Abstract
We previously reported on a new yeast strain of Clavispora sp. NRRL Y-50464 that is capable of utilizing cellobiose as sole source of carbon and energy by producing sufficient native β-glucosidase enzyme activity without further enzyme supplementation for cellulosic ethanol production using simultaneous saccharification and fermentation. Eliminating the addition of external β-glucosidase reduces the cost of cellulosic ethanol production. In this study, we present results on the isolation and identification of a β-glucosidase protein from strain Y-50464. Using Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and blast search of the NCBInr database (National Center for Biotechnology Information nonredundant), the protein from Y-50464 was identified as a β-glucosidase (BGL1) with a molecular weight of 93.3 kDa. The BGL1 protein was purified through multiple chromatographic steps to a 26-fold purity (K m = 0.355 mM [pNPG]; K i = 15.2 mM [glucose]), which has a specific activity of 18.4 U/mg of protein with an optimal performance temperature at 45 °C and pH of 6.0. This protein appears to be intracellular although other forms of the enzyme may exist. The fast growth rate of Y-50464 and its capability to produce sufficient β-glucosidase activity for ethanol conversion from cellobiose provide a promising means for low-cost cellulosic ethanol production through a consolidated bioprocessing development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Renewable bioethanol production for transportation fuels holds the promise of reduced dependent on petroleum and a more environmentally friendly energy system. Lignocellulosic biomass materials including agricultural residues are low cost feedstocks for cellulosic ethanol production [1–3]. However, the necessary deconstruction of cellulosic polymers and enzymatic hydrolysis require additional processing procedures that increase the cost of lignocellulose-to-ethanol conversion. Cellobiose is generated after cellulose depolymerization by cellulases. It needs to be further degraded into the simple sugar glucose before it can be utilized by conventional yeast for growth and subsequent ethanol fermentation. In cellulosic ethanol production using simultaneous saccharification and fermentation (SSF), β-glucosidase is required since commonly used commercial yeast cannot utilize cellobiose. To facilitate an economic and consolidated bioprocess, efforts have been made to engineer conventional ethanologenic yeast with β-glucosidase producing capability [4–9]. However, the level of the enzyme activity achieved is generally unsatisfactory. Other yeasts reported having cellobiose-utilizing activity usually produce low levels of β-glucosidase activity, and the cellobiose fermentation rate is not competitive [10–13]. Previously, we reported a temperature- and inhibitor-tolerant strain of Clavispora Y-50464 that produces sufficient native β-glucosidase enzyme activity allowing it to grow on cellobiose as sole source of carbon and energy fermenting it to ethanol [14]. Using xylose extracted corncobs residue as an example, economic cellulosic ethanol conversion was achieved without addition of β-glucosidase enzyme by SSF. In this study, we report the isolation and identification of the β-glucosidase enzyme from Y-50464. Outcomes of this research aid the development of consolidated bioprocessing for lower-cost cellulosic ethanol production.
Materials and Methods
Yeast Strains, Media, and Raw Enzyme Materials
Yeast strains Clavispora sp. NRRL Y-50464, Clavispora lusitaniae NRRL Y-5394, and Candida wickerhamii NRRL Y-2563 from the Agricultural Research Service Culture Collections, Peoria, IL, USA, were used in this study. Cell cultures were maintained and precultured using YP media consisting of 50 g glucose, 3.0 g yeast extract, and 5.0 g peptone per liter. A β-glucosidase Novo188 was provided by Novozyme (Denmark).
Growth Condition and Quantification of Metabolites
The effect of glucose on NRRL Y-50464 β-glucosidase activity was tested by fermenting cellobiose to ethanol in the presence or absence of glucose. A fleaker system with 300 mL of YM medium amended with either 5 % glucose, 5 % cellobiose, or a mixture of cellobiose, and glucose at 2.5 % each was inoculated from an overnight culture of Y-50464 grown in YM with 5 % glucose to an OD600 reading 0.1. The cultures were incubated with agitation at 250 rpm for 48 h at 37 °C. Each experiment was carried out in three replications. Samples were taken at 0, 1, 3, 6, 12, 18, 24, 30, and 48 h for evaluation of cell growth, ethanol conversion, and enzyme activity. Samples for high-performance liquid chromatography (HPLC) and enzyme assays were kept frozen at −20 °C until analyzed. HPLC assays were performed using a Shimadzu LC-20AD (Shimadzu Co., Kyoto, Japan) equipped with an HPX-87H Aminex ion exclusion column and refractive index detection as previously described [14].
