Abstract
Purpose
Non-small cell lung cancer (NSCLC) is the major subtype of lung cancer, which is the leading cause of cancer death worldwide. Tumor-associated macrophages (TAMs) are one of the main non-tumor cells in the tumor microenvironment. Here, we investigated the effect of cancer cell-derived exosomal LINC00313 on the M2 macrophage differentiation in NSCLC and clarified its underlying mechanism.
Methods
Flow cytometry, Western blotting, ELISA and immunohistochemical staining were performed to identify the macrophage phenotype by detecting the expression of M2 markers. The expression levels of LINC00313 and miR-135a-3p were measured by qRT-PCR, and luciferase reporter assay was used to validate the binding of lncRNA to miRNA, and miRNA to the target gene STAT6. The mouse-xenograft models were established by subcutaneous injection of the NCl-H1299 cells with stable overexpression or knockdown of LINC00313. GW4869 was injected intra-tumorally after tumor implantation.
Results
It was found that the cancer cells promoted M2 macrophage differentiation by secreting exosomes. LINC00313 was overexpressed in H1299-derived exosomes, and its knockdown abolished the effect of H1299-induced M2 macrophage differentiation. LINC00313 sponged miR-135a-3p to increase the STAT6 expression, resulting in the M2 macrophage differentiation. LINC00313 promoted tumor progression and promoted the expression of M2 markers in isolated tumor macrophages. A novel regulatory mechanism of M2 macrophage differentiation in NSCLC was revealed. It was found that cancer cell-derived exosomal LINC00313 promoted M2 macrophage differentiation in NSCLC by up-regulating STAT6 as miR-135a-3p sponge.
Conclusions
This study provides a new mechanism and direction to prevent the M2 macrophage differentiation in NSCLC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer death worldwide, resulting in approximately 1.6 million deaths every year [1]. Non-small cell lung cancer (NSCLC), whose major histological subtypes include adenocarcinoma, squamous cell carcinoma and larger cell carcinoma, is the main subtype of lung cancer, accounting for nearly 85% of all lung cancer cases [2]. Tumor-associated macrophages (TAMs) are primary non-tumor cells in the tumor microenvironment, which are important players in the tumor progression through influencing the immunity and inflammatory circuits [3]. Macrophages can differ into M1 and M2 phenotypes, identified by different expression levels of markers. The markers for M1 macrophages include IL-12 and tumor necrosis factor (TNF), whereas M2 macrophages produce IL-10, IL-1 receptor antagonist and type II IL-1 decoy receptor [3]. Recent studies have found that M1 and M2 macrophages show different effects on lung cancer progression, making macrophage M1/M2 polarization potential targets in the treatment of lung cancer [4, 5]. However, the specific regulatory mechanism of macrophage differentiation in NSCLC is still to be illustrated.
Long noncoding RNAs (LncRNAs) are a group of non-coding RNAs with a length > 200 nucleotides. LncRNAs have been proved to be involved in various biological processes, including cell differentiation, proliferation, and apoptosis [6,7,8]. LncRNAs play their roles by regulating protein-coding genes, mRNA processing, and maintenance of genomic integrity [9]. LINC00313, a novel lncRNA, is frequently expressed in several cancers and regulates tumor growth and development [10,11,12,13]. It is reported that the high level of LINC00313 in NSCLC is significantly correlated with a poor prognosis [10]. Although recent studies have shown that lncRNAs can act as the mediator of cellular interactions through the secretion of exosomes [14,15,16,17], whether it participates in the regulation of macrophage differentiation in NSCLC remains unclear. Here, the effect of cancer cells on the M2 macrophage differentiation in NSCLC and its underlying mechanism were investigated. The results showed that cancer cells promoted the M2 macrophage differentiation in NSCLC by secreting exosomal LINC00313.
Materials and methods
Cell culture
Human NSCLC cell lines NCl-H1299, A549 and H1975, human monocyte cell lines THP-1, and human lung fibroblast cell lines MRC-5 were purchased from the American Type Culture Collection (ATCC). All cells were cultured with RPMI-1640 medium containing 10% FBS (Gibco, NY, USA) in an 5% CO2 incubator at 37 °C.
Cell treatment and transfection
To induce macrophage differentiation, THP-1 cells were treated with 25 ng/mL PMA for 24 h, and the H1299 cells or MRC-5 cells were cultured in culture medium (CM) (1% FBS) for 48 h. Then the CM was collected.
