Abstract
Paraoxonase (PON) constitutes a family of calcium-dependent mammalian enzymes comprising of PON1, PON2, and PON3. PON family shares ~60% sequence homology. These enzymes exhibit multiple activities like paraoxonase, arylesterase, and lactonase in a substrate dependent manner. Decreased PON activity has been reported in diseases like cardiovascular disease, atherosclerosis, and diabetes. Even though, PON2 is the oldest member of the family, PON1 is the only member studied in silico. In this study, the structure of PON2 was modeled using MODELLER 9v7 and its interactions with relevant ligands and it's physiological substrate homocysteine thiolactone was performed using AutoDock 4.0. The results reveal that PON1 and PON2 share common ligand binding patterns for arylesterase and lactonase activity, whereas in case of paraoxon binding, the residues involved in the interactions were different. Interestingly, the substrate HCTL was found to have the lowest free energy of binding (ΔG) and highest affinity for PON2 than PON1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paraoxonase-1 (PON1; EC 3.1.8.1) is a calcium-dependent esterase synthesized in the liver and contained in plasma high-density lipoproteins (HDL) [1, 2]. HDL-associated proteins have been now understood to have diverse biological function in the pathogenesis of atherogenesis and inflammatory processes especially in cardiovascular disease [3]. PON1 has been found to have essential functions of ArylEsterase, organophosphatase, and lactonase activity depending on the substrate they act upon [4]. PON1 is an essential antioxidant enzyme, which possess peroxidase-like activity and also hydrolyzes platelet-activating factor, bioactive phospholipids which are involved in lipid peroxidation and vascular disease development [5]. Apart from preserving the function of HDL, PON1 has been reported to beneficially influence atherogenesis via inhibition of low-density lipoprotein (LDL) oxidation [6]. PON constitutes a family of mammalian enzymes with three members including PON1, PON2, and PON3. The members of the PON family share ~60% sequence identity. This PON enzyme has been associated with various systemic diseases like atherosclerosis [7], Diabetes [8] as well as in tissue-specific pathologies such as in ocular disease like retinal venous occlusion [9], age-related macular degeneration [10]. Amongst the PON family, PON1 is the best-studied member [11] while PON2, the oldest member of the family, has not been crystallized so far [12]. PON2 is intracellularly located and has the highest lactonase activity. The native enzyme activity of PON was recently found to be as a lactonase [12, 13]. In contrast to PON1 and PON3, PON2 is not present in serum, and it has minimal arylesterase and paraoxonase activity.
In this context, modeling and docking techniques could add insight towards structure-based analysis, pertaining to molecular interaction studies. Hence, the present study aims at predicting the three-dimensional (3D) structure of PON2 by employing in silico modeling techniques and also to characterize its interactions with ligands of biological importance.
Materials and methods
Retrieval of sequences
Human PON2 protein sequence (Swiss-Prot id Q15165) [14] was obtained at the UniportKB (http://www.uniprot.org/). Template for (human PON2) modeling the 3-D structure was identified by BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [15] search against Protein Data Bank (PDB) with default search parameters. PON 1 (1V04) [16] was selected as template to model PON2. The template structure was downloaded from PDB (http://www.pdb.org/pdb/home/home.do) [17].
Homology-modeled 3D structure generation and analysis
Since there are no three-dimensional structural data of the Human PON2, 3D structure of PON2 has been predicted using MODELLER9v7 [18].This software implements homology modeling of proteins by satisfying spatial restraints. ClustalW was used to align the target and template sequences and the resultant alignment was stored as PIR format [19].The alignment and the template atom files were given as input to MODELLER 9v7, to generate the 3D structure of PON2. Since PON family has two Ca 2+ ions [20] which is the characteristic feature of the family, the modeling process was customized to accommodate two Ca 2+ ions in the predicted structure, as present in template. The scripts “align-ligand.py” and “model-ligand.py” was used to generate ten rough 3D models. Modeller automatically derives restraints from known related structures. The restraints include distances, angles, dihedral angles, pairs of dihedral angles, and some other spatial restraints. Bond and angle values are taken from CHARMM-22 force field [21].
Model evaluation
The backbone conformation of the ten models was inspected using the Phi/Psi Ramachandran plot given by PROCHECK server (http://nihserver.mbi.ucla.edu/SAVS/) [22].The results indicate that two models out of ten generated models were perfectly fit with no residues in the disallowed region of Ramachandran plot. Furthermore, the prediction quality was assessed using Protein Quality Predictor (ProQ) based on LGscore [23].
