Abstract
We compared a series of Drosophila strains with P element insertions from −28 to −144 nucleotides 5′ to the transcription start site of the Hsp70A genes—corresponding to the range of naturally occurring P element insertion sites—to elucidate the consequences of insertion site for Hsp70A gene expression. Although all insertions reduced Hsp70A expression below that of a control strain, the magnitude of the reduction was inversely related to the number of nucleotides between the transcription start site and the insertion site. A pre-existing hypothesis is that naturally occuring transposable element insertions in Hsp promoters may be beneficial in some circumstances, which may account for their retention in natural populations. In the present study, in a control line heat shock reduced fecundity, whereas in lines with P element insertions heat shock typically increased fecundity. Finally, according to cluster-specific quantitative RT-PCR, expression of the Hsp70A cluster genes was typically greater than that of the Hsp70B gene cluster genes, although the latter are more numerous and, in this case, free of P element insertions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Transposable elements (TEs), which are DNA sequences able to move among or within chromosomes, are major components of genomes. As drivers of genome evolution, TEs can contribute to genetic and genomic variation by transposition into regulatory regions of host genes (Kazazian 2004; Volff 2006). Indeed, the regulatory regions (promoters) of heat-shock genes are remarkably prone to mutagenesis by P transposable elements both in laboratory and in natural populations of Drosophila melanogaster (Timakov et al. 2002; Lerman et al. 2003; Lerman and Feder 2005; Walser et al. 2006; Shilova et al. 2006).
We have hypothesized that the major significance of the TE insertions into heat-shock genes is that they create variation on which natural selection may act (Lerman et al. 2003; Lerman and Feder 2005; Walser et al. 2006). Molecular chaperones, the products of the genes in which the TEs have inserted, are typically pleiotropic proteins: through their deterrence of protein aggregation, they confer tolerance to stress—but they also participate in diverse processes (cell growth, apoptosis, signaling, regulation of other proteins, etc.) in the absence of stress (Lerman and Feder 2005). These latter functions typically require stringent regulation of chaperone level and activity, and high levels of chaperones can disrupt them (Krebs and Feder 1998; Krebs and Feder 1997). Accordingly TE insertions into the proximal promoter regions of heat-shock genes, which typically decrease host gene expression, can be beneficial in the absence of stress. Indeed, in several studies (Lerman et al. 2003; Lerman and Feder 2005; Walser et al. 2006; Bettencourt et al. 2002; Michalak et al. 2001), the frequency of P element insertion alleles is greater in natural populations with less heat stress than with greater heat stress.
Importantly, P element insertions are at many different sites within the proximal promoter regions of heat-shock genes, with 1–6 in any given gene (Walser et al. 2006). They may also differ in size and orientation (Walser et al. 2006). In principle, this diversity presents a wide range of variation on which natural selection may act. Not understood, however, is whether this diversity has any regularity or pattern in its phenotypes, i.e., whether the insertion site is predictably informative about the magnitude of an insertion sites’ impact on host gene expression. Such understanding is essential to making sense of the diversity of P element insertion sites in the heat-shock genes.
To address this issue, we have exploited strains created in a prior experiment on P element insertion site preference (Shilova et al. 2006). These strains share a common P element, EPgy2, and genetic background, but have the P element inserted at sites ranging from −28 bp (at the TATA box) to −240 bp nucleotides upstream of the transcription start site in the Hsp70Aa and Hsp70Ab genes. Like other Drosophila heat-shock promoters, these contain several heat-shock elements (HSEs), specific binding sites for the heat-shock transcription factor HSF. Upon heat shock, HSF trimerizes, localizes to the nucleus, and binds the HSEs, whereupon the transactivation domains are positioned to interact with the polymerase apparatus and unleash transcription (Boehm et al. 2003; Yao et al. 2006; Cartwright and Elgin 1986). As prior work has shown, the native proximal promoter sequence is critical for full-strength transcription. For example, insertion of but 1–5 nucleotides between HSEs 1 and 2 reduces promoter activity 90% (Amin et al. 1994). Precisely positioned GAGA factor binding sites [i.e., (CT)n repeats], HSEs, and nucleosomes interact with GAGA factor and HSF to juxtapose activated HSF–HSE complexes and the polymerase apparatus (Thomas and Elgin 1988; Quivy and Becker 1996; Lu et al. 1993; Cartwright and Elgin 1986). Deletion or mutation of the (CT)n elements abolishes or decreases Hsp transcription (Weber et al. 1997; Lu et al. 1993). Presumably, the insertion of a P element also interferes with this HSF-polymerase apparatus interaction. The precise physical properties of the proximal promoter regions of these genes, such as their ability to bend and so juxtapose the HSF transactivation domains and the polymerase apparatus, is presently unknown however.