Assay of β-Glucosidase Activity
Crude enzyme samples were prepared from yeast lysates using Y-PER plus dialyzable yeast protein extraction reagent (Thermo Scientific, Rockford, IL, USA) and assayed under optimal conditions using 5 mM p-nitrophenyl β-d-glucopyranoside as the substrate. Activity of β-glucosidase was assayed on a 96-well microtiter plate using a procedure as previously described [14–16]. Briefly, 100 μl of 5 mM p-nitrophenyl β-d-glucopyranoside in 100 mM citrate buffer at pH 6.0 was pipetted in each well. Then, 25 μl of enzyme was added to each sample to start the reaction. The reaction was carried out in an incubator at 45 °C for 30 min. After incubation, 125 μl ice cold 0.5 M Na2CO3 was added to stop the reaction, and the absorbance was measured at 405 nm using a Power Wavex 340 plate reader (Bio-Tek Instruments Inc., Winooski, VT, USA). Each assay reaction was carried out in triplicate. One unit of enzyme is defined as the amount of enzyme needed to release 1 μmol p-nitrophenol per minute under the defined conditions. Protein concentration was calculated using a Genesys 10UV spectrophotometer (Thermo Scientific, West Palm Beach, FL, USA) at 280 nm as previously described (http://www.strgen.org/protocols [17]).
Enzyme Localization
To determine the cellular location of the β-glucosidase enzyme, strain Y-50464 was grown in a fleaker containing 100 mL of YM medium amended with either 5 % glucose or 5 % cellobiose at 37 °C at 250 rpm. C. lusitaniae NRRL Y-5394 and C. wickerhamii NRRL Y-2563 were grown on a YM medium respectively with 5 % cellobiose at 28 °C at 250 rpm as previously described [18, 19]. All cultures were incubated for 17 h, and a 5-mL sample was taken for each and aliquoted as necessary. Cells were collected by centrifugation and resuspended to the original volume in 50 mM acetate buffer (pH 6.5). Samples of both cells and supernatants were saved for the assay. The cells were resuspended in the acetate buffer and disrupted using acid washed glass beads (Sigma Aldrich, St Louis, MO, USA) and vortexing for 3 min to break cell wall structures. The homogenate was centrifuged at 48,000×g, and the supernatant was removed and saved. Pelleted cellular debris was resuspended to its original volume using 50 mM acetate buffer (pH 6.5). Cells used for spheroplast assays were resuspended in 1 M sorbitol. Spheroplasts were formed using Zymolase 60,000 (Seikagaku Kogyo Co., Tokyo, Japan) at 37 °C for 45 min. One half of the sphereoplast sample was separated by centrifugation at 750×g and the supernatant saved. The pellet debris was resuspended in 500 μl of 1 M sorbitol (Sigma, St Louis, MO, USA) and saved for assay. Sphereoplasts from the other half of the sample were sedimented by centrifugation at 750×g and further lysed in 50 mM acetate buffer. The lysed sample was centrifuged and the supernatant saved for assay.
The pellet was resuspended to the original volume of acetate buffer and saved for assay. Each fraction collected during above described separation was assayed for its enzyme activity as previously described. All assay reactions were carried out in triplicate. Cultures for the three strains were incubated continuously, and samples were taken at 8, 24, 32, 24, and 48 h for assays of sugar consumption and ethanol production by HPLC analysis as described above.
