%) in the presence of ADP as compared to ATP (40 ± 4%) (Fig. 2a). Oligomers larger than dimers were especially enriched in the ADP sample (33 ± 5% vs. 16 ± 4%).
Oligomeric DnaK is deficient in foldase activity. a Stabilization of DnaK oligomers by chemical cross-linking. DnaK (K) was pre-incubated with either ATP or ADP and treated with 0.025% glutaraldehyde (glut.). Samples were either incubated for 20 min to allow cross-linking (cx) or quenched (0 min) immediately. Bands consistent with monomeric (m), dimeric (di), and trimeric (tri) DnaK were observed, as well as higher order structures. A representative gel is shown. This cross-linking reaction was performed nine times and quantification of the distribution of total DnaK in all conditions is reported (see Online Resource 4 and the text). The cross-linked samples were then compared in a variety of assays: b intrinsic ATPase rate, c DnaJ-mediated ATPase stimulation, and d luciferase refolding. A negative control, lacking DnaK, was tested for each experiment and subtracted. The results shown are from a single experiment performed in triplicate, and the error bars represent the standard error of the mean. e K-ADP mixtures were further purified by gel filtration, resulting in fractions enriched for monomers or oligomers (>80%). f The enriched oligomer samples had weak J stimulation, consistent with the findings from crude mixtures
Next, we probed the intrinsic ATP hydrolysis activities of these crude mixtures. We reasoned that an inactivated pool of oligomeric DnaK might have attenuated ATPase activity, reducing the apparent activity of the mixture. On the contrary, we observed that cross-linked DnaK-ADP had a 1.5-fold faster rate (0.33 min−1) than the equivalent DnaK-ATP sample (0.22 min−1; Fig. 2b, Table 1). We also observed that the cross-linked DnaK-ATP sample retained an intrinsic ATPase rate similar to the other control samples (0.20 and 0.18 min−1). Thus, ADP-promoted oligomers of DnaK appear to retain enzymatic activity, and moreover, they may have a slightly higher intrinsic rate.
Alone, DnaK has a low ATPase rate, and it depends on the co-chaperone DnaJ to greatly increase its ATPase activity. Furthermore, DnaJ is required for active refolding of denatured proteins (Liberek et al. 1991; Szabo et al. 1994; Wisen and Gestwicki 2008). Thus, we next evaluated whether the cross-linked, oligomeric DnaK could participate in DnaJ-dependent processes. By measuring ATPase activity in the presence of increasing DnaJ levels, we observed that DnaK oligomers (K-ADP cx) had significantly reduced ability to respond to this co-chaperone (Fig. 2c), with a dramatic decrease in the apparent V max for DnaJ (19 pmol Pi/μg DnaK/min) (Table 1). Conversely, the control samples (K-ATP and K-ADP) and cross-linked DnaK-ATP sample (K-ATP cx) had V max values (33, 27, and 31 pmol Pi/μg DnaK/min, respectively), similar to unmodified chaperone. Additionally, the DnaK oligomers demonstrated an increased apparent K m (0.39 μM), compared to control samples (0.20, 0.21, and 0.28 μM respectively). Similar results were seen with samples that were further enriched for oligomeric DnaK (Fig. 2e and f). Together, these results suggest that oligomeric DnaK is deficient in DnaJ-mediated ATPase stimulation. We also probed if the ATPase rate of oligomeric DnaK was deficient in stimulation by either peptide substrate or GrpE. We found that oligomeric DnaK had a decreased ability to be stimulated by the NRLLLTG peptide (Online Resource 4C), but we observed no change in its ability to be stimulated by GrpE (Online Resource 4D).
Based on these results, we hypothesized that oligomeric DnaK may also be unable to refold luciferase from a denatured state because this is a DnaJ-dependent process that is commonly termed foldase activity (Szabo et al. 1994; Wisen and Gestwicki 2008). To test this hypothesis, we evaluated whether DnaJ could restore active luciferase in combination with either the control or cross-linked DnaK samples. We found that the cross-linked DnaK-ADP had much lower refolding activity compared to the controls or cross-linked DnaK-ATP (Fig. 2d). Thus, oligomeric DnaK did not respond normally to DnaJ-mediated stimulation of either ATPase or luciferase refolding activity.
We next asked whether oligomeric DnaK could still bind a client protein, using the well-studied firefly luciferase as a model substrate (Schroder et al. 1993; Szabo et al. 1994). In these studies, we immobilized denatured luciferase in microtiter plates and measured the equilibrium binding of DnaK using anti-DnaK antibodies. In the presence of excess ADP, non-cross-linked DnaK binds substrate with an almost tenfold lower apparent K D than in the presence of ATP (Online Resource 5), consistent with the known allosteric link between nucleotide state and substrate affinity. However, in our study, we utilized an excess of ATP in the assay buffer of all tested samples; thus, pre-incubating DnaK with either ATP or ADP during the cross-linking reaction did not greatly affect the apparent affinity (Fig. 3a, Table 1). To our surprise, this method revealed that oligomeric DnaK retained full binding to luciferase (Fig. 3a, Table 1), suggesting that oligomeric DnaK is able to bind client proteins. However, we wanted to confirm this conclusion and test whether binding might arise from non-specific interactions that can sometimes be induced by cross-linking. Thus, we also tested the substrate-binding activity of the truncated nucleotide-binding domain of DnaK (NBDL). We found that the untreated NBDL had only a weak non-specific binding (apparent K D > 3 μM) and that this interaction was not changed after cross-linking with 0.025% gluteraldehyde for 20 min (Online Resource 5). This result supports the conclusion that oligomeric DnaK retains its ability to bind client proteins via its substrate-binding domain.
