Abstract
A straightforward real-time polymerase chain reaction (PCR)-based assay was designed and evaluated for the detection of Salmonella spp. in food and water samples. This new assay is based on the specific detection of the bipA gene of Salmonella, which encodes a protein of the guanosine triphosphate (GTP)-binding elongation family that displays global modulating properties, by regulating a wide variety of downstream processes. The new method correctly identified all 48 Salmonella strains used in the inclusivity test, and did not detect all 30 non-Salmonella species tested. The method was evaluated by analyzing 120 diverse food and water samples enriched in buffered peptone water. The bipA-based real-time PCR assay showed 100% efficiency, sensitivity, and specificity compared to the invA-based method previously published, which was developed as a part of a European project for the standardization of PCR methods in food microbiology. The assay includes an independent internal amplification control (IAC) in each reaction to control false negative results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salmonellosis is a major public health concern causing millions of human cases worldwide every year and resulting in thousands of deaths. This food-borne disease is by the bacteria Salmonella, a rod-shaped, gram-negative non-spore-forming bacterium. This genus is a member of the family Enterobacteriaceae, and encompasses three species: Salmonella enterica, Salmonella subterranea, and Salmonella bongori. S. enterica includes six subspecies of clinical importance for humans, and over 2,500 known types, or serotypes. Environmental sources of Salmonellae include water, soil, insects, factory and kitchen surfaces, animal feces, raw meats, raw seafood, raw poultry, eggs, milk and dairy products, sauces and salad dressings, etc. (Tirado and Schmidt 2001).
Up to 5 days are required to detect Salmonella using traditional culture-based methods (Anonymous, ISO 6539 2000). The need for new, quick, and sensitive methods to detect Salmonella is, therefore, a major concern in food safety.
Many PCR-based assays have been described to date. Both conventional and modern real-time PCR protocols have been implemented, targeting phylogenetic genes such as the 16S ribosomal subunit (Lin and Tsen 1996). However, functional genes involved in virulence and infectivity are currently the markers of choice for most PCR procedures. The most widely used gene to date to detect Salmonella spp. is invA (invasion A) (Malorny et al. 2003a, 2004). The invA gene encodes for an essential component of the invasion-associated protein secretion apparatus, and is the first gene of the inv locus (Chiu and Ou 1996). This locus allows Salmonella spp. to enter epithelial cells and cause an infection (Galan et al. 1992). Other authors use genes like tyv, prt, viaB, flic-d, or flic-a, which are O, H, and Vi antigen genes (Hirose et al. 2002) to detect and identify Salmonella enterica serovars Typhi and Paratyphi. Finally, genes in the ttrRSBCA locus, which is required for tetrathionate respiration and located near the pathogenicity island 2 of Salmonella, are also used as a target to detect Salmonella in food by real-time PCR (Malorny et al. 2004).
Most of these genes, however, present either nonspecific amplifications or inclusivity problems such as the failure in the detection of the serovar Saint Paul of Salmonella by the invA-targeted PCR (Malorny et al. 2004; Cohen et al. 1996). Furthermore, virulence genes are normally subjected to strong variability, mainly caused by silent mutations in the third base of the codon (Friis et al. 2000; Yang et al. 2000). Considering the high specificity of probes used in real-time PCR, a single mismatch can result in a dramatic decrease in the efficiency of the reaction and might therefore give rise to a false negative result.
Regulatory genes are less subjected to spontaneous silent mutations than regular functional genes and an inverse correlation has been demonstrated between the rate of evolution of transcription factors and the number of genes they regulate (Rajewsky et al. 2002). Therefore, we sought to develop a PCR tool to detect Salmonella targeting one of these genes. The bipA (or typA) gene is a member of the “GTP-binding elongation” family of genes, and belongs to the category N. This gene exhibits global modulating properties and acts as an essential translation factor for the efficient expression of fis, thus regulating a wide variety of downstream processes, such as type III secretion and DNA metabolism (Owens et al. 2004). BipA was first revealed as a strongly upregulated protein when Salmonella enterica was exposed to the host defense protein BPI (Qi et al. 1995). Nevertheless, further studies showed that null Escherichia coli mutants of bipA (or typA) are pleiotropic, with deficiencies in crucial processes such as the expression of the K5 capsule system (Rowe et al. 2000), poor growth at low temperatures (Grant et al. 2003; Pfennig and Flower 2001), defects in flagella-mediated cell motility (Grant et al. 2003; Farris et al. 1998) and loss of resistance to some antimicrobial peptides (Qi et al. 1995; Barker et al. 2000). BipA binds to ribosomes at a site that concurs with that of elongation factor G, and has a GTPase activity that is sensitive to high guanosine diphosphate (GDP):guanosine triphosphate (GTP) ratios and stimulated by ribosomes programmed with mRNA and aminoacylated tRNAs.