Docking
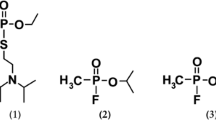
The set of biologically relevant ligand molecules studied in this study include homocysteine thiolactone [Pubchem, 107712], γ-thiobutyro lactone[Pubchem, 13852], Δ-valero lactone[Pubchem, 10953], benzyl acetate[Pubchem, 8785], 2-naphthyl acetate[Pubchem, 73709], phenyl acetate [Pubchem, 31229], and paraoxon[Pubchem, 9395] .The structure of these ligand molecules were retrieved from NCBI-Pubchem Compound database [24] (Fig. 1).
The geometry of the ligands were optimized via geometry optimization protocol, (Broyden-Fletcher-Golfarb-Shanno line search method set to 1,000 steps) using ArgusLab [25], a popular free suite of molecular modeling and simulation solutions for drug discovery. Each of the minimization was carried out using Universal Force Field (UFF) [25] (Table 1).
Docking calculations for the selected compounds with PON2 were carried out using AutoDock 4.0 suite, a comprehensive software for performing automated docking of ligands to their macromolecular receptors [26]. In this docking simulation, we used semi-flexible docking protocols [27], in which the target protein PON2 was kept rigid and the ligands being docked were kept flexible in order to explore an arbitrary number of torsional degrees of freedom. Graphical User Interface program “AutoDock Tools” was used to prepare, run, and analyze the docking simulations. Kollman United atom charges [28] and polar hydrogens were added into the receptor PDB file for the preparation of protein in docking simulation. Gasteiger charges were also assigned prior to docking. The rigid roots for each ligand were defined automatically. The amide bonds were made non-rotatable and the peptide backbone bonds were made rotatable. The modified ligand molecules were saved as PDBQT. The grid parameter files are to be set before running a docking calculation, as AutoDock 4.0 requires pre-calculated grid maps for each atom type present in the ligand being docked. The grid which is set must cover the area of interest in the protein. Since, the active binding site is unknown; the grid box was set to cover the entire protein. The grid box size in x-, y-, and z-axes was set to 126 × 126 × 126 Å and was kept constant for all the ligands. The spacing between the grid points was 0.375 Å. Autogrid 4.0 program, provided along with AutoDock 4.0 was used to produce grid maps. To search for the best conformers, Lamarckian Genetic Algorithm was chosen. A maximum of ten conformers were considered for each compound and a maximum of 2,500,000 energy evaluations, maximum number of generations set to 27,000, maximum number of top individual that automatically survived was set to 1, rate of mutation was set to 0.02 and the rate of crossover was set to 0.8. Finally, the protein-ligand complexes were analyzed using DS Visualizer [29] and PyMOL visualization tool [30].
Results
Human PON2 shares 81% sequence similarity to recombinant variant PON1 (1V04), this close homology indicates the fold equivalence of the 3D structures. Hence, the homology model predicted in this study is highly plausible (Fig. 2).
The best refined model generated had a ground state energy value of −40,123.82 Kcal/mol. The structural alignment of Human PON2 model to the template using Combinatorial Extension of Polypeptides [31] exhibited 0.4 Å of RMSD of the backbone superimposition. This confirms that the folds shared are highly similar. The backbone conformation of the model generated was inspected using Phi/Psi Ramachandran plot obtained in PROCHECK server (Fig. 3).
Eighty- nine percent of the residues lie in the most favorable regions of the Ramachandran plot (100% in allowed region; Table 2). Moreover, the ProQ prediction for the model also indicates significant predictive accuracy, wherein, the LGscore [32] obtained was 5.486 (LGscore range for excellent model > 5). Docking simulation of ten runs of GA-LA was performed for a set of seven minimized ligands into PON2. The best docked conformation of each ligand with PON2 was determined as the one having the lowest binding free energy among the ten different poses generated. The binding free energy and binding site residues of PON2 which are involved in hydrogen bond and hydrophobic interactions with the ligands used for the study are given in Table 3.
Discussion
Homology modeling of PON2
Owing to the high degree of similarity between recombinant variant PON 1 and human PON2, as expected, after homology modeling, the folds of its ground state model remained very similar to that of its template with an RMSD of 0.4 Å computed over the backbone atomic coordinates. Superposition of the 3D structures of the two isoforms reveals non-conserved residues at the N-terminal region. The analysis of human PON2 model revealed that, the six-bladed β-propeller scaffolds with each blade containing four strands and the Velcro closure of N- and C-termini typical of PON1, was conserved in PON2 also. Two calcium ions, 7.3 Å apart, are seen in the central tunnel of the propeller, one at the top interacting with Asp 168, Asp 54, and Ile 116 residues of PON2 and one in central section interacting with Glu 53, Asp 268, Asn 269, Asn 167, and Asn 223 residues of PON2.