Oligomeric DnaK retains holdase activity. The same samples and color scheme as described in Fig. 2 were used to evaluate the ability of oligomeric DnaK to bind the model substrate luciferase using two independent assays. a An ELISA-like platform to probe the binding of DnaK to denatured luciferase and b the holdase activity to probe the binding of DnaK to unfolding intermediates of luciferase. A negative control, lacking luciferase, was tested for each experiment and subtracted. All of the experiments were performed in triplicate, and the error bars represent the standard error of the mean
To confirm this interaction in a different experimental platform, we evaluated the ability of stabilized DnaK oligomers to protect native luciferase from terminal heat denaturation, which is a chaperone function that is commonly called holdase activity. In this assay, native luciferase was exposed to elevated temperatures, a process that is sufficient to heat inactivate the enzyme in the absence of chaperone. DnaK is known to limit this terminal denaturation, allowing luciferase to spontaneously reactivate when returned to room temperature. We found that both cross-linked DnaK-ATP and DnaK-ADP retained full holdase activity (Fig. 3b, Table 1). Again, we tested cross-linked NBDL under the same conditions and found that it lacked this activity (Online Resource 5). Furthermore, the DnaK substrate (NRLLLTG) inhibited the holdase activity of both cross-linked DnaK-ATP and DnaK-ADP samples (Online Resource 6). Together, these results suggest that oligomeric DnaK retains the ability to bind client proteins through its substrate-binding domain. Taken together, these biochemical findings suggest that oligomeric DnaK retains many chaperone functions, including holdase ability but that it is functionally distinct from monomeric DnaK because it partially loses some J-stimulated functions.
DnaK-ADP is multimeric by electron microscopy
In theory, oligomerization of DnaK could result from unfolded monomers being bound as substrates, resulting in relatively disordered multimers. Conversely, these structures could result from the ordered assembly of intact monomers. To better differentiate between these possibilities, we visualized DnaK-ADP samples by EM (Online Resource 7). In doing so, we were able to make several important conclusions. First, we found that monomeric DnaK was readily visible and its two-domain architecture could be resolved, despite its relatively small size (70 kDa). Second, we observed numerous structures that appeared to be small, oligomeric forms of DnaK (Online Resource 7). Importantly, these oligomeric forms were not disordered aggregates; rather, they seemed to represent an ensemble of structurally defined multimers, with sizes roughly corresponding to dimers, trimers, and other small oligomers.
To better understand the protein architectures present in the DnaK-ADP samples, we applied reference-free alignment and classification of 6,090 particle projections into 150 classes (Online Resource 7). Representative class averages are shown, and these clearly demonstrate the presence of monomeric, dimeric, and small multimeric forms of DnaK (Fig. 4b). For reference, a ribbon representation of the homology model of DnaK-ATP, based on the yeast Hsp110 (Sse1) crystal structure (PDB: 2QXL) (Liu and Hendrickson 2007), and the monomeric DnaK-ADP structure (PDB 2KHO) (Bertelsen et al. 2009) are shown (Fig. 4a).
EM visualization of oligomeric DnaK. a DnaK-ATP homology model based off the Sse1-ATP crystal structure (pdb code 2QXL) and the DnaK-ADP + substrate NMR structure (pdb code 2KHO) serve as a reference for the EM images. b Representative 2D class averages of DnaK-ADP and DnaK-ADP + substrate. The box size is ∼27 × 27 nm. The number of particles in each class average is shown in the bottom left of the box, while in the bottom right, we indicate whether the class average was interpreted as monomeric (m), dimeric (d), or oligomeric DnaK greater than dimer (>d). c Quantification of the number of dimeric and monomeric DnaK particles reveals that addition of substrate leads to monomerization. Some class averages could not be clearly classified and were therefore left unassigned (see Online Resources 7 and 8 for details)
If the dimeric and oligomeric forms of DnaK that we visualized by EM were similar to those seen in the previous biochemical studies (e.g., gel filtration), we expected that they would monomerize in the presence of excess peptide substrate. To test this idea, we added the NRLLLTG peptide substrate (400 μM), collected 6,098 particles, and, as previously, applied reference-free alignment and classification into 150 classes (Online Resource 8). As expected, we observed a dramatic increase in the number of particles classified as monomers and a corresponding decrease in those classified as dimers (Fig. 4b and c; Online Resources 7 and 8). Thus, we conclude that reversible multimers of DnaK are assembled from individual, folded monomers.
Discussion
Oligomerization of DnaK and other Hsp70s has been observed in vitro and in cellular studies (Angelidis et al. 1999; Blond-Elguindi et al. 1993; Carlino et al. 1992; Freiden et al. 1992; Schonfeld et al. 1995). However, the possible roles of these oligomers have not been clear. One hypothesis is that oligomeric DnaK may represent an inactive pool that can be rapidly monomerized and activated when needed. However, this model had not been directly tested. Furthermore, the structure of these oligomers was not clear, and so it was not certain if they were assembled from folded or unfolded monomers. Herein, we aimed to better understand nucleotide-dependent changes in DnaK oligomerization to clarify the possible roles of these structures in the chaperone cycle.