A final issue is that the Hsp70Aa and Hsp70Ab genes, clustered in opposite orientation at chromosomal locus 87A7, are only 2 of the 5–6 Hsp70 genes in D. melanogaster (Bettencourt and Feder 2001; Maside et al. 2002); the others are clustered at 87C1. All Hsp70 genes encode a protein of identical sequence. Thus, potentially, a P element insertion into the proximal promoter region of 1 or 2 Hsp70 genes might be inconsequential for overall Hsp70 expression, especially if activated HSF can redistribute to the other Hsp70 genes and increase their expression in compensation. The similarity among the Hsp70 genes in coding sequence, however, is not as extensive for regulatory sequence, and the Hsp70 genes at the two clusters differ in the kinetics and tissue specificity of their expression (Lakhotia and Prasanth 2002). Hence, a P element insertion into an 87A7 Hsp70 might have a considerable phenotype. Distinguishing among these alternatives has previously been challenging because of the near-identity of the gene products. Here, however, we use cluster-specific quantitative real-time PCR to distinguish the products in simulataneous measurements.
2 Materials and methods
2.1 Drosophila strains
A Drosophila melanogaster strain (hereafter “US-4”) obtained from the Bloomington Drosophila Stock Center was used to generate 23 lines with EPgy2 P transposable elements independently inserting in the proximal promoter regions of the Hsp70A genes, as described (Shilova et al. 2006). We examined 10 of these lines (Table 1) with various P element insertion sites, as well as the US-4 parental strain. EPgy2 is longer in line 332II than in other lines (see Table 1).
2.2 Pretreatment to induce gene expression
Three days after eclosion, groups of 20 virgin females in glass vials with fresh medium were used in following experimentation.
First, to determine a regime for maximum induction of Hsp70 mRNA in the US-4 line, flies were pretreated at 36.0 or 37.0°C for 30 min, and duration of recovery at 25.0°C varied (0, 30, 60 and 90 min). Second, pretreatment temperature was varied (from 35.0 to 37.5°C for 30 min), with recovery at 25.0°C for a standard time (30 min).
According to the preliminary results, flies were thereafter exposed to 36.5°C for 30 min, typically followed by 30 additional min at 25°C. Additionally, US-4, 111 IIa and 30 IIb flies were exposed to 36.5°C for 30 min, and duration of recovery at 25°C varied (0, 30, 60, 90, 120, 180 min). After these thermal treatments, flies were frozen in liquid nitrogen and stored at −80°C.
2.3 mRNA quantification by quantitative real-time PCR
As previously described (Shilova et al. 2006), total RNA was extracted from previously treated or control flies with an RNeasy Protect kit (Qiagen) according to the manufacturer’s instructions. Equal amounts (1 μg) of RNA were used to synthesize first-strand cDNA with Oligo(dT) in the reverse transcription reaction (ABgene 1st Strand Synthesis kit) according to the manufacturer’s instructions. Hsp70 mRNA was determined by quantitative real-time PCR with an MJ Opticon detector, SYBR green detection method, and ABgene reagents (Rochester, NY). The housekeeping gene Rp49 was used as a loading control. All reactions were performed in triplicate.