The hydrogen bond donors, acceptors, bond length, van der Waals interaction residues, binding free energy, and docking energy of each ligand with PON2 is given in Table 3
Docking Homocysteine thiolactone into PON2
After docking homocysteine thiolactone (HCTL) into PON2, hydrogen bond interactions between hydrogen atom of the hydroxyl group of Thr170, oxygen atom of carboxyl group of Ile169, oxygen atom of carboxyl group of Ile225, and oxygen atom of carboxyl group of Asp168 with oxygen and hydrogen atoms of HCTL were observed. The residues involved in van der Waals interaction were Glu53, Asp54, His114, Ile116, Ser117, Asn167, Asp168, Ile169, Ala171, Asn223, Asn226, Asp268, Asn269, and Leu270 and Ca2+ ion was found to be involved actively. The binding free energy and docking energy of the complex was observed to be −6.63 and −7.12 Kcal/mol, respectively which was found to be the lowest amongst all the ligands used in the study (Fig. 4a).
Docking γ-thiobutyro lactone into PON2
Docking simulation of γ-thiobutyro lactone (GTBL) into PON2 resulted in the formation of single hydrogen bond with the bond distance of 2.004 Å and it was observed that hydrogen atom of hydroxyl group of Thr170 acts as hydrogen bond donor to interact with oxygen atom of GTBL. The residues taking part in van der Waals interactions are Glu53, Asp54, His114, Ile116, Ser117, Thr118, Asn167, Asp168, Ile169, Ala171, Asn223, Asn226, Asp268, Asn269, and Leu270. Also, a Ca2+ ion was found to be actively involved in the interaction. The binding free energy and docking energy of the complex was observed to be −3.69 Kcal/mol each (Fig. 4b).
Docking Δ-valero lactone into PON2
Upon assessing position and orientation of PON2–Δ-valero lactone (DVL)-docked complex, hydrogen atom of amine group of Ala171 interacts with an oxygen atom of DVL and hydrogen atom of hydroxyl group of Thr170 of PON2 interacts with oxygen atom of DVL. The van der Waals interactions between PON2 and DVL were also observed in the residues Glu53, Asp54, His114, Ile116, Ser117, Thr118, Asn167, Asp168, Ile169, Asn223, Asn226, Asp268, Asn269, and Leu270and a Ca2+ ion was found to be actively involved in the interaction. Both binding free energy and docking energy of the complex was observed to be −4.06 Kcal/mol (Fig. 4c).
Docking phenyl acetate into PON2
Phenyl acetate (PA), arylester substrate for PON2, is hydrolyzed by PON2. The binding modes of PA with PON2 were analyzed through docking studies. On examining the position and orientation of PA in PON2 predicted by our docking procedure, it was observed that hydrogen atom of the amine group of Thr118 and hydrogen atom of Ala171 acts as hydrogen bond donors to make hydrogen bond interaction with oxygen atoms of PA with bond length of 2.062 and 1.947 Å, respectively. Also, the oxygen atom of Thr118 formed a covalent bond with oxygen atom of PA. In addition, the residues Glu53, Asp54, His114, Ile116, Ser117, Asn167, Asp 168, Ile169, Thr170, Asn223, Ile225, Asn226, Asp268, Asn269, and Leu270 were involved in van der Waals interactions. The Ca2+ ion at the top of the central tunnel has greater accessibility to the ligand binding site for making van der Waals contacts into PA. The binding free energy and docking energy calculated by AutoDock for PA was −4.73 and −5.38 KCal/mol, respectively (Fig. 4d).
Docking benzyl acetate into PON2
Docking simulation of benzyl acetate (BA) into PON2 resulted in the formation of two hydrogen bonds and it was observed that hydrogen atom in the hydroxyl group of Thr170 and hydrogen atom of the amine group of Ala171 acts as hydrogen bond donor to interact with oxygen atom of BA. The amino acid residues Glu53, Asp54, Ile55, Asp56, His114, Ile116, Ser117, Thr118, Asn167, Asp 168, Ile169, Thr170, Asn223, Ile225, Asn226, Asp268, Asn269, and Leu270 were involved in van der Waals interaction with active participation of Ca2+ ion. The binding free energy and docking energy of the complex was observed to be −5.21 and −6.16 Kcal/mol, respectively (Fig. 4e).