To explore the role of exosomes, 10 μM GW4869 (Sigma, USA) was added into H1299 cells to reduce the secretion of exosomes. H1299 cells were treated with DMSO (Sigma, USA) as a negative control (NC).
For the transfection of LINC00313, H1299 cells were infected with 100 MOI LINC00313 overexpression or knockdown lentivirus for 2 days before other treatment (Genepharma, Shanghai, China). In addition, H1299 cells were infected with 100 MOI negative control overexpression or knockdown lentivirus. MiR-135a-3p mimic, miR-135a-3p inhibitor, NC mimic, or NC inhibitor (Ribobio, Guangzhou, China) was transfected using Lipofectamine 3000 (Invitrogen, USA). The sequences are as follows: miR-135a-3p mimic: 5′-UAUGGCUUUUUAUUCCUAUGUGAACAUAGGAAUAAAAAGCCAUAUU-3′, NC mimic: 5′-UUCUCCGAACGUGUCACGUTT-3′, miR-135a-3p inhibitor: 5′-UCACAUAGGAAUAAAAAGCCAUA-3′, NC inhibitor: 5′-CAGUACUUUUGUGUAGUACAA-3′.
Flow cytometry
Flow cytometry was performed to analyze the phenotype of macrophages. THP-1 cells (1 × 105) were stained with 5 μL of fluorescence-labeled anti-human monoclonal antibodies against CD-206-PE, CD163 PE, and CD68-FITC. All the antibodies were purchased from BD Biosciences (CA, USA). Fluorescence was measured by flow cytometry.
qRT-PCR
The total RNA was extracted by the RNA extraction kit (Thermo Fisher Scientific, MA, USA), and reversely transcribed to cDNA using the PrimeScript RT reagent kit with gDNA Eraser (Takara, Tokyo, Japan), followed by real-time PCR using SYBR Premix Ex TaqII (Takara, Tokyo, Japan) with the housekeeping gene U6 used for normalization. The primer sequences are shown in Table 1.
Western blotting
The protein samples were collected from the cells or tissues, lysed with RIPA buffer (high) (R0010, Solarbio, Shanghai, China) on ice for 20 min, and quantified by the BCA protein assay kit (PC0020, Solarbio, Shanghai, China). Then they were incubated with the following antibodies (1:1000) purchased from Abcam (anti-iNOS, ab178945; anti-CD86, ab220188; anti-CD163, ab182422; anti-CD206, ab125028; anti-STAT6, ab32520; anti-CD63, ab59479; anti-CD81, ab79559) as well as anti-GAPDH and horseradish peroxidase-labeled goat anti-rabbit/mouse antibodies (1:5000). The primary antibody Fizz1 (Cat.sc-80324, 1:1000) was purchased from Santa Cruz. The protein bands were visualized by ECL Western blotting substrate (PE0010, Solarbio, Shanghai, China) and the intensities of protein bands were quantified by ImageJ software.
Detection of markers
The levels of TNF-α, IL-1β and TGF-β in the CM or serum were measured by the commercialized ELISA Kit (Elabscience, TX, USA). The absorbance was measured at 450 nm through Multiskan SkyHigh (Thermo Fisher Scientific, MA, USA).
Dual luciferase reporter assay
The wild type (WT) or mutant (Mut) sequences of 3′UTR of STAT6 and LINC00313 were synthesized by Genepharma (Shanghai, China) to construct the luciferase reporter plasmids including LINC00313-WT, LINC00313-Mut, STAT6-WT and STAT6-Mut. Upon reaching 70% confluence, the constructed luciferase reporter vectors were transfected into NCl-H1299 cells along with miR-135a-3p mimic or inhibitor by using Lipofectamine 6000™ (C0526, Beyotime Biotechnology, Shanghai, China). After 48 h, dual-luciferase activities in all groups were measured by dual-luciferase reporter assay (Promega, Shanghai, China) and normalized to Renilla signals.
RNA immunoprecipitation assay
The RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA) was applied in RNA immunoprecipitation with the Ago2 antibody (Abcam, USA). 2 × 107 NCl-H1299 cells were collected and resuspended in 200 μL RIPA lysis buffer. The protein A/G beads (BioWorld, USA) were coated with 5 μg control rabbit IgG antibody and Ago2 antibody. Then the cell lysates were incubated with the antibody-coated beads at 4 °C overnight. The RNA complexes pulled down were purified by TRIzol reagent. The levels of miR-135a-3p and LINC00313 RNA in the precipitates were measured by qRT-PCR.