Results obtained with this new bipA real-time PCR-based method have been compared to those obtained by the invA-based PCR method described by Hoorfar (1999) and Malorny et al. (2003a), which was developed as a part of a European project for the standardization of PCR methods in food microbiology. This new assay displays the requirements of a diagnostic PCR assay, and meets the requirements to become a standardized method for the quick identification of Salmonella spp.-contaminated water and food samples.
Material and Methods
Bacterial Reference Strains Used in this Study
A total of 48 Salmonella strains were used for inclusivity tests (Table 1). These strains were either obtained from the Spanish Type Culture Collection (CECT), or isolated by the Public Health Laboratory of Girona, and the Laboratory of Microbiology of the Hospital Universitari Dr. Josep Trueta of Girona. In addition, 30 different bacterial species belonging to all major phylogenetic lineages were used for exclusivity tests. Some of them were specifically selected because of the close phylogenetic relation to Salmonella spp. or because they grow in the same environments and conditions (Table 1).
DNA Purification
For inclusivity and exclusivity tests, bacterial strains were grown in nutritive medium as previously described (Miller 1972). Isolated colonies were resuspended in bidistilled sterile water and DNA was extracted and purified using the NucleoSpin® Tissue kit (Macherey-Nagel, Düren, Germany) as specified by the manufacturer. DNA from food and water samples was extracted from 1-ml aliquot of a pre-enriched culture in buffered peptone water using the same kit.
DNA concentration was determined by the PicoGreen™ method (Molecular Probes, Carlsbad, CA, USA), which compares fluorescence values with those of a calibration curve built from a dilution series of salmon sperm DNA (Sigma Chemicals, Madrid, España).
Genomic copy numbers were calculated based on the Salmonella enterica serotype Typhimurium genome size (4,857 kb) (McClelland et al. 2001), and considering that the average base pair weight is 652 g mol−1.
Primer Design
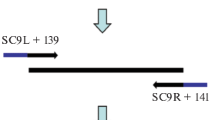
All sequences of the bipA gene from Salmonella spp. available in the databases were retrieved and aligned using ClustalX version 1.81 (Thompson et al. 1997). The consensus sequence was then used to design a set of primers and a probe for the quantitative determination of Salmonella spp. by real-time PCR using the software Primer Express™ v. 2.0 (Applied Biosystems, Foster City, CA). Both forward SAL1410f and reverse SAL1494r primers, and the probe SAL1441pr (Table 2) were evaluated with the NetPrimer software (PREMIER Biosoft International, Palo Alto, CA, USA) for the formation of primer-dimer structures and hairpins. The oligonucleotides for the amplification of the internal amplification control were manually designed based on the IAC sequence (Table 2).
The Salmonella and IAC probes were labeled at the 5′ end with 6-FAM and VIC, respectively. TAMRA was used as a quencher at the 3′ end in both cases.
Real-Time PCR Conditions
Real-Time PCR was carried out in 20-μl (total volume) reaction mixtures using an ABI Prism 7300 SDS RTi-PCR system (PE Applied Biosystems, Foster City, CA, USA). A positive reaction and a non-template reaction were included in all tests.
An artificial, chimeric 65-bp DNA fragment was designed and used as an internal amplification control (IAC) in all reactions (Table 2). This DNA fragment was synthesized by TIB MOLBIOL (Syntheselabor GmbH, Berlin, Germany) and used at an optimal concentration of 103 amplicon copies per reaction.