Docking 2- naphthyl acetate into PON2
Docking of 2-naphthylacetate into PON2 resulted in the formation of two hydrogen bonds and it was observed that hydrogen atom of amine group of Ile57 and hydrogen atom of amine group of Ile272 were the hydrogen bond donors to oxygen atoms of 2- naphthyl acetate (2-NA) and residues involved in van der Waals interaction were Glu53, Asp54, Ile55, Asp56, His114, Ile116, Ser117, Thr118, Asn167, Asp 168, Ile169, Thr170, Asn223, Ile225, Asn226, Asp268, Asn269, Leu270, Ser271, and Ser334 and Ca2+ ion was found to be involved actively. The binding free energy and docking energy of the complex was observed to be −6.34 and −7.08 Kcal/mol, respectively (Fig. 4f).
Docking paraoxon into PON2
The binding of paraoxon into PON2 resulted in the formation of hydrogen bonds and it was observed that hydrogen atom of amine group of Lys46 acts as hydrogen bond donor and oxygen atom of Pro95 acts as hydrogen bond acceptor. Van der Waals interaction with paraoxonase (PAR) was formed by His43, Leu44, Ile45, Met88, Glu93, Lys94, Arg96, Leu341, and Tyr351 residues of PON2. The binding mode of PAR to PON2 was different from the other substrates. PAR exhibited binding, in a region outlying from the substrate binding site which was in coherence with the already documented experimental report wherein, PON2 was also shown to exhibit negligible paraoxonase activity [12] (Fig. 4g).
Comparing PON 1 and PON2
The residues Glu 53, His 114, Asn168, Asn 223, Asp 270, and Asn269 exhibit hydrogen bonding interaction for both lactonase and arylesterase activity in PON1 [33], including the natural substrate HCTL, whereas, in PON2 the same residues Glu 53, His 115, Asn167, Asn 224, Asp 269, and Asn270 were found to have hydrophobic interactions for the same activity. The paraoxon binding site was found to overlap with the lactonase and arylesterase binding sites of PON1, but similar interaction was not observed in PON2 and it was found to bind to a region away from the central Ca 2+ ion, at residues His43, Leu44, Ile45, Lys46, Met88, Glu93, Lys94, Pro95, Arg96, Leu341, and Tyr351 forming the hydrogen bonding and hydrophobic interactions. It was observed that the PON2 had lowest binding energy and highest affinity for the physiological substrate HCTL (−6.63 Kcal/mol [34]) than PON1 which showed a binding energy of (−5.72 Kcal/mol).
The docked structures of PON2 exhibit relatively a large number of amino acid residues involved in ligand interactions and the involvement of one of the Ca 2+ ions were observed in each PON2–ligand interaction except paraoxon. This shows that the calcium present in the central section of the central tunnel has catalytic activity. As shown in the Table 3, it is obvious that out of the seven ligands tested, six ligands share the same binding modes in PON2 enzyme; whereas, paraoxon exhibits a completely different binding mode. There are several reports which show homocysteine (Hcys) to be a risk factor for cardiovascular disease and atherosclerosis [35], and recent reports have shown that the concentration of Hcys is increased in the serum and vitreous of diabetic patients [34, 36]. In all these conditions, the level of PON is found to be lowered [37, 38]. In addition to the known ligands, this study also gives the molecular interaction for the physiological substrate namely, HCTL which is formed in all cell types when there is excess homocysteine, as a result of error-editing met-tRNA synthetase [39].
In summary, this is the first molecular modeling study that proposes the structure of the PON2 isoform. Unlike PON1, which is mainly associated with HDL, PON2 is not found in the circulation and acts as an intracellular antioxidant [40, 41] and may provide an innate antioxidant activity in most of the cells, independent of secretory PON. PON2 is ubiquitously present in all tissues and is also reportedly present intracellularly in the three major vascular cell types namely, cultured human umbilical vein endothelial cells, smooth muscle vascular cells, and aortic adventitial fibroblasts with the major function of reducing the reactive oxygen species-mediated endothelial cell dysfunction [42]. PON2-deficient mice exhibit elevated tissue levels of lipid hydroperoxides and increased migration of macrophage into the artery wall, leading to formation of atherosclerotic lesion as compared to their wild-type controls [43]. It is suggested that PON2 protects macrophages from foam cell formation and thus contributes to the prevention of atherogenesis [44]. Importantly, a decreased PON2 expression has been observed in hypercholesterolemic patients [45] and during progression of atherogenesis [46].