Except as noted, the PCR reaction used primers complementary to both Hsp70Aa and Hsp70Ab but not to the other Hsp70 genes, and thus reported aggregate Hsp70A mRNA but not Hsp70B mRNA. A final experiment re-analyzed the same samples with primers complementary to Hsp70B genes but not Hsp70A genes. Specificity was established in preliminary experiments with Hsp70A and Hsp70B deletion strains (Shilova et al. 2006). Primers are:
-
- Hsp70A (QAF3: CCTGGAGAGCTACGTCTTCAAT; QAR3: GTCGTTGCACTTGTCCAA),
-
- Hsp70B (QBF3: GTCTACCAGCATGGCAAGGTT; QBR3: ACCAATGAGGCGTTCCGAG),
-
- Rp49 (RPF2: CGCACCAAGCACTTCATCC, RPR2: AGCGGCGACGCACTCTGT).
2.4 Reproductive success
Virgin females and males were collected within eight hours of eclosion and maintained separately on standard medium for 3 days. Females to be heat-shocked were placed in glass vials, which were submerged in a circulating water bath at 36.5°C for 30 min; otherwise, all flies were maintained at 25°C. For days 4–6 post-eclosion, both heat-shocked and control females (12 for each strain) were placed individually in vials containing standard medium and two males. Each trio was transferred to a fresh vial for days 7–9 and 10–12, and then discarded. All adults eclosing from each vial were counted (Silbermann and Tatar 2000).
2.5 Statistical analysis
After assessing among-group variance homogeneity (Levene’s test) and normality (Kolmogorov-Smirnov-Lilliefors test), independent sample t tests were used to compare the mean mRNA levels. Fecundity was compared in heat-shocked and control flies of the same strain with a Mann–Whitney U test.
3 Results
3.1 Maximum induction of Hsp70A gene expression
In pilot determinations of maximal Hsp70A gene expression (i.e., the total of Hsp70Aa and Hsp70Ab mRNA), Hsp70A mRNA was negligible in controls not exposed to heat shock (Figs 1, 2). Hsp70A mRNA increased during 30 min of heat shock at 36°C, and continued to increase for at least 30 min thereafter. More intense heat shock (37°C) prolonged this increase. For flies heat-shocked for 30 min at 35.0–37.5°C and analyzed after an additional 30 min at 25°C, mRNA levels were greatest after 36.5°C heat shock. Accordingly, subsequent experimentation typically used 36.5°C for 30 min followed by 25° for 30 min.
3.2 Effect of P element insertion site on Hsp70A expression
Relative to the US-4 line, from which the Hsp70A P element lines were derived, P element insertion into the Hsp70A proximal promoters decreased Hsp70A mRNA levels. The magnitude of the decrease was inversely related to the distance of the insertion from the transcription site, ranging from 56% of US-4 levels at nucleotide −28 to 3% at nucleotide −144 (Fig. 3, Table 1). Whether the insertion was in Hsp70Aa or Hsp70Ab had no obvious impact on the magnitude of the decrease. Interestingly, in the one line with P elements in both Hsp70Aa and Hsp70Ab, the magnitude of the decrease was similar to lines with insertions in only one gene.
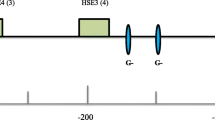
Inducible thermotolerance and relative expression of Hsp70A mRNA as a function of P element insertion site. Top, inducible thermotolerance, defined as the difference in survival between flies with and without a mild heat pretreatment (36.5° for 30 min + 25° for 30 min) before heat shock (39.5°C for 30 min), is replotted from Fig. 7 (bottom right) in Shilova et al. (2006). US-4 is the control line without any P elements in Hsp70A. Middle: Three-day-old virgin flies were pretreated at 36.5°C for 30 min following with 30 min recovery at 25°C before RNA extraction. US-4 is the control line without any P elements in Hsp70A. Right axis indicates expression as copies normalized against Rp49 expression, and left axis as normalized against Hsp70A expression in the US-4 strain. Error bar indicates standard error for four separate extractions of mRNA from a pool of 20 virgin females. Bottom, cartoon indicating approximate location of TATA box (T), heat-shock elements (H), and GAGA factor binding sites (G). Arrows indicate sites of naturally occuring P element insertions (Walser et al. 2006)
For two lines (111 IIA, with a P element insertion at −97 in Hsp70Aa, and 30 IIb, with a P element insertion at −96 in both Hsp70Aa and Hsp70Ab), we compared Hsp70A mRNA levels with those in US-4 at several times after heat shock (Fig. 4). In contrast to the data in Fig. 3, the 30IIb double mutant had lower Hsp70A mRNA levels than the 111 IIA single mutant. In both lines, however, the Hsp70A mRNA levels were lower than in the US-4 control line. All three lines exhibited approximately the same relative kinetics of Hsp70A expression, although the absolute amounts of expression differed.