Extraction and identification of Exosomes
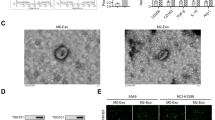
H1299 cells were plated in a cell culture dish (10 cm) at a density of 1 × 107, and cultured for 48 h. Then the medium was collected, and centrifuged three times at different speeds (300 g, 10 min, room temperature; 3000 g, 20 min, 4 °C; 13,000 g, 50 min, 4 °C). The resulting supernatant was harvested, centrifuged at 100,000g for 3 h at 4 °C and resuspended with 1 × PBS (50 μL) to collect the exosomes, and the exosomes were stored at − 80 °C. The morphology of the exosomes was identified by transmission electron microscopy (TEM). Specifically, the exosome pellets were fixed by 2.5% glutaraldehyde, dehydrated with increasing concentrations of alcohol, and observed under a TEM (JEM-1400Flash, JEOL, Tokyo, Japan). The mobility of exosomes was recorded and analyzed by NTA analytical software (NanoSight, version 3.0).
Exosome uptake analysis
To analyze exosome uptake, PKH67 (Sigma, CA, USA) was used to label exosomes [18]. Briefly, the PKH67 dye working solution was added to the exosomes. The samples were mixed gently for 4 min and then washed 3 times with 1 × PBS to remove the redundant PKH67. Then 10 μg of PKH67-labeled exosomes were used to incubate the THP-1 cells (2 × 105) for 2 h at 37 °C. After that, the THP-1 cells were incubated with Hoechst 33,342 staining solution (C1029, Beyotime Biotechnology, Shanghai, China) at 37 °C, and the nuclei were observed. The exosome uptake by the THP-1 cells was visualized under a fluorescence microscope (Carl Zeiss, Jena, Germany).
Mouse-xenograft experiments
BALB/c nude mice (6-week-old, male) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China), and used to establish the human NSCLC transplantation model. The NCl-H1299 cells were transfected with LINC00313 overexpression or knockdown lentivirus to construct the stable expression cell lines. The NCl-H1299 cells were collected and counted. Then they (2 × 107) were resuspended in 100 μL PBS and subcutaneously injected into the mice. The exosome inhibitor GW4869 or negative control DMSO was injected intra-tumorally at 2.5 mg/g the next day after tumor implantation [19]. The subcutaneous tumor volume was analyzed every 5 days. The mice were sacrificed at the 20th day after the injection and the tumors in all groups were removed for weight analysis.
Immunohistochemistry staining
The tumor tissues were fixed with 4% paraffin, embedded, and sectioned. The sections were treated with 3% hydrogen peroxide (Beyotime Biotechnology, Shanghai, China) for 10 min, and blocked by 5% bovine serum albumin (Thermo Fisher Scientific, MA, USA), followed by incubation with the primary antibody against CD68 (ab182422, Abcam, Cambridge, UK) and CD206 (ab125028, Abcam, Cambridge, UK) at 4 °C overnight, and with horseradish peroxidase-labeled goat anti-mouse IgG antibody (ab205719, Abcam, Cambridge, UK) at room temperature for 1 h. The color was developed using diaminobenzidine chromogen solution (Beyotime Biotechnology, Shanghai, China). Finally, the sections were observed under the microscope (Olympus, Tokyo, Japan).
Isolation of tumor macrophages
The macrophages were isolated by the tumor macrophage isolation fluid (Sangon Biotech, Shanghai, China). In brief, the tumor tissues were digested by collagenase (Thermo Fisher Scientific, MA, USA) into cell suspension. Then, 1 mL of cell suspension (2 × 108 cells) was added with 1 mL of macrophage isolation fluid, and centrifuged at 1500 r/min for 20 min. Milky white macrophages in the second layer were added with 4 mL of washing solution, and centrifuged at 1800 r/min for 20 min. Finally, the resulting precipitates were considered macrophages.
Statistical analysis
All values were presented as mean ± SEM. Statistical analysis was performed by using one-way ANOVA, and the difference was compared by Bonferroni test using GraphPad Prism 7the. p < 0.05 was considered statistically significant.