DnaK oligomerization in vivo
One of our questions was whether DnaK oligomers are present in cells and whether any common stresses might influence their relative abundance. Using gel filtration and native gel electrophoresis on E. coli lysates, we observed bands consistent with monomeric, dimeric, and trimeric forms of DnaK under resting conditions (Fig. 1c). It is possible that these bands could represent DnaK–substrate complexes, but the repeating separation values of 70 kDa suggest that these bands are homo-multimers. Interestingly, oligomeric DnaK levels increased in response to heat stress (Fig. 1c). This result is consistent with previous work on human cells, where the stress inducible Hsp70 isoform (Hsp72) and the constitutively active isoform (Hsc70) have been found to form oligomeric structures in response to heat stress (Angelidis et al. 1999). Thus, we conclude that DnaK, like other Hsp70s, samples both monomeric and oligomeric forms in vivo.
A variety of mechanisms could explain the increase in oligomeric DnaK in response to heat stress. For example, heat stress is known to increase expression of this chaperone (Cowing et al. 1985); thus, a simple model is that increases in protein concentration lead to enhanced oligomerization. Consistent with this idea, we found that overexpression of DnaK increases oligomerization (Online Resource 2D). Another explanation is that heat stress might change the ratio of ATP and ADP, resulting in increased abundance of the more oligomerization prone, DnaK-ADP form. At high temperatures, the co-chaperone GrpE is known to be partially unfolded and become less active (Grimshaw et al. 2003, 2005; Siegenthaler and Christen 2006). Because GrpE normally catalyzes ADP release, its inactivation might be expected to favor accumulation of the ADP-bound state. In what could be a related finding, it has also been shown that the human ER resident Hsp70, BiP, self-oligomerizes in response to glucose deprivation (Freiden et al. 1992). Glucose deprivation depletes cellular ATP, and thus, these conditions might also be expected to promote the ADP-bound (or nucleotide free) conformation. However, we did not observe an increase in DnaK oligomerization under hydrogen peroxide-induced oxidative stress or during stationary phase (see Online Resource 3). Much like glucose deprivation, both of these stress conditions increase the expression of DnaK and deplete cellular ATP (Delaney 1990; Dukan and Nystrom 1998; Osorio et al. 2003; Spence et al. 1990; Tran and Unden 1998; Winter et al. 2005). Clearly, further work is required to fully understand the mechanism by which oligomeric DnaK increases in response to heat stress.
It is important to note that DnaK oligomerization in response to specific types of oxidative stress has been well characterized by Jakob and colleagues (Hoffmann et al. 2004; Winter et al. 2008, 2005). In response to a combination of heat stress and hypochlorous acid, they found that DnaK becomes oligomeric and inactivated in a reversible fashion. At the same time, Hsp33 is activated, and this chaperone becomes responsible for cellular protection. Thus, in response to these stress conditions, E. coli clearly inactivates DnaK and transitions to an Hsp33-based protection mechanism. In our study, we focused instead on different stress conditions, namely, heat stress alone or hydrogen peroxide treatment under normal growth temperatures, which are known to activate the DnaK chaperone system and, moreover, rely on its molecular chaperone activity to maintain viability (Delaney 1990; Winter et al. 2005). We took this approach in an effort to understand potential differences in the way that DnaK responds to stresses that have distinct requirements for its activity. Combined with the findings of the Jakob group, the results suggest that DnaK may be able to form multiple types of oligomers, some of which have partial activity while others do not. Taking this conclusion further, the increase in oligomerization observed during heat shock may confer a selective advantage (under some conditions) by creating an active form possessing distinct chaperone activities from the monomer.
DnaK oligomers are small, defined multimers
A major goal of this effort was to determine whether oligomeric DnaK was a product of non-specific interactions or whether these higher molecular weight structures represented oligomeric states assembled from well-behaved and properly folded monomers. Electron microscopy has recently been applied to the study of other Hsp70-related processes (Moreno-del Alamo et al. 2010; Schuermann et al. 2008; Xing et al. 2010). Therefore, we envisioned that this method could be used to explore DnaK oligomer structures, despite their small size. In these experiments, we found that, for most of the monomeric DnaK particles, the two-domain architecture is clearly present. Moreover, we found that DnaK-ADP samples contained small, defined oligomers (see Fig. 4). We also found that monomers could be released from the oligomers by substrate, suggesting a dynamic exchange between monomers and discrete multimers. Together, these findings are supportive of multimers being composed of individual folded monomers. In further support of this idea, we found that cross-linked DnaK-ADP samples retained ATPase and holdase activity, which is expected of functional monomers assembled into multimers.