Conclusion
In this work, molecular modeling and docking studies were performed to explore possible binding modes of known substrates HCTL, GTBL, DVL, PA, BA, 2-NA, and PAR into PON2 enzyme. MODELLER 9v7 software was used to model the enzyme PON2 and AutoDock 4.0 software was used to dock the ligands into PON2. The binding pattern of the amino acid residues participating in the interaction were identified and validated by site of reaction and calculated interaction energies of the docked complexes. It was found that Thr170 made an important contribution in terms of hydrogen bond formation for lactonase activity which needs to be further validated by wet lab studies. The binding energy for HCTL, which is the physiological substrate for this enzyme, is −6.63 Kcal/mol and therefore has the maximal affinity when compared to other lactones (GTBL and DVL) and ester substrates (PA, BA, and 2- NA) used for this study. This is indicative of elevated lactonase activity of PON2 when compared to its esterase activity. Since, this study reveals the higher lactonase activity, characteristic of PON2. Thereby, factors augmenting PON2 activity will be of significant therapeutic value.
References
Aviram M, Rosenblat M, Bisgaier CL, Newton RS, Primo-Parmo SL, La Du BN. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J Clin Invest. 1998;101:1581–90.
Domagala TB, Lacinski M, Trzeciak WH, Mackness B, Mackness MI, Jakubowski H. The correlation of homocysteine-thiolactonase activity of the paraoxonase (PON1) protein with coronary heart disease status. Cell Mol Biol (Noisy-le-Grand, France). 2006;52:4–10.
Scanu AM, Edelstein C. HDL: bridging past and present with a look at the future. FASEB J. 2008;22:4044–54.
Durrington PN, Mackness B, Mackness MI. Paraoxonase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2001;21:473–80.
Yeung DT, Josse D, Nicholson JD, Khanal A, McAndrew CW, Bahnson BJ, et al. Structure/function analyses of human serum paraoxonase (HuPON1) mutants designed from a DFPase-like homology model. Biochim Biophys Acta. 2004;1702:67–77.
Mackness MI, Arrol S, Abbott C, Durrington PN. Protection of low-density lipoprotein against oxidative modification by high-density lipoprotein associated paraoxonase. Atherosclerosis. 1993;104:129–35.
Seo D, Goldschmidt-Clermont P. The paraoxonase gene family and atherosclerosis. Curr Atheroscler Rep. 2009;11:182–7.
Nowak M, Wielkoszynski T, Marek B, Kos-Kudla B, Swietochowska E, Sieminska L, et al. Antioxidant potential, paraoxonase 1, ceruloplasmin activity and C-reactive protein concentration in diabetic retinopathy. Clin Exp Med. 2009;10:185–92.
Angayarkanni N, Barathi S, Seethalakshmi T, Punitham R, Sivaramakrishna R, Suganeswari G, et al. Serum PON1 arylesterase activity in relation to hyperhomocysteinaemia and oxidative stress in young adult central retinal venous occlusion patients. Eye (London, England). 2008;22:969–74.
Ates O, Azizi S, Alp HH, Kiziltunc A, Beydemir S, Cinici E, et al. Decreased serum paraoxonase 1 activity and increased serum homocysteine and malondialdehyde levels in age-related macular degeneration. Tohoku J Exp Med. 2009;217:17–22.
Tavori H, Khatib S, Aviram M, Vaya J. Characterization of the PON1 active site using modeling simulation, in relation to PON1 lactonase activity. Bioorg Med Chem. 2008;16:7504–9.
Draganov DI, La Du BN. Pharmacogenetics of paraoxonases: a brief review. Naunyn-Schmiedebergs Arch Pharmacol. 2004;369:78–88.
Khersonsky O, Tawfik DS. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44:6371–82.
The Universal Protein Resource (UniProt) 2009. Nucleic Acids Research. 2009;37:D169–74.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004;11:412–9.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42.
Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–91.
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500.
Horke S, Witte I, Wilgenbus P, Altenhofer S, Kruger M, Li H, et al. Protective effect of paraoxonase-2 against endoplasmic reticulum stress-induced apoptosis is lost upon disturbance of calcium homoeostasis. Biochem J. 2008;416:395–405.
Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815.
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86.
Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–86.
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–33.
Thompson MA ArgusLab 4.0.1.
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–62.