Results
NSCLC cells promoted M2 macrophage differentiation through secreting exosomes in vitro
To explore the role of NSCLC cells on M2 macrophage differentiation, the CM of MRC-5 cells and H1299 cells was used to incubate the PMA-stimulated THP-1 cells. Then, M2 markers (CD163 and CD206) and macrophage marker CD68 were detected by flow cytometry. As shown in Fig. 1A, the CM of H1299 induced an increased number of CD68+CD163+ cells and CD68+CD206+ cells, indicating an increased number of M2 macrophages. In addition, M2 markers (CD163 and CD206) were quantified by Western blotting. The results further identified the increased expression of M2 markers (CD206 and CD163) in THP-1 cells incubated with H1299-CM (Fig. 1B). Considering that exosomes are one of the main ways of signal exchange among cells [20], the secretion of exosomes was inhibited by using GW4869 to examine the roles of exosomes in the induction of M2 macrophage by H1299. H1299 cells were manipulated with GW4869 or DMSO. As shown in Fig. 1C, compared to the CM of H1299 cells treated with DMSO, GW4869 treatment inhibited the number of CD68+CD163+ cells and CD68+CD206+ cells, indicating that H1299 promotes the M2 macrophage differentiation through exosome secretion. Furthermore, the exosomes of H1299 were extracted and the homogeneous and spherical vesicles of typical exosomal structures were observed (Fig. 1D). The particle diameter was 99.8 ± 6.2 nm for the exosomes (Fig. 1E). It was confirmed by Western blotting that the exosomes marker CD63 and CD81 were enriched in the exosomes extracted from H1299 compared with those from H1299-CM (Fig. 1F), indicating that exosomes were fully collected from the CM.
H1299 cells promoted M2 macrophage differentiation through secreting exosomes in vitro. CM of NCl-H1299 cells and MRC-5 cells were used to incubate the THP-1 cells for 24 h. A M2 macrophages markers (CD206 and CD163) and macrophage marker CD68 were detected by flow cytometry. B Western blotting showed increased expressions of M2 markers (CD206 and CD163). The expression levels of indicated proteins were analyzed by ImageJ. C H1299 cells were treated with GW4869 (10 μM) for 24 h or not, and the CM was collected to incubate THP-1 cells. M2 markers (CD206 and CD163) and macrophage marker CD68 were detected by flow cytometry. D TEM demonstrated the round vesicle feature of H1299-derived exosomes. Bar = 200 nm. E The diameter for H1299-derived exosomes was tracked by NTA. F CD63 and CD81 (representative exosomal markers) in H1299-derived exosomes were detected by Western blotting. G PKH67-labeled exosomes derived from H1299 cells were incubated with THP-1 at 37 °C for 12 h, and taken up by THP-1. H The THP-1 cells were incubated with the H1299-exosomes and CM from H1299 pre-treated with GW4869. The TNF-α and IL-1β levels were detected by ELISA. I The TGF-β level was detected by ELISA. n = 3, data were described by mean ± SEM. * (**p < 0.01; ***p < 0.001) was considered different from control group
Then the THP-1 cells were treated with exosomes. PKH67-labeled exosomes were absorbed by THP-1 (Fig. 1G). ELISA was used to assess the levels of M1 makers (TNF-α and IL-1β) and M2 marker TGF-β. The results revealed that H1299-derived exosomes decreased the expression of TNF-α and IL-1β, and increased the expression of TGF-β, suggesting the decrease of the M1 macrophage differentiation and increase of the M2 macrophage differentiation. In contrast, pretreatment with GW4869 on the H1299 showed no effect on the expressions of TNF-α, IL-1β and TGF-β (Fig. 1H, I).
Tumor cell-derived exosomal LINC00313 promoted M2 macrophage differentiation in vitro
The microarray data sets (GSE50081, n = 124; and GSE30219, n = 84) were downloaded from the Gene Expression Omnibus (GEO) to screen the differentially expressed lncRNAs (LINC01116, LINC00968, LINC00313, lncRNA C1orf132 and lncRNA TMPO-AS1) in NSCLC. In addition, TCGA database also showed that the five lncRNAs (LINC01116, LINC00968, LINC00313, lncRNA C1orf132 and lncRNA TMPO-AS1) were related to the prognosis of NSCLC (Fig. S1A). Then the expressions of the five lncRNAs in exosomes from NSCLC cells (H1299, A549, and H1975) were detected by qRT-PCR. It was found that compared to those in exosomes from MRC-5 cells, the levels of LINC01116 and LINC00313 were significantly up-regulated in exosomes from NSCLC cells (Fig. S1B). Furthermore, LINC00313 had the maximum change multiple (Fig. S1B). Then, the PMA-stimulated THP-1 cells were cultured with H1299-CM. Compared to the control group, the LINC00313 expression was increased in THP-1 cells treated with H1299-CM (Fig. 2A). Therefore, it