DnaK oligomers are functional holdases but inactive foldases
Another major question is whether DnaK oligomers retain any of the chaperone functions of the well-studied monomer. This question is important because the current models for ATPase cycling and chaperone activity do not take into account any change in DnaK’s oligomerization, yet it appears to form in cells. In fact, it was assumed that the oligomers may serve as a reservoir of inactive chaperone. To test this idea, we examined a number of chaperone functions in vitro and found that DnaJ could not stimulate DnaK oligomers in either ATPase assays (see Fig. 2c) or luciferase refolding experiments (see Fig. 2d). However, DnaK oligomers retained many other chaperone functions, such as stimulation by GrpE. Furthermore, it retained the ability to bind luciferase and protect it from terminal heat denaturation (“holdase” activity). Consistent with this finding, oligomeric DnaK is known to bind lambdaP (Osipiuk et al. 1993), and oligomeric BiP still binds its peptide substrates (Blond-Elguindi et al. 1993). Furthermore, while studying pre-steady state kinetics, Farr and Witt (1997) observed results consistent with the formation of an oligomeric DnaK–peptide complex. While one can never fully exclude the possibility of cross-linking artifacts, our results and these previous studies (Osipiuk et al. 1993; Blond-Elguindi et al. 1993; Farr and Witt 1997) support the idea that oligomeric DnaK is capable of binding to client proteins. Thus, we hypothesize that oligomerization largely converts DnaK from a co-chaperone dependent foldase to a co-chaperone independent holdase (Fig. 5). This finding could have important consequences on DnaK biology because, under heat stress conditions, the enzyme might be required to predominately act in a holdase capacity until conditions improve. Such a model would be consistent with oligomerization providing a selective advantage under conditions of heat stress.
Proposed model of the DnaK chaperone cycle Oscillation of DnaK between its ATP- and ADP-bound states, in collaboration with DnaJ and GrpE, has been shown to be required for iterative substrate binding and refolding. In addition to this well-known cycling, our data shows that DnaK-ADP additionally samples multimeric structures, discussed in detail within the text. These oligomers are biochemically distinct from monomers because they have a limited foldase activity while maintaining holdase activity. We speculate that DnaK oligomers are an active part of the chaperone cycle
DnaK is structurally heterogeneous
Recent NMR studies have suggested that the NBD and SBD of DnaK sample a number of relative orientations in the ADP + peptide state (Bertelsen et al. 2009). Consistent with this idea, we were able to visualize the two domain structure of DnaK by EM (Fig. 4b) and found that the domains are present in a variety of relative orientations (e.g., some DnaK molecules were elongated, while others were compact and more reminiscent of the Hsp110 crystal structure (Liu and Hendrickson 2007)). Although some of this diversity could certainly be attributed to stain drying artifacts or deformations on the carbon support of the EM grid, recent studies using EPR and FRET have also observed multiple inter-domain conformations of Hsp70s in both the ATP- and ADP-bound state (Marcinowski et al. 2011; Schlecht et al. 2011). Moreover, the work of Schlecht et al. (2011) further highlights the functional importance of this conformational heterogeneity. They found that DnaK adopts a variety of open and closed forms that permit flexibility during binding to client proteins with different topologies. Thus, a recurring theme is that Hsp70-class chaperones sample a variety of possible conformations. While their substrates, co-chaperones, and nucleotide ligands can shift this balance, they rarely trap the chaperone in a uniform state. For DnaK, our results suggest that this structural heterogeneity should further include allowances for formation of higher ordered multimers.
DnaK oligomerization interface and its potential for small molecule modulation
The EM work has clearly demonstrated that oligomeric DnaK has a defined architecture made up of intact monomers. Furthermore, these oligomers retain some chaperone activities, including substrate binding, while losing the ability to refold substrates or be stimulated by the co-chaperone DnaJ. Thus, we conclude that oligomeric DnaK is formed via a definable interaction surface that could potentially be exploited to regulate chaperone activity. For example, promoting oligomerization of human Hsp70s might block some chaperone functions, perhaps rendering cancer cells more susceptible to apoptosis (Powers et al. 2009). However, what is the nature of this intermonomer interface? Although molecular structures have remained elusive, previous work and our studies provide some clues. For example, Ladjimi and colleagues found that the SBD, but not the NBD, of a human Hsp70 retains the ability to oligomerize (Benaroudj et al. 1996, 1997). Other evidence also points to the SBD being the major site of intermonomer contacts in BiP (Gaut 1997). Interestingly, deletion of the 10-kDa C-terminus of Hsp70 does not affect oligomerization (Fouchaq et al. 1999), suggesting that the entire SBD is not required. The fact that oligomerization seems to be primarily mediated through the SBD, along with the observation that many of the factors that promote monomerization also promote peptide release, has appropriately lead to the hypothesis that DnaK oligomerization may be mediated through the substrate-binding pocket (Benaroudj et al. 1996, 1997). However, several groups have demonstrated that oligomeric DnaK can still bind substrates (Blond-Elguindi et al. 1993; Farr and Witt 1997; Osipiuk et al. 1993), suggesting that any SBD-mediated interface must leave a free substrate-binding region available in at least one DnaK molecule within the oligomer. In support of this model, our results again demonstrate that oligomeric DnaK is still able to bind substrate (see Fig. 3). This result suggests that some of the substrate-binding pockets within DnaK oligomer are left unoccupied. Further work is required to define the interface(s), but these results suggest that this contact surface might be a good place to target with chemical probes. Recent success in targeting protein–protein interfaces in the chaperone complex support this idea (Wisen et al. 2010; Chang et al. 2011).
Materials and methods
Materials
Reagents utilized throughout this study were obtained as follows: ATP-agarose column (Sigma, St. Louis, MO, USA), NRLLLTG peptide (University of Michigan Peptide Core) (Buczynski et al. 2001), luciferase, and Steady-Glo Reagent (Promega, Madison, WI, USA). The optical density and luminescence measurements were performed using a SpectraMax M5 multimode plate reader (Molecular Devices, Sunnyvale, CA, USA).