Purification of β-Glucosidase Protein
A Y-50464 strain culture was incubated in 500 mL YM containing 3 % cellobiose in a 1-L flask at 37 °C with agitation at 170 rpm for 17 h. Cells were then pelleted by centrifugation and lysed using Y-PER plus dialyzable yeast protein extraction reagent following manufacturer’s instructions. Briefly, the cell pellet was resuspended to an appropriate amount of Y-PER reagent and incubated with agitation at 250 rpm at 30 °C for 30 min. Then, the suspension was centrifuged and the supernatant saved for protein purification use. Purification of the protein was performed using a previously reported procedure [12] with modifications. The cell lysate was treated with 80 % ammonium sulfate at 4 °C overnight. The precipitate was collected by centrifugation at 48,000×g for 20 min. The pellet was dissolved in 50 mM imidazole buffer (pH 6.5) and dialyzed overnight. The dialyzed protein solution was loaded on a diethylaminoethyl (DEAE) Bio-Gel A agarose column (2.5 by 6.5 cm) pre-equilibrated with 50 mM imidazole buffer (pH 6.5). The column was washed with 5 volumes of the buffer and eluted by NaCl with a gradient of 0 to 0.5 M using the same buffer (100 mL each). Fractions showing β-glucosidase activity were pooled together and dialyzed overnight against 50 mM acetate buffer (pH 5.0) at 4 °C. A cellobiose-Sepharose affinity matrix was prepared by coupling cellobiose to epoxy-activated Sepharose 6B following manufacturer’s instructions (affinity chromatography; Pharmacia Fine Chemicals, Uppsala, Sweden). The dialyzed protein solution containing β-glucosidase obtained after DEAE Bio-Gel column was subject to affinity chromatography on cellobiose-Sepharose 6B column (2.5 × 12 cm) pre-equilibrated with 50 mM acetate buffer (pH 5.0). The column was washed extensively with 50 mM acetate buffer (pH 5.0). The protein was eluted with a gradient of 0–2.0 M NaCl in acetate buffer (pH 5.0). Fractions with active β-glucosidase enzyme activity were pooled and dialyzed against 50 mM acetate buffer (pH 5.0) at 4 °C. Samples taken at each step were verified for purity and size of the protein by running a 10 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was used for subsequent identification and characterization.
Confirmation of Protein Activity
Native PAGE was also used to confirm the protein activity. A protein sample was loaded on a 12 % Tris–HCl gel (Bio-Rad, Hercules, CA, USA) electrophoresis at 4 °C without SDS in the running buffer or loading buffer. Upon completion, the gel was washed in100 mM citrate buffer (pH 6.0) and then covered in 50 mL of 10 mM p-nitrophenyl β-d-glucoside in 100 mM citrate buffer (pH 6.0) and incubated at 45 °C for 5 min. An equal volume of cold 0.5 M Na2CO3 was added to the reaction to detect a positive band.
Identification of β-Glucosidase Protein
Protein identification was performed using mass spectrometry by Applied Biomics, Inc (Hayward, CA, USA). Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF MS) and TOF/TOF tandem MS/MS were performed on AB SCIEX TOF/TOF™ 5800 System (AB SCIEX). MALDI-TOF mass spectra were acquired in reflectron positive ion mode, averaging 4,000 laser shots per spectrum. TOF/TOF tandem MS fragmentation spectra were acquired for each sample, averaging 4,000 laser shots per fragmentation spectrum on each of the 10 most abundant ions present in the sample (excluding trypsin autolytic peptides and other known background ions). Both of the resulting peptide mass and the associated fragmentation spectra were submitted to a GPS Explorer workstation equipped with MASCOT search engine (Matrix science) to search the database of National Center for Biotechnology Information non-redundant (NCBInr). Based on the identification of amino acid sequence and its β-glucosidase activity, a blast search was performed using NCBI database. High confidence matched proteins were aligned with selective candidates of peptide sequences using Biological Workbench v3.2 (http://workbench.sdsc.edu/).