Gowthaman U, Jayakanthan M, Sundar D. Molecular docking studies of dithionitrobenzoic acid and its related compounds to protein disulfide isomerase: computational screening of inhibitors to HIV-1 entry. BMC Bioinformatics. 2008;9 Suppl 12:S14.
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012.
Talley TT, Harel M, Hibbs RE, Radic Z, Tomizawa M, Casida JE, et al. Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore. Proc Natl Acad Sci USA. 2008;105:7606–11.
DeLano WL (2002) The PyMOL molecular graphics system. In ed^eds. Palo Alto, CA, USA: Delano Scientific.
Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–47.
Cristobal S, Zemla A, Fischer D, Rychlewski L, Elofsson A. A study of quality measures for protein threading models. BMC Bioinform. 2001;2:5.
Hu X, Jiang X, Lenz DE, Cerasoli DM, Wallqvist A. In silico analyses of substrate interactions with human serum paraoxonase 1. Proteins. 2009;75:486–98.
Aydemir O, Turkcuoglu P, Guler M, Celiker U, Ustundag B, Yilmaz T, et al. Plasma and vitreous homocysteine concentrations in patients with proliferative diabetic retinopathy. Retina (Philadelphia, Pa). 2008;28:741–3.
Kazemi MB, Eshraghian K, Omrani GR, Lankarani KB, Hosseini E. Homocysteine level and coronary artery disease. Angiology. 2006;57:9–14.
Coral K, Angayarkanni N, Gomathy N, Bharathselvi M, Pukhraj R, Rupak R. Homocysteine levels in the vitreous of proliferative diabetic retinopathy and rhegmatogenous retinal detachment: its modulating role on lysyl oxidase. Investig Ophthalmol Vis Sci. 2009;50:3607–12.
Ikeda Y, Suehiro T, Itahara T, Inui Y, Chikazawa H, Inoue M, et al. Human serum paraoxonase concentration predicts cardiovascular mortality in hemodialysis patients. Clin Nephrol. 2007;67:358–65.
Jayakumari N, Thejaseebai G. High prevalence of low serum paraoxonase-1 in subjects with coronary artery disease. J Clin Biochem Nutr. 2009;45:278–84.
Jakubowski H, Zhang L, Bardeguez A, Aviv A. Homocysteine thiolactone and protein homocysteinylation in human endothelial cells: implications for atherosclerosis. Circ Res. 2000;87:45–51.
Ng CJ, Wadleigh DJ, Gangopadhyay A, Hama S, Grijalva VR, Navab M, et al. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low density lipoprotein. J Biol Chem. 2001;276:44444–9.
Aviram M, Rosenblat M. Paraoxonases 1, 2, and 3, oxidative stress, and macrophage foam cell formation during atherosclerosis development. Free Radic Biol Med. 2004;37:1304–16.
Mackness B, Hunt R, Durrington PN, Mackness MI. Increased immunolocalization of paraoxonase, clusterin, and apolipoprotein A-I in the human artery wall with the progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 1997;17:1233–8.
Ng CJ, Bourquard N, Grijalva V, Hama S, Shih DM, Navab M, et al. Paraoxonase-2 deficiency aggravates atherosclerosis in mice despite lower apolipoprotein-B-containing lipoproteins: anti-atherogenic role for paraoxonase-2. J Biol Chem. 2006;281:29491–500.
Ng CJ, Hama SY, Bourquard N, Navab M, Reddy ST. Adenovirus mediated expression of human paraoxonase 2 protects against the development of atherosclerosis in apolipoprotein E-deficient mice. Mol Genet Metab. 2006;89:368–73.
Rosenblat M, Hayek T, Hussein K, Aviram M. Decreased macrophage paraoxonase 2 expression in patients with hypercholesterolemia is the result of their increased cellular cholesterol content: effect of atorvastatin therapy. Arterioscler Thromb Vasc Biol. 2004;24:175–80.
Fortunato G, Di Taranto MD, Bracale UM, Del Guercio L, Carbone F, Mazzaccara C, et al. Decreased paraoxonase-2 expression in human carotids during the progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:594–600.
Acknowledgements
We thank the Vision Research Foundation, Sankara Nethralaya, for providing us the Center for Bioinformatics facility.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Barathi, S., Charanya, M., Muthukumaran, S. et al. Comparative modeling of PON2 and analysis of its substrate binding interactions using computational methods. j ocul biol dis inform 3, 64–72 (2010). https://doi.org/10.1007/s12177-011-9057-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12177-011-9057-7