PCR conditions were 95°C for 10 min, then 50 cycles consisting of 95°C for 15 s and 60°C for 1 min. Reaction mixtures contained 1× Universal Master Mix (PE Applied Biosystems, Foster City, CA, USA), 1-μl DNA template, 300 nM of SAL1410f, 300 nM SAL1494r, 100 nM of SAL1441pr, 300 nM of IACf, 300 nM of IACr, 50 nM of IACpr, and 103 copies of IAC.
Fluorescence values in PCR reactions were analyzed using the ABI Prism 7300 System SDS software v1.2.3 (PE Applied Biosystems, Foster City, CA, USA). An automatic threshold setting of 0.2 was used for all samples.
Determination of the Linearity and Efficiency of the Reaction
The linearity and the efficiency of the PCR assay were determined using purified DNA from the S. enterica subsp. enterica LT2 CECT 878 strain within a concentration range of 101 to 106 genomic DNA copies per reaction. One microliter of each dilution was added to five replicas in PCR tubes and run in the presence of 103 copies of the IAC as described above. The linearity of the PCR reaction was assessed by plotting the obtained threshold cycle (Ct) against the logarithm of the number of genomes in the reaction. The efficiency of the bipA-based real-time PCR assay was calculated using the formula: Efficiency = [10(−1/slope)] − 1.
Food and Water Samples
A total of 120 samples including 15 fish samples (monkfish, hake, smoked salmon), 10 egg samples (fresh egg, potato omelet, pasteurized egg, boiled egg, refrigerated omelet), 26 meat samples (lamb, minced beef and pork, chicken and minced chicken, hamburger meat, duck meat, and sausages), 20 dairy products (milk, cream, powdered milk, sour cream, cheese, fromage frais, yogurt), 26 prepared food samples (tuna pies, salads, lasagna, puff pastry, muffins, black pepper), and 23 water samples (bottled, from water supply systems, from a well) were analyzed using both the invA-based PCR method (Friis et al. 2000; Rowe et al. 2000) and the bipA-based real-time PCR. The samples were chosen to find the highest number of naturally positive samples. All samples were enriched in BPW for 16–20 h at 36 ± 2°C, prior to being processed.
Results
Selectivity Test
Inclusivity and exclusivity tests were performed with a total of 48 Salmonella and 30 non-Salmonella strains (Table 1). All 48 tested strains of Salmonella were correctly identified, and all non-Salmonella strains presented a threshold cycle number (Ct) above 30, whereas the IAC threshold cycle number (Ct) was around 28–29 in all reactions.
Linearity and Efficiency of the Reaction
Genomic DNA of the strain S. enterica subsp. enterica LT2 CECT 878 was used as a template for linearity and efficiency tests The real-time PCR described here detected as little as 102 genomes of Salmonella per reaction in all cases, whereas in only two of the five replicas 101 genomes of Salmonella were detectable (Table 3). The linearity of the assay was calculated using the equation Ct = 38.268 – 3.3224 log (N of genomes) with a R 2 correlation value of 0.9993.
The efficiency of the bipA-based real-time PCR was 99.98%.
Detection Limit of the Assay
To calculate the detection limit of our real-time PCR method, enrichments of 22 food and water negative samples were inoculated with S. enterica subsp. enterica Serovar Typhimurium CECT 4594. Food samples were inoculated with 6 cfu/25 g, whereas bottled water samples were inoculated with 3 cfu l−1, and well or water supply system samples with 3 cfu/100 ml. The Salmonella suspension used for the inoculation was quantified by plate counting in nutritive agar plates.
The probability of detection of Salmonella in the samples was calculated as a percentage, resulting in a 100% in the case of water samples contaminated with 3 cfu, whereas in the case of food samples, the probability of detecting the Salmonella in a sample inoculated with 6 cfu/25 g was 95.45%.