Characterization of β-Glucosidase from Y-50464
Enzyme kinetic parameters for the purified β-glucosidase from strain NRRL Y-50464 were determined using the substrate p-nitrophenyl β-d-glucopyranoside at a series of concentrations from 0.1 to 5 mM in 100 mM citrate buffer at pH 6.0. A commercial source of Novo188 was used as a reference. Each enzyme reaction was started with 20 μl of enzyme sample for a total volume of 2 mL in a cuvette. The 20-μl enzyme sample represents respectively 0.35 U for BGL1 from Y-50464 and 1.6 U for Novo188 at the highest possible dilution from the original sample with a detectable enzyme activity. Absorbance was measured at 405 nm for 5 min at 45 °C using an Agilent 8453 spectrophotometer. Each assay was performed in triplicate. A Lineweaver–Burk plot was constructed and V max and K m calculated [20–23]. Product inhibition (K i) by glucose was measured using the same procedure with addition of glucose in the reaction at concentrations ranging from 0.1 to 100 mM as previously described [11]. The substrate concentration for these reaction mixtures contained either 2.5 or 5 mM of p-nitrophenyl β-d-glucopyrandoside.
The optimal temperature for the purified β-glucosidase activity was determined by the standard enzyme assay as described above at a temperature range of 35–65 °C with a 5 °C increment for each assay. The optimal pH for the enzyme activity was determined at 45 °C over a pH ranging from pH 3.0 to 7.0 with an increased interval of one using a citrate buffer as described above. All enzyme reaction assays were carried out in triplicate.
Results and Discussion
Growth and Ethanol Conversion on Cellobiose
When grown on glucose, cell growth of strain Y-50464 was observed to reach the maximum 18 h after incubation. When cellobiose was used as sole source of carbon and energy, it took about 30 h for the cell density to reach the same level (Fig. 1). The complete consumption of cellobiose apparently took a longer time than glucose (Fig. 2). This delayed growth reflected the extra steps of cellobiose conversion to glucose, which involves β-glucosidase. On a medium containing a mixture of cellobiose and glucose, cells grew quickly at the beginning but slowed down before the density reached its highest level at 30 h. Cellobiose started to be consumed at 12 h only after depletion of glucose in the medium (Fig. 2c). This observation further confirmed that the immediate availability of glucose supported early cell growth and cellobiose was utilized for subsequent growth and fermentation. However, the final ethanol production by the yeast was not affected by the different media (Fig. 2).
Ethanol conversion from cellobiose and/or glucose. Recovery of ethanol (open circle) conversion from YP medium containing cellobiose (filled square, a) and glucose (filled circle, b) each at 5 % or in combination of the sugars with 2.5 % each (c) for Clavispora NRRL Y-50464. Values are means of three replications
When cellobiose was used as sole source of carbon and energy, strain Y-50464 showed the fastest growth and reached log phase within 8 h followed by C. lusitaniae Y-5394. C. wickerhamii Y-2563 exhibited the slowest growth rate as observed at 8 and 24 h (Table 1). As shown by HPLC assay, Y-2563 consumed less than a half the amount of cellobiose in the medium and resulted in a poor ethanol production of only 9.8 g/L at 48 h. Both Clavispora strains completely utilized cellobiose and produced ethanol of 22.7 and 21.4 g/L for Y-50464 and Y-5349, respectively, at the end of fermentation. However, the rate of cellobiose consumption and ethanol production for strain Y-50464 was significantly faster than that of Y-5394 (Table 1). As demonstrated in a previous study, application of Y-50456 achieved a successful lower-cost corncob-to-ethanol conversion by not needing additional β-glucosidase enzyme [14]. The capability of Y-50464 to produce sufficient β-glucosidase activity and conversion of ethanol from cellobiose is a desirable characteristic for its application in cellulosic ethanol production.
Beta-Glucosidase Activity
When cultured on medium containing only glucose, crude cell extracts from Y-50464 did not show significant β-glucosidase activity over a 48-h time course study (Fig. 3). In contrast, on cellobiose medium, β-glucosidase activity was observed to increase overtime with the highest activity observed at 18 h after incubation. As anticipated, the β-glucosidase from cellobiose-glucose mixed medium was not induced until after 12 h when glucose was depleted. This is consistent and supported by the diauxic lag observed in cell growth in the mixed sugars (Fig. 2c).