Plasmids and protein purification
Wild-type E. coli DnaK, the DnaK mutant T199A, E. coli DnaJ and E. coli GrpE were purified as previously described (Chang et al. 2010). When eluting DnaK from the ATP-agarose column, samples to be tested in the ADP-bound state were eluted using ADP. All proteins were concentrated and exchanged into EM buffer [25 mM HEPES buffer, 20 mM KCl (150 mM KCl for DnaJ), 5 mM MgCl2, pH 7.4] and stored at −80°C until use. DnaK to be used for EM was never frozen and further purified by Superdex 200 gel filtration column (GE Healthcare, Piscataway, NJ, USA). DnaK NBDL (residues 1–392) was purified by the same approach. The nucleotide state of DnaK was established by pre-incubating with either 1 mM ATP or ADP for 30 min at 4°C, which gave the expected change in tryptophan fluorescence (Online Resource 1).
Gel filtration
Samples were prepared with 14 μM DnaK (unless otherwise indicated), 1 mM nucleotide, and the indicated amount of NRLLTG (NR) peptide, J-domain, or GrpE in EM buffer. Samples were allowed to equilibrate at 4°C for >30 min. A sample of 300 μL was separated into fractions (1 mL) using a Superdex 200 gel filtration column (GE Healthcare, Piscataway, NJ, USA) in EM buffer at a flow rate of 0.6 mL/min. Absorbance at 285 nm was monitored and normalized according to the nucleotide peak to control for variations in injection volumes.
Gel electrophoresis
For denaturing gel electrophoresis, samples were denatured with 3× sodium dodecyl sulfate (SDS) denaturing loading dye (1×: 80 mM Tris–HCl, 10% v/v glycerol, 5.3% v/v β-mercaptoethanol, 2% w/v SDS, 0.01% w/v bromophenol blue, pH 6.8) and heating to 95°C for 2 min. Samples were then separated for 1 h at 200 V using a 10% SDS-PAGE gel. For native gel electrophoresis, samples were prepared by adding 3× native loading dye (1×: 80 mM Tris–HCl, 10% v/v glycerol, 0.01% w/v bromophenol blue, pH 6.8). Samples were separated for 4–6 h at 120 V and 4°C using a 4–20% Tris–HCl native gel (Biorad, Hercules, CA, USA) in Tris–acetate running buffer (30 mM Tris–HCl, 0.1% v/v acetic acid, 1 mM EDTA, pH 8.0).
Monitoring DnaK oligomerization in lysates
W3110 cells were grown to mid-log phase at 30°C in Luria–Bertani (LB). Cells were then either left at 30°C in LB or subject to a specific stress for 1 h. Heat stress conditions were achieved by moving the mid-log phase culture to 43°C (Paek and Walker 1987). Oxidative stress was applied with the addition of 4 mM H2O2 (Delaney 1990; Winter et al. 2005). To probe the effect of DnaK over-expression, Δdnak (DE3) cells transfected with wild-type DnaK under a T7-promoter were utilized Chang et al. 2011. Cells were grown to mid-log phase at 30°C, and DnaK expression was induced by isopropyl β-d-1-thiogalactopyranoside for 1 h. Following growth, cultures were immediately moved to an ice-water bath (4°C) and diluted to an OD600 of 0.4 and pelleted. Cells were lysed by resuspending the pellet in 500 μL of lysozyme reaction buffer [1 mg/mL lysozyme in 50 mM Tris–HCl, 2 mM EDTA, 200 μM phenylmethanesulfonylfluoride (PMSF), pH 8.0] for 30 min at 4°C, followed by the addition of 500 μL of 2× DNAase I reaction buffer (60 μg/mL DNAse I, 50 mM Tris–HCl, 4 mM MgCl2, 12 mM CaCl2, 200 μM PMSF, pH 8.0). After 10 min, cells were sonicated using a microtip (ThermoFisher Sci.) at 60% for 20 s. Cell debris was pelleted by centrifugation at 13.2 rpm for 15 min, and the resulting supernatant was passed through a 20-μm filter. Lysates were subsequently analyzed as described above. For the Western blots, gels were transferred to 0.2 μm Protran nitrocellulose (Whatman, Piscataway, NJ, USA). The membrane was blocked with 5% w/v milk in TBS-T (25 mM Tris–HCl, 140 mM NaCl, 0.1% Tween-20) and probed with 1:500 mouse monoclonal anti-DnaK antibody (Assay Designs/Enzo Life Sciences, SPA-880F, Plymouth Meeting, PA, USA) followed by an horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Anaspec, 28173, Fremont, CA, USA). Western blots were developed using the Western Lightning Plus-ECL kit according to the manufacturer’s protocol (PerkinElmer, Waltham, MA, USA).
Cross-linking
Samples were prepared with 14 μM DnaK (or DnaK NBDL) and 1 mM nucleotide in EM buffer and allowed to equilibrate at 4°C for >30 min. Samples were then moved to room temperature and 0.025% v/v glutaraldehyde in EM buffer was added to each. The reactions were quenched by addition of 100 mM glycine (pH 8.0). This quenching step was either performed immediately (for the control samples) or after incubating the solutions for 20 min at room temperature. To evaluate the cross-linking efficiency, 10 μL of each sample was evaluated by denaturing gel electrophoresis as described above. Furthermore, cross-linked samples were consistently tested for activity in DnaJ-mediated ATPase stimulation as a quality control measure. When testing these samples in the biochemical assays, they were diluted into assay buffer containing 1 mM nucleotide. Each biochemical assay was carried out on at least two independently cross-linked samples. The results depicted in Figs. 2 and 3 are from a single, representative experiment performed in triplicate, while the values reported in Table 1 are an average of all the replicates.