Efficiency of the Assay Using Food and Water Samples
Sensitivity, specificity, and efficiency of the bipA-based real-time PCR assay were calculated after analyzing 97 diverse food samples and 23 water samples by both the invA-based (Malorny et al. 2004; Hoorfar 1999) PCR, and the real-time PCR method presented here (Table 4). To increase the number of positive samples to analyze, some food and water samples initially yielding a negative result for Salmonella with both PCR methods were spiked with 50 cfu of S. enterica subsp. enterica Serovar Typhimurium CECT 4594 per analytical portion (food 25 g, water 100 ml or 1 l) and then analyzed again.
Results were in agreement in all cases, which resulted in a sensitivity, specificity, and efficiency of the bipA-based real-time PCR assay of 100% for all types of food matrices and waters. Accordingly, neither false positives nor false negatives were obtained in the 120 samples analyzed (Table 4).
To avoid PCR inhibition, 1:10 and 1:100 dilutions were used.
Discussion
The new real-time PCR-based assay for the detection of Salmonella spp. in food and water samples through the detection of the bipA gene of Salmonella was designed and evaluated against the procedure of the European project FOOD-PCR based on the detection of the invA-gene (Friis et al. 2000; Rowe et al. 2000). As it happens with conventional microbiological analysis, this method requires previous enrichment incubation in buffered peptone water. After genomic DNA extraction using a column-based extraction kit, a bipA-targeted real-time PCR is performed in the presence of an internal amplification control. The described procedure detects Salmonella in food and water samples in less than 24 h, drastically reducing the time needed by the conventional culture-based methods. Most PCR-based methods for the detection of Salmonella used to date target either phylogenetic or functional genes. However, most of the assays used until now present either specificity or inclusivity problems (Malorny et al. 2004; Cohen et al. 1996). For instance, the S. enterica subsp. enterica serovar Saint Paul is not detected by the invA-based assay. As previously mentioned (Table 1), this strain was satisfactorily tested with our system. Moreover, critical variations in the target DNA, which can hamper the specificity and accuracy of the analysis, are produced by either the codon usage or silent mutations (Cohen et al. 1996). This means that, for example, in a 21-bp oligonucleotide up to seven positions are at risk of being nonspecific because of the natural genetic variability of bacterial populations, which can compromise the selectivity of the PCR assay. For some reason, the bipA (or typA) gene does not seem to show this variability, which makes it an ideal target for a PCR. Besides, another pitfall of some of the PCR-based methods reported so far is that they do not include an internal amplification control in the reaction.
The use of an internal amplification control (IAC) in the reactions helps to control false negatives resulting from PCR inhibition or reaction malfunction because of either spoiled reactives or a damaged thermocycler (Hoorfar et al. 2003). In these reactions, the IAC consists of an artificially constructed DNA sequence, which is detected by two primers and a probe labeled with the VIC reporter. The optimal IAC concentration was set as low as 103 copies per reaction. The IAC was amplified with Ct values of 28–29 in all the samples except those containing high amounts of genomic DNA of Salmonella, in which the IAC was usually undetectable or appeared at Ct values above 30 (Table 3) because of template competence.
Selectivity of the bipA-targeted primers and probe was first tested using the NCBI Blastn program (Altschul et al. 1997). An inclusivity/exclusivity test was then performed using 48 Salmonella and 30 non-Salmonella strains, revealing 100% selectivity of the primers and probe presented in this work. Besides, no false negatives or false positives were detected when comparing the results obtained in the analysis of 120 food and water samples with the results obtained by the PCR method described by the European Project Food-PCR (Malorny et al. 2003a, b Hoorfar 1999).
The real-time PCR presented here allows detection of as few as 102 genomes of Salmonella per reaction in the presence of an internal amplification control when using a quantified Salmonella suspension. Besides, the detection probability of 6 and 3 cfu in food and water sample rinses was 95.45% and 100%, respectively. Considering that the samples are subjected to an overnight BPW enrichment before DNA extraction, an initial contamination of one viable cell of Salmonella in a given sample would normally produce at least 105–107 cfu ml−1, assuming 16–20 h at doubling times of 0.45 to 1 h, which is detectable by the bipA-based real-time PCR. Typically, in real analyses, a BPW overnight incubation of a 10-cfu inoculum would produce a positive real-time PCR result at Ct values between 25 and 28.