Beta-glucosidase activity over time. Comparison of β-glucosidase activity for Clavispora NRRL Y-50464 on YP medium containing cellobiose (black bar) and glucose (blank bar) each at 5 % or in combination of the sugars with 2.5 % each (gray bar) in a time course study till 48 h after incubation. Values are means of two replications
Enzyme Location
Using a medium containing glucose as a background and negative reference for a crude enzyme localization assay, little β-glucosidase activity was observed in any cell culture fractions from strain Y-50464 (Table 2). When induced by cellobiose, strain Y-50464 displayed a similar profile but significantly higher levels of β-glucosidase activity in all culture fractions compared to C. lusitaniae Y-5394. In all cases (Y-50464, Y-5394, and Y-2563), only a minor fraction of β-glucosidase activity was released to the culture medium. Both for Y-50464 and Y-5394, most of the activity remained within spheroplasts after zymolyase treatment of the cells, which, together with the lysed spheroplast assays, indicates a cytoplasmic and membrane location of the enzyme. β-Glucosidase for yeast C. lusitaniae Y-5394 was reported to be associated with cytoplasm [18–20], and our results are in agreement with this. A notable amount of β-glucosidase activity was also observed in other fractions, which suggests more than one form of β-glucosidase may exist. In contrast, C. wickerhamii Y-2563 showed a different profile in spheroplast assays. The β-glucosidase from Y-2563 was reported to be extracytoplasmatic but cell associated [20, 24]. The current study confirms this main location, with most of the cell-associated β-glucosidase being released after zymolyase-treatment. Y-2563 produced high levels of β-glucosidase activity; however, it did not produce sufficient ethanol or consume cellobiose efficiently after 48 h incubation as described above. Whether the substrate analogue pNPG affected the efficiency of the enzyme assay or strain Y-2563 required a different temperature for optimal enzyme expression remain to be clarified. Βeta-glucosidase has been observed as an extracellular, intercellular, or membrane-bounded enzyme associated with wide ranges of microbes [10, 12, 21, 25–27]. Based on our assays, an intracellular protein from Y-50464 was isolated, purified, and characterized in this study.
Protein Purification
A purified protein was obtained from the soluble extract of a cellobiose induced culture of Y-50464 through a series of chromatographic steps. The protein was recovered at 17 % of its original total enzyme content with a purification fold of 26 (Table 3). It was estimated with an approximate molecular mass of 93.3 kDa using SDS-PAGE gel electrophoresis (Fig. 4a). A native gel showed a single active band suggesting that the protein band observed in the SDS-PAGE gel (Fig. 4b).
Beta-glucosidase on gel. An sodium dodecyl sulfate polyacrylamide gel electrophoresis gel (a) showing a purified β-glucosidase from Clavispora NRRL Y-50464 with an estimated molecular size at 93 kD. A protein band of β-glucosidase (b) shown on a native gel confirmed the protein as the active component of the cellobiose reduction assay
Identification of β-Glucosidase
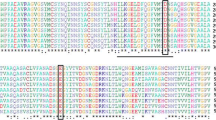
Using MALDI-TOF and TOF/TOF tandem MS/MS, 13 peptides were obtained with a total of 442 amino acid residues. Database search with NCBI non-redundant resulted in a high confidence prediction matching a hypothetical protein of CLUG_01181 from C. lusitaniae ATCC 42720 with a protein score of 100% CI. An amino acid alignment between the peptides and the hypothetical protein CLUG_01181 displayed no variations of amino acid for all covered peptides (Fig. 5). Phylogenetic analysis based on amino acid sequence indicated that β-glucosidase from Y-50464 is distinct from any known β-glucosidase associated with Candida tenuis, Debaryomyces hansenii, Meyerozyma guilliermondii, Scheffersomyces stipitis, Schwanniomyces etchellsii, and Spathaspora passalidarum (Fig. 6). The isolated protein from Y-50464 was closely related to a hypothetical protein from C. lusitaniae xp_002620022.1. Based on amino acid confirmation and its high level of specific enzyme activity, this protein from Y-50464 was identified as a β-glucosidase. CLUG_01181 is a predicted hypothetical protein based on computation annotation. This report provides the first evidence to confirm its function as a β-glucosidase. Since the β-glucosidase activity was also observed in other culture fractions of Y-50464, additional β-glucosidase proteins may exist. We designate this enzyme as BGL1 from Clavispora NRRL Y-50464. Its amino acid sequence suggests BGL1 from Y-50464 is associated with the glycoside hydrolase family 3.