is speculated that the tumor cell-derived exosomal LINC00313 may be involved in M2 macrophage differentiation. Then, to explore the role of LINC00313 on macrophages, the cultured THP-1 cells were divided into 6 groups: control group (without any treatment), H1299-CM group (co-cultured with the CM of H1299 cells), H1299 (LINC00313 KD)-CM group (co-cultured with the CM of LINC00313-knockdown H1299 cells), NC group (infected with negative control lentivirus), LINC00313 KD group (infected with LINC00313 knockdown lentivirus), and LINC00313 OE group (infected with LINC00313 overexpression lentivirus). The results of qRT-PCR showed that compared to the H1299-CM group, LINC00313-knockdown H1299-CM decreased the LINC00313 expression in THP-1 cells (Fig. 2A). Moreover, compared to the NC group, overexpression of LINC00313 markedly increased the LINC00313 level, while knockdown of LINC00313 suppressed the LINC00313 level (Fig. 2A). Then, M2 markers (CD206 and CD163) of THP-1 cells were detected by flow cytometry. As shown in Fig. 2B, compared to the control group, H1299-CM treatment increased the number of M2 markers (CD206 and CD163) of THP-1 cells. Compared to the H1299-CM group, LINC00313-knockdown H1299-CM further suppressed the levels of CD206 and CD163 of THP-1 cells (Fig. 2B). Compared to the NC group, overexpression of LINC00313 induced increased levels of D206 and CD163, while knockdown of LINC00313 in THP-1 cells had the opposite effect (Fig. 2B). Moreover, Western blotting was performed to assess the expressions of M1 markers (iNOS and CD86) and M2 markers (CD206, CD163, and Fizz1). Both H1299-CM treatment and overexpression of LINC00313 suppressed the levels of M1 markers (iNOS and CD86), and increased the levels of M2 markers (CD206, CD163, and Fizz1) (Fig. 2C). In addition, knockdown of LINC00313 increased the levels of M1 markers (iNOS and CD86), and decreased the levels of M2 markers (CD206, CD163, and Fizz1) (Fig. 2C). Furthermore, qRT-PCR and ELISA were performed to assess the levels of M1 markers (TNF-α and IL-1β) and M2 marker (TGF-β). Overexpression of LINC00313 reduced the levels of TNF-α and IL-1βand increased the level of TGF-β, while knockdown of LINC00313 in THP-1 cells or treatment with LINC00313-knockdown H1299-CM in THP-1 cells increased the levels of TNF-α and IL-1βand reduced the level of TGF-β (Fig. 2D, E). All the results indicated that exosomal LINC00313 secreted by H1299 induces M2 macrophage differentiation.
H1299-derived exosomal LINC00313 promoted M2 macrophage differentiation in vitro. THP-1 cells were incubated with CM-H1299 or LINC00313-knockdown H1299-CM. A qRT-PCR was used to detect the LINC00313 expression. B M2 markers (CD206 and CD163) and macrophage marker CD68 were detected by flow cytometry. C Western blotting was used to identify the expressions of M2 markers (CD206, CD163, Fizz1) and M1 markers (CD86, iNOS). D The mRNA levels of M1 markers (TNF-α and IL-1β) and M2 marker (TGF-β) were measured by qRT-PCR. E ELISA was used to detect the M2 marker released by THP-1. n = 3, data were described by mean ± SEM. * (*p < 0.05; **p < 0.01; ***p < 0.001) was considered different from control group
LINC00313 promoted M2 macrophage differentiation through up-regulating STAT6
It is reported that STAT6 activates M2 macrophage-specific target gene transcription, leading to the M2 macrophage differentiation [21, 22]. Then the effect of LINC00313 on STAT6 expression was examined by qRT-PCR and Western blotting. As shown in Fig. 3A, B, LINC00313 contributed to the expression of STAT6, which was inhibited by LINC00313 knockdown. To further explore whether LINC00313 played its role by STAT6, the cultured THP-1 cells were divided into 3 groups: control group (infected with negative control lentivirus), LINC00313 KD group (infected with LINC00313 knockdown lentivirus), and LINC00313 KD + STAT6 OE group (infected with LINC00313 knockdown lentivirus and STAT6 overexpression lentivirus). It was found that knockdown of LINC00313 increased the levels of M1 markers (iNOS and CD86), and decreased the levels of M2 markers (CD206, CD163, and Fizz1), which was reversed by overexpression of STAT6 (Fig. 3C). The number of CD206- and CD163-positive M2 cells decreased in LINC00313 knockdown cells but increased in STAT6 overexpression cells (Fig. 3D). Knockdown of LINC00313 enhanced the expressions of M1 markers TNF-α and IL-1βbut decreased the expressions of M2 markers TGF-β, which were all reversed by overexpression of STAT6 (Fig. 3E). All these data suggested that LINC00313 regulates M2 macrophage differentiation through up-regulating STAT6 expression.