The bipA-based real-time PCR assay presented in this work is linear and highly selective, specific, and efficient. It is also fast because it permits a sample for the presence of Salmonella to be analyzed in no more than 24 h. Therefore, this method might substitute the previous invA-based method for the rapid detection of Salmonella in food and water samples.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anonymous. ISO 6539 (2000) Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella spp
Barker HC, Kinsella N, Jaspe A, Friedrich T, O’Connor CD (2000) Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol Microbiol 35:1518–1529
Chiu CH, Ou JT (1996) Rapid identification of salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 34:2619–2622
Cohen HJ, Mechanda SM, Lin W (1996) PCR amplification of the fimA gene sequence of Salmonella typhimurium, a specific method for detection of salmonella spp. Appl Environ Microbiol 62:4303–4308
Farris M, Grant A, Richardson TB, O’Connor CD (1998) BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol Microbiol 28:265–279
Friis C, Jensen LJ, Ussery DW (2000) Visualization of pathogenicity regions in bacteria. Genetica 108:47–51
Galan JE, Ginocchio C, Costeas P (1992) Molecular and functional characterization of the salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol 174:4338–4349
Grant AJ, Farris M, Alefounder P, Williams PH, Woodward MJ, O’Connor CD (2003) Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol Microbiol 48:507–521
Hirose K, Itoh K, Nakajima H et al (2002) Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars typhi and paratyphi A. J Clin Microbiol 40:633–636
Hoorfar J (1999) EU seeking to validate and standardize PCR testing of food pathogens. ASM news 65,799. ASM News 65:799
Hoorfar J, Cook N, Malorny B et al (2003) Making internal amplification control mandatory for diagnostic PCR. J Clin Microbiol 41:5835
Lin CK, Tsen HY (1996) Use of two 16S DNA targeted oligonucleotides as PCR primers for the specific detection of salmonella in foods. J Appl Bacteriol 80:659–666
Malorny B, Hoorfar J, Bunge C, Helmuth R (2003a) Multicenter validation of the analytical accuracy of salmonella PCR: Towards an international standard. Appl Environ Microbiol 69:290–296
Malorny B, Tassios PT, Radstrom P, Cook N, Wagner M, Hoorfar J (2003b) Standardization of diagnostic PCR for the detection of foodborne pathogens. Int J Food Microbiol 83:39–48
Malorny B, Paccassoni E, Fach P, Bunge C, Martin A, Helmuth R (2004) Diagnostic real-time PCR for detection of salmonella in food. Appl Environ Microbiol 70:7046–7052
Mc Clelland M, Sanderson KE, Spieth J et al (2001) Complete genome sequence of salmonella enterica serovar typhimurium LT2. Nature 413:852–856
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Owens RM, Pritchard G, Skipp P et al (2004) A dedicated translation factor controls the synthesis of the global regulator fis. EMBO J 23:3375–3385
Pfennig PL, Flower AM (2001) BipA is required for growth of escherichia coi K12 at low temperature. Mol Genet Genomics 266:313–317
Qi SY, Li Y, Szyroki A, GilesI G, Moir A, O’Connor CD (1995) Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol Microbiol 17:523–531
Rajewsky N, Socci ND, Zapotocky M, Siggia ED (2002) The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res 12:298–308
Rowe S, Hodson N, Griffiths G, Roberts IS (2000) Regulation of the escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J Bacteriol 182:2741–2745
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tirado C, Schmidt K (2001) WHO surveillance programme for control of foodborne infections and intoxications: preliminary results and trends across greater Europe. J Infect 43:80–84
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Acknowledgments
Thanks are due to Jordi Batlle from the Laboratory of Microbiology of the Hospital Universitari Dr. Josep Trueta for providing some of the Salmonella strains used in this study. Laia Calvó is the recipient of a Beatriu de Pinós BP2005 fellowship from the Generalitat de Catalunya.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Calvó, L., Martínez-Planells, A., Pardos-Bosch, J. et al. A New Real-Time PCR Assay for the Specific Detection of Salmonella spp. Targeting the bipA Gene. Food Anal. Methods 1, 236–242 (2008). https://doi.org/10.1007/s12161-007-9008-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-007-9008-x