Phylogenetic tree. A phylogenetic tree showing relationships of a β-glucosidase from Clavispora NRRL Y-50464 and closely related enzymes based on amino acid sequence obtained from NCBI protein database. Legends of NCBI protein accession number ACF93471 is from Schwanniomyces etchellsii, EDK38776.2 and XP_001485145.1 from Meyerozyma guilliermondii ATCC 6260, EGV61510.1 and EGV65580.1 from Candida tenuis ATCC 10573, EGW32194.1 and EGW31177.1 from Spathaspora passalidarum NRRL Y-27907, XP_001383273.1, XP_001385159.1, XP_001387766.1, and XP_001387350.1 from Scheffersomyces stipitis CBS 6054, XP_002620022.1from Clavispora lusitaniae ATCC 42720, and XP_461831.2 and XP_457283.2 from Debaryomyces hansenii CBS767
Characterization of BGL1 from Y-50464
The optimal temperature of BGL1 activity was estimated at 45 °C as measured by the enzyme assay (Fig. 7a). The optimal pH for the best enzyme performance was observed at pH 6 at 45 °C (Fig. 7b). Enzyme kinetic analysis indicated a K m of 0.355 mM for BGL1 from Y-50464 using pNPG as substrate (Table 4). It appeared to have a higher specific activity than Novo188 at a K m of 0.448 mM. End product inhibition of β-glucosidase activity by glucose was estimated by a K i of 15.2 mM for BGL1 from Y-50464. In contrast, Novo188 was more sensitive to glucose inhibition with a K i of 0.735 mM. Since Novo188 is not a pure commercial enzyme, it is unclear if its impurity affected the sensitivity to the glucose inhibition. Among reported native β-glucosidases produced by other fungal species [28–37], the K m of BGL1 from Y-50464 is ranked in the middle of the category. However, it is superior to other recently characterized fungal β-glucosidases (K m = 0.57 mM, K i = 2.70 mM and K m = 0.38, K i = 3.25 mM, respectively) [38] both in K m [pNPG] and product inhibition caused by glucose.
Most fungal species producing native β-glucosidases are nonethanol producers such as species of Aspergillus, Trichoderma, and Penicillium. To facilitate cellulosic ethanol production, supplemental enzyme can be added to common cellulolytic enzyme preparations to enhance cellulosic conversion [34, 39]. However, it is not cost effective and requires additional processing steps. Among yeast strains selected for enzyme productivity, ethanol production is often not satisfactory. For example, C. wickerhamii produces a higher level of β-glucosidase activity; however, its ethanol fermentation capability using cellobiose is low. Two Clavispora strains tested in this study showed promising ethanol fermentation from cellobiose. Strain Y-50464 was the most efficient with regard to growth and ethanol conversion on cellobiose.
Consolidated bioprocess is economically desirable for fermentation based technology. This concept and practice have become an increasingly popular target for sustainable biofuels productions. We previously demonstrated a successful example of cellulosic ethanol conversion using Y-50464 without addition of exogenous β-glucosidase [14]. It appeared that the native β-glucosidase activity produced by Y-50464 was sufficient for ethanol production from xylose extracted corncob residues by SSF. Isolation and identification of a β-glucosidase in this study provided additional evidence to support the function of cellobiose utilization by strain Y-50464. These results will advance the development of consolidated bioprocesses for lower-cost cellulosic ethanol production.