LINC00313 promoted M2 macrophages through up-regulating STAT6. A, B THP-1 cells were incubated with CM-H1299 or LINC00313-knockdown H1299-CM. STAT6 expression was detected by qRT-PCR and Western blotting. C LINC00313 was knocked down in THP-1 cells with or without overexpression of STAT6. M2 markers (CD206, CD163, Fizz1) and M1 markers (CD86, iNOS) were assessed through Western blotting. D The M2 phenotype was detected by flow cytometry. E The mRNA levels of M1 markers (TNF-α, IL-1β) and M2 marker (TGF-β) were evaluated through qRT-PCR. n = 3, data were described by mean ± SEM. * (*p < 0.05; **p < 0.01; ***p < 0.001) was considered different from control group
LINC00313 regulated STAT6 through sponging miR-135a-3p
As CeRNA is one of the main mechanisms of lncRNA regulating the protein level [23], the miRNA which could be regulated by LINC00313 and could regulate STAT6 expression was predicted by using TarBase, LncBase and TargetScan. Four candidate miRNAs which might be the mediator between LINC00313 and STAT6 were found (Fig. 4A). Among the four miRNAs, only miR-135a-3p showed a marked decrease in the cells overexpressing LINC00313 (Fig. 4B). The potential binding sites of miR-135a-3p and LINC0033 and 3'UTR of STAT6 mRNA were predicted (Fig. 4C–H). Luciferase reporter assay further showed that miR-135a significantly decreased the luciferase activity of the WT rather than the Mut LINC00313 and Mut STAT6 3'UTR, indicating that miR-135a specifically binds to the LINC00313 and STAT6 3'UTR (Fig. 4D–I). RIP assay showed that treatment with LINC00313 could significantly increase the association of miR-135a, and treatment with miR-135a could increase the association of LINC00313 (Fig. 4E). H1299 exosomes could significantly decrease the miR-135a level in THP-1 cells (Fig. 4F), and the same was true for overexpression of LINC00313 (Fig. 4G). Both qRT-PCR and Western blotting verified that miR-135a negatively regulates the STAT6 expression (Fig. 4J, K).
LINC00313 regulated STAT6 through sponging miR-135a-3p. A Schematic illustration showing the overlapping miRNAs predicted by TarBase, LncBase, and TargetScan. B The relative miRNA expression in cells overexpressing LINC00313 was detected by qRT-PCR. C The predicted binding sites of LINC00313 and miR-135a. D After 24-h transfection into THP-1 cells with luciferase reporter plasmids containing WT or Mut LINC00313 along with miR-135a-3p mimic or inhibitor, luciferase activities were evaluated in all groups. E The RNA immunoprecipitation assay was performed. The RNA levels of LINC00313 and miR-135a in the precipitates were detected by qRT-PCR. F, G The miR-135a-3p level was measured by qRT-PCR. H The putative binding sequence for miR-135a-3p in the 3'UTR of STAT6 mRNA. I After 24-h transfection into THP-1 cells with luciferase reporter plasmids containing WT or Mut STAT6 3'UTR along with miR-135a-3p mimic or inhibitor, luciferase activities were evaluated in all groups. J, K THP-1 cells were treated with miR-135a-3p mimic or inhibitor for 24 h, and the expression of STAT6 was detected through qRT-PCR and Western blotting. n = 3, data were described by mean ± SEM. * (*p < 0.05; **p < 0.01; ***p < 0.001) was considered different from control group
Exosomal LINC00313 enhanced tumor progression and macrophage polarization in vivo
The effect of exosomal LINC00313 was then validated in vivo. The NCl-H1299 cells were transfected with LINC00313 overexpression or knockdown lentivirus to construct the stable expression cell lines. The transfection efficiency of LINC00313 was detected by RT-qPCR (Fig. 5A). Then, to explore the role of LINC00313 in tumor growth and M2 macrophage differentiation, the mouse-xenograft models were established by subcutaneous injection of the NCl-H1299 cells with stable overexpression or knockdown of LINC00313. To explore whether exosomes played roles on tumor growth and M2 macrophage differentiation, the exosomes inhibitor GW4869 was intra-tumorally injected after tumor implantation. The mice were divided into 4 groups: H1299 group (subcutaneously injected with H1299 cells with negative control lentivirus infection and intra-tumorally injected with DMSO), H1299 + GW4869 group (subcutaneously injected with H1299 cells with negative control lentivirus infection and intra-tumorally injected with GW4869), H1299 + LV-LINC00313 OE group (subcutaneously injected with H1299 cells with LINC00313 overexpression infection and intra-tumorally injected with DMSO), and H1299 + LV-LINC00313 KD group (subcutaneously injected with H1299 cells with LINC00313 knockdown infection and intra-tumorally injected with DMSO). As shown in Fig. 5B–D, compared to the H1299 group, inhibition of exosomes by GW4869 inhibited tumor growth. Meanwhile, overexpression of LINC00313 promoted tumor progression, while knockdown of LINC00313 suppressed tumor growth (Fig. 5B–D).