ATPase activity
This procedure was adapted from a previously described protocol (Chang et al. 2008). Briefly, the final concentration of DnaK was 0.5 μM, unless otherwise noted. In each experiment, the signal from non-specific ATP hydrolysis in a control lacking DnaK was subtracted. In testing GrpE stimulation, 0.5 μM DnaK was tested in the presence of 0.5 μM DnaJ and 200 μM NRLLLTG peptide. DnaJ, NRLLLTG, and GrpE stimulation curves were evaluated by fitting the data using a hyperbolic fit with a non-zero intercept. The non-linear fit was performed using GraphPad Prism version 5.0 for Windows (GraphPad Software San Diego, CA, USA). For clarity, graphs in Online Resource 4 were transformed to obtain a zero y-intercept for all samples.
Luciferase binding
The procedure for DnaK binding to luciferase was adapted from a previous report (Miyata et al. 2010). Briefly, firefly luciferase (61.3 μM) was denatured by incubation with 6 M GuHCl for 1 h at room temperature and diluted to 100 nM for storage. Aliquots (50 μL) were added to each well of 96-well plates (Thermo Fisher, clear, nonsterile, flat bottom, Waltham, MA, USA), and these plates were then incubated for 2 h at 37°C. The wells were washed with 150 μL of TBS-T (3 × 3 min, rocking). To these wells, 50 μL of a DnaK or DnaK NBDL solution (at indicated concentrations) in binding buffer (25 mM HEPES, 150 mM NaCl, 20 mM KCl, 5 mM MgCl2, pH 7.2) with 1 mM ATP was added to each well and allowed to bind at room temperature while rocking overnight. After washing (3 × 3 min, rocking) with TBS-T, 5% w/v milk in 100 μL of TBS-T was added to each well for 5 min. Next, the primary antibody was added (1:5,000 dilution of rabbit anti-DnaK (Abcam, ab80161, Cambridge, MA, USA) in TBS-T, 50 μL/well), and the plates were incubated for 1 h at room temperature. Wells were again washed with TBS-T, followed by addition of the secondary antibody [1:5,000 dilution HRP-conjugated goat anti-rabbit (Anaspec, 28177, Fremount, CA) in TBS-T, 50 μL/well], and the plates were incubated for 1 h at room temperature. Finally, wells were washed one last time with TBS-T (3 × 3 min, rocking) and developed for 2 min using a TMB substrate kit (Cell Signaling Technology, Danvers, MA, USA). The absorbance was measured at 450 nM. In each experiment, the signal from non-specific binding of DnaK to empty control wells was subtracted. Binding curves were fit using hyperbolic fit with a non-zero intercept with GraphPad Prism version 5.0 for Windows (GraphPad Software, San Diego, CA, USA).
Luciferase holdase
Holdase activity was evaluated as described with minor changes (Chang et al. 2010). The final concentrations per well were 0.016 μM luciferase and 1 mM ADP. After the reaction mixture was heated to 39.5°C for 8 min, 10 μL of the samples were transferred into 96-well, opaque white assay plate (Thermofisher, Waltham, MA, USA) and then 10 μL of 5% v/v SteadyGlo reagent in 50 mM glycine buffer (30 mM MgSO4, 10 mM ATP, and 4 mM dithiothreitol, pH 7.8) was added into each well. Results were again fit using a hyperbolic fit with a non-zero intercept.
Luciferase refolding
Luciferase refolding activity was evaluated as described with minor changes (Wisen and Gestwicki 2008). The final concentrations were 1 μM DnaK, 100 nM denatured luciferase, and 1 mM ATP. After 1 h of incubation at 37°C, 14 μL of 2% v/v SteadyGlo reagent was added into each well, and the luminescence was measured. For each experiment, the signal from a negative control containing everything, but DnaK was subtracted.