Expression of Hsp70A genes in Drosophila melanogaster upon heat-shock at 36.5°C for 30 min following different time of recovery at 25°C. Relative gene expression was indicated as normalized copies against Rp49 gene. Samples from different recovery time were distributed on the same PCR plate and against the same standard curve. US-4 is the control line with no P element insertion in Hsp70A
3.3 Does Hsp70B expression increase when Hsp70A expression decreases?
As explained in the Introduction, we sought to determine whether the expression of the Hsp70B cluster genes increased when insertion of P elements decreased Hsp70A expression. Such an increase occurred in only one of the 3 lines examined, 134Ib, in which overall Hsp70A expression was the lowest (Fig. 5). Curiously, although the Hsp70B genes are more numerous than the Hsp70A genes, their contribution to overall Hsp70 mRNA expression was less than that of the Hsp70A genes in three of the four comparisons.
Relative expression of Hsp70A and Hsp70B gene in transgenic lines of Drosophila melanogaster, standardized by normalization against Rp49 gene copies. The flies were pretreated at 36.5°C for 30 min following with 30 min recovery at 25°C before RNA extraction. US-4 is the control line with no P element insertion in Hsp70A. Error bar indicates standard error from seven replicates for Hsp70A and four replicates for Hsp70B
3.4 Effect of transposition into Hsp70 on fecundity
All mated females produced progeny, and were more fecund 7–9 days after eclosion than before (Fig. 6). Strains differed in fecundity in the absence of heat shock, so each strain’s fecundity after heat shock has been compared with its own pre-heat-shock level. Heat shock delivered 3 days after eclosion increased fecundity relative to controls in 3 of 4 of the lines with P element insertions in Hsp70A. In the last, the increase was relatively large but not statistically significant (P = 0.09). By contrast, in the US-4 line, the control line in which the P element insertion was outside Hsp70A, heat shock initially had no effect on fecundity but subsequently (7–9 days after eclosion) decreased fecundity. In the same time period, the lines with P element insertions in Hsp70 either increased fecundity or showed no change for heat-shocked females.
P element insertions in Hsp70A increase fecundity after heat shock. Heat-shock was delivered to experimental females 3 days after eclosion, and progeny counted for three successive 3-day periods thereafter (shaded bars, mean + SE of 10–12 females). Open bars indicate progeny of non-heat-shocked controls. In the heat shock 3 days after, fecundity was significantly greater (*P < 0.05, Mann–Whitney U test) than in controls for three of four lines with P element insertions in Hsp70A (P < 0.09 in the final line). In the US-4 line, a control in which the P element insertion was outside Hsp70A, fecundity was initially unchanged after heat shock but decreased in the ensuing 3-day period
4 Discussion
In natural populations of Drosophila, transposable elements commonly interrupt the proximal promoter region of heat-shock genes (Walser et al. 2006). In the Hsp70 genes, P elements have inserted in at least 20 different sites (Fig. 3, bottom; see also Walser et al. 2006). In Hsp22 are 9 such sites; 4 are in Hsp83 and 11 are in Hsrω. One explanation for these sites is that they are permissive of transposable element insertion due to their nucleotide sequence, chromatin structure, and lack of protection by nucleosomes and bound proteins (Walser et al. 2006, Shilova et al. 2006). Indeed, the naturally occurring insertion sites often coincide with nucleosome-free and DNAse-hypersensitive regions (Walser et al. 2006, Shilova et al. 2006). In addition, in the laboratory, P elements tend to insert at these same sites (Walser et al. 2006, Shilova et al. 2006). A second, nonexclusive explanation is that insertions at these sites have beneficial phenotypes and thus are preserved by natural selection, whereas deleterious insertions are rapidly purged (Walser et al. 2006). As suggested in the Introduction, the numerous insertions may thus represent a palette of naturally occurring variation on which natural selection can act to fine-tune Hsp gene expression.