Exosomal LINC00313 promoted tumor progression and macrophage polarization in vivo. The NCl-H1299 cells were transfected with LINC00313 overexpression or knockdown lentivirus to construct the stable expression cell lines. The NCl-H1299 cells were collected and counted. NCl-H1299 cells (2 × 107) were resuspended in 100 μL PBS and subcutaneously injected into the mice. The exosomes inhibitor GW4869 was intra-tumorally injected at 2.5 mg/g the next day after tumor implantation. A The transfection efficiency of LINC00313 was detected by RT-qPCR. B Pictures of tumors isolated. C The subcutaneous tumor volume was analyzed every 5 days. D The weight of tumor isolated from the mice was measured. E CD206 and CD163 expression levels were analyzed by immunohistochemical staining. F Expressions of LINC00313 and miR-135a-3p in isolated tumor macrophages were detected by qRT-PCR. G STAT6 expression in isolated tumor macrophages was detected by Western blotting. H The subpopulations of CD206 and CD163 in isolated tumor macrophages were analyzed by flow cytometry. I The expressions of M1 markers (iNOS and CD86) and M2 markers (CD206, CD163 and Fizz1) in isolated tumor macrophages were assessed by Western blotting. J The mRNA levels of M1 markers (TNF-α and IL-1β) and M2 marker (TGF-β) in isolated tumor macrophages were measured by qRT-PCR. n = 3, data were described by mean ± SEM. * (*p < 0.05; **p < 0.01; ***p < 0.001) was considered different from control group
To further clarify the distribution of macrophages in NSCLC, the expression levels of CD206 and CD163 were analyzed by immunohistochemical staining. The result showed that overexpression of LINC00313 increased the levels of CD206 and CD163, while knockdown of LINC00313 suppressed the levels of CD206 and CD163 (Fig. 5E). Then, macrophages were isolated from NSCLC. The levels of LINC00313, miR-135a-3p in isolated macrophages were detected by qRT-PCR and the STAT6 expression was assessed by Western blotting. Overexpression of LINC00313 increased the expressions of LINC00313 and STAT6 and suppressed the level of miR-135a-3p, while knockdown of LINC00313 exerted the opposite effect (Fig. 5F–G). Furthermore, the subpopulations of M2 macrophages were estimated via flow cytometry. The results showed a lower ratio of CD68+CD206+ and CD68+CD163+ after knockdown of LINC00313, while CD68+CD206+ and CD68+CD163+ levels were higher after overexpression of LINC00313 (Fig. 5H). The results of Western blotting showed that knockdown of LINC00313 increased the levels of M1 markers (iNOS and CD86) and decreased the levels of M2 markers (CD206, CD163 and Fizz1) (Fig. 5I). On the contrary, overexpression of LINC00313 repressed iNOS and CD86 and promoted CD206, CD163 and Fizz1 (Fig. 5I). Meanwhile, qRT-PCR suggested that GW4869 and knockdown of LINC00313 increased the levels of M1 markers (TNF-α and IL-1β) and decreased the level of M2 marker (TGF-β) (Fig. 5J). Overexpression of LINC00313 decreased the levels of M1 markers (TNF-α and IL-1β) and increased the level of M2 marker (TGF-β) (Fig. 5J).
Discussions
TAMs are important players in cancer metastasis and immunosuppression [24, 25]. Most TAMs are M2 macrophages which are considered to promote cancer progression, while M1 macrophages often predict a good prognosis [26, 27]. Therefore, the M1/M2 macrophage differentiation can be a potential direction for cancer treatment. In this paper, it was revealed that LINC00313 acted a key role in M2 macrophage differentiation in NSCLC. The results showed that cancer cell-derived LINC00313 promoted M2 macrophage differentiation in NSCLCs by sponging miR-135a-3p, resulting in the up-regulation of STAT6.