References
Bothast R, Saha B (1997) Ethanol production from agricultural biomass substrate. Adv Appl Microbiol 44:261–286
Wheals AE, Basso LC, Alves DM, Amorim HV (1999) Fuel ethanol after 25 years. Trends Biotechnol 17:482–487
Zaldivar J, Nielsen J, Olsson L (2001) Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol 56:17–34
Adam AC, Rubio-Texeira M, Polina J (1995) Induced expression of bacterial β-glucosidase activity in Saccharomyces. Yeast 11:395–406
Skory CD, Freer SN, Bothast RJ (1996) Expression and secretion of the Candida wickerhamii extracellular β-glucosidase gene bglB in Saccharomyces cerevisiae. Curr Genet 30:417–422
van Rooyen R, Hahn-Hägerdal B, La Grange DC, van Zyl WH (2005) Construction of cellobiose-growing and fermenting Saccharomyces cerevisae strains. J Biotechnol 120:284–295
Shen Y, Zhang Y, Ma T, Bao X, Du F, Zhuang G et al (2008) Simultaneous saccharification and fermentation of acid-pretreated corncobs with a recombinant Saccharomyces cerevisiae expressing β-glucosidase. Bioresour Technol 99:5099–5103
Saitoh S, Hasunuma T, Tanaka T, Kondo A (2010) Co-fermentation of cellobiose and xylose using β-glucosidase displaying diploid industrial yeast strain OC-2. Appl Microbiol Biotechnol 87:1975–1982
Gurgu L, Lafraya A, Polaina J, Marin-Navarro J (2011) Fermentation of cellobiose to ethanol by industrial Saccharomyces strains carrying the β-glucosidase gene (BGL1) from Saccharomycopsis fibuligera. Bioresour Technol 102:5229–5236
Freer SN (1985) Purification and characterization of the extracellular β-glucosidase produced by Candida wickerhamii. Arch Biochem Biophys 243:515–522
Saha BC, Freer SN, Bothast RJ (1994) Production purification and properties of a thermostable β-glucosidase from a color variant strain of Aureobasidium pullulans. Appl Environ Microbiol 60:3774–3780
Saha BC, Bothast RJ (1996) Production, purification, and characterization of a highly glucose-tolerant novel β-glucosidase from Candida peltata. Appl Environ Microbiol 62:3165–3170
Barbosa AM, Giese EC, Dekker RFH, Borsato D, Briones Pérez AI, Úbeda Iranzo JF (2010) Extracellular β-glucosidase production by the yeast Debaryomyces pseudopolymorphus UCLM-NS7A: optimization using response surface methodology. New Biotechnol 27:374–381
Liu ZL, Weber SA, Cotta MA, Li S-Z (2012) A new β-glucosidase producing yeast for lower-cost cellulosic ethanol production from xylose-extracted corncob residues by simultaneous saccharification and fermentation. Bioresour Technol 104:410–416
Grover AK, Macmurche DD, Cushley RJ (1977) Studies on almond emulsion β-D-glucosidase I Isolation and characterization of a bifunctional isozyme. Biochem Biophys Acta 482:89–108
Saha BC, Biswas A, Cotta MA (2008) Microwave pretreatment, enzymatic saccharification and fermentation of wheat straw to ethanol. J Biobased Mater Bioenergy 2:210–217
Liu ZL, Moon J (2009) A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10
Freer SN, Detroy RW (1982) Direct fermentation of cellodextrins to ethanol by Candida wickerhamii and C. lusitaniae. Biotechnol Lett 4:453–458
Freer SN, Detroy RW (1983) Characterization of cellobiose fermentation to ethanol by yeasts. Biotechnol Bioeng 25:541–557
Freer SN, Greene RV (1990) Transport of glucose and cellobiose by Candida wickerhamii and Clavispora lusitaniae. J Biol Chem 265:12864–12868
Mahajan PM, Desai KM, Lele SS (2010) Production of cell membrane-bound α- and β-glucosidase from Lactobacillus acidophilus. Food Bioprocess Technol 5:706–718
Skory CD, Freer SN, Bothast RJ (1996) Properties of an intercellular β-glucosidase purified from cellobiose-fermenting yeast Candida wickerhamii. Appl Microbiol Biotechnol 46:353–359