Deducing the phenotypic consequences of naturally occurring P element insertions is challenging because extensive additional genetic variation segregates in the background of these populations. While we have compared such populations nonetheless, the present study represents our exploitation of a set of lines differing in P element insertion site but with otherwise similar but nonidentical genetic backgrounds. We thus view these lines as a proxy for naturally occurring variation. An alternative approach would be to compare variants against a homogeneous genetic background as have, for example, Gong and Golic (2006), but with some cost to the relevance of the findings to natural populations (Feder and Krebs 1997).
Expectedly and as previously shown (Lerman et al. 2003; Lerman and Feder 2005), P element insertions in the proximal promoter of Hsp70 genes typically reduce gene expression. This reduction, moreover, is evident throughout the time course of gene expression after heat shock (Fig. 4). Interestingly, the magnitude of the reduction is inversely related to the distance between the corresponding P element insertion and the transcription start site, ranging from 3 to 56% of levels in the control (US-4) strain. The greater impact of the more proximal insertions could have at least two explanations. First, as they occur in or near the TATA box, they may directly interfere with the orderly engagement of the polymerase machinery with its normal binding site. Second, as explained (see Introduction), the interaction of the polymerase machinery with transactivation domains of bound HSF trimers is normally essential for the expression of these genes. Potentially, the interposition of P elements between these interacting protein complexes may interfere with their interaction. If so, P elements near the transcription start site may interfere with all bound HSFs, whereas more distal P elements may interfere with only some.
The inducible thermotolerance phenotypes of the P element insertions, by contrast, seem related mainly to their presence/absence rather than their specific location [although the strain with the insertion at -28 exhibited both the lowest Hsp70 mRNA level (present study) and Hsp70 protein level (Shilova et al. 2006) of any strain]. All insertion strains tested had lower thermotolerance than the US-4 control strain (Fig. 3 top, replotted from Shilova et al. 2006), but differed little among one another. Strains differing in Hsp70 copy number (rather than in regulatory polymorphisms) however, in general, vary in basal and/or inducible tolerance in proportion to Hsp70 copy number (Solomon et al. 1991; Gong and Golic 2006; Krebs and Feder 1998; Feder et al. 1996; Welte et al. 1993), although some strains depart from this generalization under specific experimental conditions (Gong and Golic 2006; Welte et al. 1993).
Although P element insertions reduce inducible thermotolerance (Fig. 3), they can be beneficial for reproduction. In the control (US-4) strain, heat shock delivered 3 days after eclosion reduced average fecundity, significantly so 7–9 days after eclosion (Fig. 6). By contrast, in the 4 Hsp70 P element insertion lines examined, an identical regime never decreased average fecundity, but increased average fecundity in 11 of 12 comparisons (significantly in 4 comparisons). Silbermann and Tatar (2000) also documented that heat shock reduces fecundity, and further that the reduction stems from decreased hatching success rather than increased post-hatching mortality. Interestingly, Silberman and Tatar (2000) compared strains in which P-element transformation was used to increase (rather than decrease) Hsp70 expression. In their work, as in the present study, heat shock reduced fecundity more in lines with high Hsp70 expression than in lines with lower Hsp70 expression. Similarly, Lerman et al. (2003) showed that lines with naturally occuring P elements in the proximal promoter region of Hsp70Ba can have greater fecundity than wild-type lines. These findings are consistent with a general finding that reduction of Hsp70 expression is often beneficial except when acute thermal stress necessitates it for survival (see Introduction). Hsp70-null flies, by contrast, undergo a decrease in fecundity (Gong and Golic 2006).