Electron microscopy
Samples were prepared containing 14 μM DnaK, 1 mM ADP, with/without 400 μM NRLLTG peptide (substrate), and allowed to equilibrate at 4°C for >30 min. Samples were then separated using a Superdex 200 gel filtration column in EM buffer as described above. A fraction from between 13.8 and 14.0 mL was collected and immediately prepared for EM using a conventional negative staining protocol (Ohi et al. 2004). Briefly, eluted fractions were diluted 100× into EM buffer containing 1 mM ADP or 1 mM ADP + 400 μM substrate, as indicated, and 2 μL of the sample was adsorbed to a glow-discharged carbon-coated copper grid (Electron Microscopy Sciences, Hatfield, PA, USA) and stained with 0.75% uranyl formate (Polysciences Inc, Warrington, PA, USA) solution. Samples (see Fig. 4, Online Resources 7, 8) were imaged at room temperature with a Tecnai T12 electron microscope equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on a Gatan 4 × 4 k pixel charge-coupled device camera using low-dose procedures at a magnification of ×52,000 and a defocus value of about–1.5 μm. The images were binned (2 × 2 pixels) to obtain a pixel size of 4.16 Å on the specimen level, and particles were manually excised using Boxer (part of the EMAN 1.9 software suite; Ludtke et al. 1999). 2D analysis of DnaK-ADP and DnaK-ADP + 400 μM substrate were carried out using the SPIDER image processing suite (Frank et al. 1996). For the 2D analysis of DnaK-ADP, we interactively selected particles windowed into 64 × 64 pixel images. A total of 6,313 particles from 49 images were subjected to ten cycles of reference-free alignment specifying 150 classes. Based on the first classification, we observed a heterogeneous sample containing monomeric two-lobe particles of DnaK, small more compact monomeric particles, dimeric assembles of DnaK, and some higher-ordered assembles of DnaK (Online Resource 7). We then selected only the particles that were well resolved, removing 223 particles. The remaining 6,090 particles were subjected to a second reference-free alignment to improve their classification (Fig. 4, Online Resource 7). For the 2D analysis of DnaK-ADP + 400 μM substrate, we followed the same procedure. A total of 6,689 particles were selected from 84 images, excluding underrepresented large oligomers (Online Resource 8, arrow) and initially subjected to ten cycles of reference-free alignment specifying 150 classes. Based on this first classification, we observed an increase in monomeric DnaK species as compared to the DnaK-ADP sample (Online Resource 8). We removed poorly resolved particles or larger (Online Resource 8, arrow) underrepresented species (591 particles in total). The remaining 6,098 particles were subjected to a second multi-reference alignment to improve their classification (Fig. 4, Online Resource 8). The number of particles belonging to each class as well as whether that class was interpreted as a monomer (m), dimer (d), assembly larger than a dimer (>d), or ambiguous (−) is recorded in Online Resources 7 and 8.
References
Angelidis CE, Lazaridis I, Pagoulatos GN (1999) Aggregation of hsp70 and hsc70 in vivo is distinct and temperature-dependent and their chaperone function is directly related to non-aggregated forms. Eur J Biochem 259:505–512
Benaroudj N, Batelier G, Triniolles F, Ladjimi MM (1995) Self-association of the molecular chaperone HSC70. Biochemistry 34:15282–15290
Benaroudj N, Triniolles F, Ladjimi MM (1996) Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone HSC70. J Biol Chem 271:18471–18476
Benaroudj N, Fouchaq B, Ladjimi MM (1997) The COOH-terminal peptide binding domain is essential for self-association of the molecular chaperone HSC70. J Biol Chem 272:8744–8751
Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A 106:8471–8476
Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJ (1993) Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem 268:12730–12735
Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B (1995) Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem 270:16903–16910
Buczynski G, Slepenkov SV, Sehorn MG, Witt SN (2001) Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-rate constants. J Biol Chem 276:27231–27236
Carlino A, Toledo H, Skaleris D, DeLisio R, Weissbach H, Brot N (1992) Interactions of liver Grp78 and Escherichia coli recombinant Grp78 with ATP: multiple species and disaggregation. Proc Natl Acad Sci U S A 89:2081–2085
Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE (2008) High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem 372:167–176
Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE (2010) Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK). J Biol Chem 285:21282–21291
Chang L, Miyata Y, Ung PMU, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ERP, Gestwicki JE (2011) Chemical screens against a reconstituted multi-protein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol 18:210–221
Cowing DW, Bardwell JC, Craig EA, Woolford C, Hendrix RW, Gross CA (1985) Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci U S A 82:2679–2683
Delaney JM (1990) Requirement of the Escherichia coli dnaK gene for thermotolerance and protection against H2O2. J Gen Microbiol 136:2113–2118
Dukan S, Nystrom T (1998) Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev 12:3431–3441
Evans CG, Chang L, Gestwicki JE (2010) Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem 53:4585–4602
Farr CD, Witt SN (1997) Kinetic evidence for peptide-induced oligomerization of the molecular chaperone DnaK at heat shock temperatures. Biochemistry 36:10793–10800
Fouchaq B, Benaroudj N, Ebel C, Ladjimi MM (1999) Oligomerization of the 17-kDa peptide-binding domain of the molecular chaperone HSC70. Eur J Biochem 259:379–384
Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116:190–199
Freiden PJ, Gaut JR, Hendershot LM (1992) Interconversion of three differentially modified and assembled forms of BiP. EMBO J 11:63–70
Gaut JR (1997) In vivo threonine phosphorylation of immunoglobulin binding protein (BiP) maps to its protein binding domain. Cell Stress Chaperones 2:252–262
Grimshaw JP, Jelesarov I, Siegenthaler RK, Christen P (2003) Thermosensor action of GrpE. The DnaK chaperone system at heat shock temperatures. J Biol Chem 278:19048–19053
Grimshaw JP, Siegenthaler RK, Zuger S, Schonfeld HJ, Z’Graggen BR, Christen P (2005) The heat-sensitive Escherichia coli grpE280 phenotype: impaired interaction of GrpE(G122D) with DnaK. J Mol Biol 353:888–896