Wallecha A, Mishra S (2003) Purification and characterization of two β-glucosidases from a thermo-tolerant yeast Pichia etchellsii. Biochim Biophys Acta 1649:74–84
Yapi D, Yapi A, Gnakri D, Niamke SL, Kouame LP (2009) Purification and biochemical characterization of a specific β-glucosidase from the digestive fluid of larvae of the palm weevil, Rhynchophorus palmarum. J Insect Sci 9:4
Freer SN (1993) Kinetic Characterization of a β-glucosidase from a yeast Candida wickerhamii. J Biol Chem 268:9337–9342
Skory CD, Freer SN (1995) Cloning and characterization of a gene encoding a cell-bound extracellular β-glucosidase in the yeast Candida wickerhamii. Appl Environ Microbiol 61:518–525
Lee K-M, Jeya M, Joo A-R, Singh R, Kim I-W, Lee J-K (2010) Purification and characterization of a thermostable endo β-1, 4-glucanase from novel strain Penicillium purpurogenum. Enzyme Microb Technol 46:206–211
Orkman WE, Day DF (1982) Purification and properties of beta-glucosidase from Aspergillus terreus. Appl Environ Microbiol 44:1289–1295
Srivastava SK, Gopalkrishnan KS, Ramachandran KB (1984) Kinetic characterization of a crude beta-d-glucosidase from Aspergillus wentii Pt 2804. Enzyme Microb Technol 6:508–512
Kantham L, Jagannathan V (1985) Beta-Glucosidase of Penicillium funiculosum. II. Properties and mycelial binding. Biotechnol Bioeng 23:786–791
Kitpreechavanich V, Hayashi M, Nagai S (1986) Purification and characterization of extracellular beta-xylosidase and beta-glucosidase from Aspergillus fumigates. Agric Biol Chem 50:1703–1711
Chen H, Hayn M, Esterbauer H (1992) Purification and characterization of two extracellular beta-glucosidases from Trichoderma reesei. Biochim Biophys Acta 1121:54–60
Ohnishi M, Okada G, Yazaki T (1998) Characterization of the subsite structure of the beta glucosidase from Aspergillus niger, an aspect of the mechanism of carbohydrate recognition. Carbohydr Res 308:201–205
Hong J, Tamaki H, Kumagai H (2006) Unusual hydrophobic linker region of beta-glucosidase (BGLII) from Thermoascus aurantiacus is required for hyper-activation by organic solvents. Appl Microbiol Biotechnol 73:80–88
Langston J, Sheehy N, Xu F (2006) Substrate specificity of Aspergillus oryzae family 3 beta glucosidase. Biochim Biophys Acta 1764:972–978
Chen M, Qin Y, Liu Z, Liu K, Wang F, Qu Y (2010) Isolation and characterization of a β-glucosidase from Penicillium decumbens and improving hydrolysis of corncob residue by using it as cellulose supplementation. Enzyme Microb Technol 46:444–449
Decker CH, Visser J, Schreier P (2000) Beta-Glucosidases from five black Aspergillus species: study of their physico-chemical and biocatalytic properties. J Agric Food Chem 48:4929–4936
Chauve M, Mathis H, Huc D, Casanave D, Monet F, Ferreira NL (2010) Comparative kinetic analysis of two fungal β-glucosidases. Biotechnol Biofuels 3:3
Kristensen JB, Felby C, Jørgensen H (2009) Yield-determining factors in high-solids enzymatic hydrolysis of lignocelluloses. Biotechnol Biofuels 2:11
Acknowledgment
The authors are grateful to Chris Skory and Kristina Glenzinski for technical assistance in enzyme kinetic analysis. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Z.L., Weber, S.A. & Cotta, M.A. Isolation and Characterization of a β-Glucosidase from a Clavispora Strain with Potential Applications in Bioethanol Production from Cellulosic Materials. Bioenerg. Res. 6, 65–74 (2013). https://doi.org/10.1007/s12155-012-9236-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-012-9236-9