Two outcomes of evolution in Drosophila resonate with this general finding. First, Drosophila species have repeatedly undergone duplication of the Hsp70 genes themselves (Bettencourt and Feder 2001; Evgen’ev et al. 2004). Second, in Drosophila species Hsp gene expression has incorporated features that effectively suppress gene expression, some uniformly (as with insertion and retention of transposable elements) and some active primarily in the absence of heat shock (Lindquist 1993; Lakhotia and Prasanth 2002). The result, at least for Hsp70 proper, is that the protein can be undetectable in the absence of heat shock (Velazquez et al. 1983) and sometimes even after heat shock (Wang and Lindquist 1998; although at least mRNA is detectable during development, particularly in a subset of late spermatogonial cells; Lakhotia and Prasanth 2002). With respect to Hsp70 gene duplication, the multiple copies provide both redundancy and an opportunity to evolve distinctive copy-specific gene expression. Indeed, Lakhotia and Prasanth (2002) have demonstrated differences in the spatial and temporal expression of the 2 Hsp70A and 3–4 Hsp70B copies, and their tissue specificity. They and Hochstrasser (1987) have also reported variation in the size of chromosome puffs associated with the Hsp70A and Hsp70B gene clusters. Previously, however, quantitation of the relative expression of these clusters has been impossible because their protein coding sequence is identical. Here we have used quantitative RT-PCR with Hsp70A- or Hsp70B-specific primers to characterize the mRNA expression of the Hsp70A and Hsp70B clusters independently. Surprisingly, expression of the Hsp70A cluster, even with transposable elements present in the proximal promoter of one or both genes, appears quantitatively more significant than that of the Hsp70B cluster. The basis for this difference, presumably due to cluster-specific variation in proximal promoter sequence, awaits elaboration.
References
Amin J, Fernandez M, Ananthan J, Lis JT, Voellmy R (1994) Cooperative binding of heat shock transcription factor to the hsp70 promoter in vivo and in vitro. J Biol Chem 269:4804–4811
Bettencourt BR, Feder ME (2001) Hsp70 duplication in the Drosophila melanogaster species group: how and when did two become five. Mol Biol Evol 18:1272–1282
Bettencourt BR, Kim I, Hoffmann AA, Feder ME (2002) Response to natural and laboratory selection at the Drosophila hsp70 genes. Evolution 56:1796–1801
Boehm AK, Saunders A, Werner J, Lis JT (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23:7628–7637
Cartwright IL, Elgin SCR (1986) Nucleosomal instability and induction of new upstream protein-dna associations accompany activation of 4 small heat-shock protein genes in Drosophila melanogaster. Mol Cell Biol 6:779–791
Evgen'ev MB, Zatsepina OG, Garbuz D, Lerman DN, Velikodvorskaya V, Zelentsova E, Feder ME (2004) Evolution and arrangement of the hsp70 gene cluster in two closely related species of the virilis group of Drosophila. Chromosoma 11:3223–232
Feder ME, Cartaño NV, Milos L, Krebs RA, Lindquist SL (1996) Effect of engineering hsp70 copy number on Hsp70 expression and tolerance of ecologically relevant heat shock in larvae and pupae of Drosophila melanogaster. J Exp Biol 199:1837–1844
Feder ME, Krebs RA (1997) Ecological and evolutionary physiology of heat-shock proteins and the stress response in Drosophila: complementary insights from genetic engineering and natural variation. In: Bijlsma R, Loeschcke VS (eds) Stress, adaptation, and evolution. Birkhäuser Verlag, Basel, pp 155–173
Gong WJ, Golic KG (2006) Loss of hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics 172:275–286
Hochstrasser M (1987) Chromosome structure in four wild-type polytene tissues of Drosophila melanogaster: The 87A and 87C heat shock loci are induced unequally in the midgut in a manner dependent on growth temperature. Chromosoma 95:197–208
Kazazian HH (2004) Mobile elements: drivers of genome evolution. Science 303:1626–1632
Krebs RA, Feder ME (1997) Deleterious consequences of Hsp70 overexpression in Drosophila melanogaster larvae. Cell Stress Chaperones 2:60–71