Hesterkamp T, Bukau B (1998) Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E.coli. EMBO J 17:4818–4828
Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U (2004) Identification of a redox-regulated chaperone network. EMBO J 23:160–168
King C, Eisenberg E, Greene L (1995) Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J Biol Chem 270:22535–22540
Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A 88:2874–2878
Liu Q, Hendrickson WA (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131:106–120
Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97
Marcinowski M, Holler M, Feige MJ, Baerend D, Lamb DC, Buchner J (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol 18:150–158
Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684
Miyata Y, Chang L, Bainor A, McQuade TJ, Walczak CP, Zhang Y, Larsen MJ, Kirchhoff P, Gestwicki JE (2010) High-throughput screen for Escherichia coli heat shock protein 70 (Hsp70/DnaK): ATPase assay in low volume by exploiting energy transfer. J Biomol Screen 15:1211–1219
Moreno-del Alamo M, Sanchez-Gorostiaga A, Serrano AM, Prieto A, Cuellar J, Martin-Benito J, Valpuesta JM, Giraldo R (2010) Structural analysis of the interactions between Hsp70 chaperones and the yeast DNA replication protein Orc4p. J Mol Biol 403:24–39
Ohi M, Li Y, Cheng Y, Walz T (2004) Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6:23–34
Osipiuk J, Georgopoulos C, Zylicz M (1993) Initiation of lambda DNA replication. The Escherichia coli small heat shock proteins, DnaJ and GrpE, increase DnaK’s affinity for the lambda P protein. J Biol Chem 268:4821–4827
Osorio H, Carvalho E, del Valle M, Gunther Sillero MA, Moradas-Ferreira P, Sillero A (2003) H2O2, but not menadione, provokes a decrease in the ATP and an increase in the inosine levels in Saccharomyces cerevisiae. An experimental and theoretical approach. Eur J Biochem 270:1578–1589
Paek KH, Walker GC (1987) Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol 169:283–290
Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL (1993) ATP-induced protein-Hsp70 complex dissociation requires K + but not ATP hydrolysis. Nature 365:664–666
Palleros DR, Shi L, Reid KL, Fink AL (1994) hsp70-protein complexes. Complex stability and conformation of bound substrate protein. J Biol Chem 269:13107–13114
Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27:437–496
Patury S, Miyata Y, Gestwicki JE (2009) Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem 9:1337–1351
Powers MV, Clarke PA, Workman P (2009) Death by chaperone: HSP90, HSP70 or both? Cell Cycle 8:518–526
Rist W, Graf C, Bukau B, Mayer MP (2006) Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J Biol Chem 281:16493–16501
Schlecht R, Erbse AH, Bukau B, Mayer MP (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol 18:345–351
Schonfeld HJ, Schmidt D, Schroder H, Bukau B (1995) The DnaK chaperone system of Escherichia coli: quaternary structures and interactions of the DnaK and GrpE components. J Biol Chem 270:2183–2189
Schroder H, Langer T, Hartl FU, Bukau B (1993) DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J 12:4137–4144
Schuermann JP, Jiang JW, Cuellar J, Llorca O, Wang LP, Gimenez LE, Jin SP, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R (2008) Structure of the Hsp110: Hsc70 nucleotide exchange machine. Molecular Cell 31:232–243
Shi L, Kataoka M, Fink AL (1996) Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry 35:3297–3308
Siegenthaler RK, Christen P (2006) Tuning of DnaK chaperone action by nonnative protein sensor DnaJ and thermosensor GrpE. J Biol Chem 281:34448–34456
Slepenkov SV, Witt SN (1998) Peptide-induced conformational changes in the molecular chaperone DnaK. Biochemistry 37:16749–16756
Spence J, Cegielska A, Georgopoulos C (1990) Role of Escherichia coli heat shock proteins DnaK and HtpG (C62.5) in response to nutritional deprivation. J Bacteriol 172:7157–7166
Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell 26:27–39
Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU (1994) The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A 91:10345–10349
Szabo A, Korszun R, Hartl FU, Flanagan J (1996) A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J 15:408–417
Tilly K, McKittrick N, Zylicz M, Georgopoulos C (1983) The dnaK protein modulates the heat-shock response of Escherichia coli. Cell 34:641–646
Toledo H, Carlino A, Vidal V, Redfield B, Nettleton MY, Kochan JP, Brot N, Weissbach H (1993) Dissociation of glucose-regulated protein Grp78 and Grp78-IgE Fc complexes by ATP. Proc Natl Acad Sci U S A 90:2505–2508
Tran QH, Unden G (1998) Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem 251:538–543
Winter J, Linke K, Jatzek A, Jakob U (2005) Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell 17:381–392
Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U (2008) Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135:691–701
Wisen S, Gestwicki JE (2008) Identification of small molecules that modify the protein folding activity of heat shock protein 70. Anal Biochem 374:371–377
Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung P, Chang L, Evans CG, Walter GAM, Wipf P, Carlson HA, Brodsky JL, Zuiderweg ERP, Gestwicki JE (2010) Binding of a small molecule at a protein–protein interface regulates chaperone activity in the Hsp70-Hsp40 complex. ACS Chem Biol 5:611–622
Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ (2003) The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry 42:4937–4944
Xing Y, Bocking T, Wolf M, Grigorieff N, Kirchhausen T, Harrison SC (2010) Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J 29:655–665
Young JC, Barral JM, Ulrich Hartl F (2003) More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci 28:541–547
Acknowledgments
The authors thank Erik Zuiderweg and Ursula Jacob for helpful comments. We would also like to thank Min Su for technical training on the electron microscope, Brinae Bain for experimental support, and Murray Murkoff for sharing ideas. A.D.T. was supported by a pre-doctoral fellowship from the National Institute of Health (F30AG035464). This work was additionally supported by a grant from the National Institute of Health (NS059690) to J.E.G.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 36272 kb)
Rights and permissions
About this article
Cite this article
Thompson, A.D., Bernard, S.M., Skiniotis, G. et al. Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK). Cell Stress and Chaperones 17, 313–327 (2012). https://doi.org/10.1007/s12192-011-0307-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-011-0307-1