Krebs RA, Feder ME (1998) Hsp70 and larval thermotolerance in Drosophila melanogaster: how much is enough and when is more too much. J Insect Physiol 44:1091–1101
Lakhotia SC, Prasanth KV (2002) Tissue- and development-specific induction and turnover of hsp70 transcripts from loci 87A and 87C after heat shock and during recovery in Drosophila melanogaster. J Exp Biol 205:345–358
Lerman DN, Feder ME (2005) Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol Biol Evol 22:776–783
Lerman DN, Michalak P, Helin AB, Bettencourt BR, Feder ME (2003) Modification of heat-shock gene expression in Drosophila melanogaster populations via transposable elements. Mol Biol Evol 20:135–144
Lindquist S (1993) Autoregulation of the heat-shock response. In: Ilan JT (ed) Translational regulation of gene expression 2, Vol. 2. Plenum Press, New York, pp 279–320
Lu Q, Wallrath LL, Granok H, Elgin SCR (1993) (CT)n.(GA)n repeats and heat-shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila Hsp26 gene. Mol Cell Biol 13:2802–2814
Maside X, Bartolome C, Charlesworth B (2002) S-element insertions are associated with the evolution of the hsp70 genes in Drosophila melanogaster. Curr Biol 12:1686–1691
Michalak P, Minkov I, Helin A, Lerman DN, Bettencourt BR, Feder ME, Korol AB, Nevo E (2001) Genetic evidence for adaptation-driven incipient speciation of Drosophila melanogaster along a microclimatic contrast in “Evolution Canyon,” Israel. Proc Natl Acad Sci U S A 98:13195–13200
Quivy JP, Becker PB (1996) The architecture of the heat-inducible Drosophila hsp27 promoter in nuclei. J Mol Biol 256:249–263
Shilova VY, Garbuz DG, Myasyankina EN, Chen B, Evgen’ev MB, Feder ME, Zatsepina OG (2006) Remarkable site specificity of local transposition into the Hsp70 promoter of Drosophila melanogaster. Genetics 173:809–820
Silbermann R, Tatar M (2000) Reproductive costs of heat shock protein in transgenic Drosophila melanogaster Evolution 54:2038–2045
Solomon JM, Rossi JM, Golic K, McGarry T, Lindquist S (1991) Changes in Hsp70 alter thermotolerance and heat-shock regulation in Drosophila New Biologist 3:1106–1120
Thomas GH, Elgin SC (1988) Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J 72:191–201
Timakov B, Liu X, Turgut I, Zhang P (2002) Timing and targeting of P-element local transposition in the male germline cells of Drosophila melanogaster. Genetics 160:1011–1022
Velazquez JM, Sonoda S, Bugaisky G, Lindquist S (1983) Is the major Drosophila heat shock protein present in cells that have not been heat shocked. J Cell Biol 96:286–290
Volff JN, 2006 Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28:913–922
Walser JC, Chen B, Feder ME (2006) Heat-shock promoters: Targets for evolution by P transposable elements in Drosophila. PLoS Genetics 2:1541–1555
Wang Z, Lindquist S (1998) Developmentally regulated nuclear transport of transcription factors in Drosophila embryos enable the heat shock response. Development 125:4841–4850
Weber JA, Taxman DJ, Lu Q, Gilmour DS (1997) Molecular architecture of the hsp70 promoter after deletion of the TATA box or the upstream regulation region. Mol Cell Biol 173:799–808
Welte MA, Tetrault JM, Dellavalle RP, Lindquist SL (1993) A new method for manipulating transgenes: engineering heat tolerance in a complex, multicellular organism. Curr Biol 3:842–853
Yao J, Munson KM, Webb WW, Lis JT (2006) Dynamics of heat shock factor association with native gene loci in living cells. Nature 442:1050–1053
Acknowledgement
The work was supported by Russian Academy Grant for Molecular and Cellular Biology to M.B.E., Russian Grants for Basic Science 06-04-48993-a and 06-04-48854-a, and National Science Foundation grant IBN03-16627. Bing Chen was supported by the China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, B., Shilova, V.Y., Zatsepina, O.G. et al. Location of P element insertions in the proximal promoter region of Hsp70A is consequential for gene expression and correlated with fecundity in Drosophila melanogaster . Cell Stress and Chaperones 13, 11–17 (2008). https://doi.org/10.1007/s12192-007-0002-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-007-0002-4