In recent years, numerous studies have used THP-1 to induce macrophages [28, 29]. Compared with human peripheral blood mononuclear cells (PBMCs), THP-1 cell lines are easier to be cultured and expanded in the laboratory, and they have a more stable gene background. In addition, there is no individual difference in THP-1 cell line, which is conducive to the reproduction of experimental results. Therefore, in this study, 25 ng/mL PMA was added to THP-1 cells for 24 h to induce macrophage differentiation. M1/M2 polarization brought about the distinct surface marker profiles. Koelink et al. suggested thatCD206 is critical to maintaining macrophage differentiation [30]. Interestingly, Jaguin et al. in 2013 observed no difference in the CD206 expression between M1 and M2 macrophages and proposed that the specificity of M2 macrophages was upregulation of CD200R membrane glycoprotein [31]. CD163 is considered a marker for M2. Murray et al. supported an approach that the markers (or lack of marker expression used) are combined to assess the activation of macrophages [32]. Therefore, flow cytometry, Western blotting and ELISA were performed to detect the levels of iNOS, CD86, TNF-α, IL-1β, CD206, CD163, Fizz1, and TGF-β. Studies have shown there are high expression levels of iNOS, CD86, TNF-α and IL-1β in M1 macrophages. Meanwhile, mannose-binding lectin receptors (CD206 and CD163), Fizz1, and TGF-β are found in M2 macrophages [33].
In the present study, it was found that treatment with CM of NCl-H1299 induced a significant increase of the expression of M2 marker in THP-1 cells, indicating that cancer cells can induce M2 macrophage differentiation. Exosomes are small membrane vesicles of endocytic origin that are secreted by most cells in culture, which functions as the mediator of the cell–cell signal exchange [34]. M2 macrophage differentiation could be induced through the mediating function of exosomes. For example, exosomes from adipose-derived stem cells polarize M2 macrophages, thereby mitigating adipose inflammation and obesity [35]. In the injured spinal cord, the exosomes released by mesenchymal stem cells specifically target M2 macrophages to improve the recovery [36]. In the present study, it was found that GW4869, an inhibitor of exosome production, could inhibit the effect of CM of H1299 on M2 macrophage differentiation. Exosomes isolated from H1299 CM showed the same effect as the CM-H1299. All these data indicated that H1299 influences M2 macrophages through secreting exosomes.
LncRNAs can act as a type of exosomal content to regulate various biological processes [14, 37]. However, the role of lncRNAs in the cancer cell-induced M2 macrophage differentiation has been rarely investigated. LINC00313 has been shown to be correlated with a poor prognosis in cancers, indicating its important roles in tumor progression [10,11,12,13]. For instance, Zhai et al. showed that LINC00313 increases the CDK6 expression through competitive binding to miR-4677-3p, facilitating the development of cervical carcinoma [11]. Besides, Chen et al. suggested that LINC00313 is increased in osteosarcoma, and its LINC00313 suppresses osteosarcoma cell proliferative, migratory and invasive abilities while promoting apoptosis and autophagy [12]. In addition, Wang et al. reported that LINC00313 is significantly up-regulated in NSCLC, which may provide a novel biomarker for NSCLC treatment [10]. In this study, the level of LINC00313 in the exosomes extracted from NSCLC cells was detected, and a significant increase of LINC00313 level in exosomes from NSCLC cells was found. It is reported that exosomes release lncRNAs to macrophages to mediate macrophage differentiation [38, 39]. In this study, H1299-CM was cultured with the PMA-stimulated THP-1 cells. H1299-CM treatment increased the LINC00313 expression in THP-1 cells. All these data indicated that LINC00313 from NSCLC cells participates in M2 macrophage differentiation. To explore the role of LINC00313 in macrophages, LINC00313 in THP-1 cells was overexpressed or knocked down, and it was found that LINC00313 could promote the M2 differentiation of THP-1 cells. Knockdown of LINC00313 in THP-1 cells inhibited the M2 differentiation and promoted the M1 differentiation of THP-1 cells. The LINC00313-knockdown H1299-CM lost its effect of promoting M2 macrophage differentiation. All these data suggested that H1299 cells promote M2 macrophage differentiation through secreting exosomal LINC00313.
MiRNAs are a class of small non-coding RNA molecules of 21–25 nucleotides, which post-transcriptionally regulate the protein expression [40]. LncRNAs act as the sponge for miRNAs in gene expression regulation [41]. In this study, the results of bioinformatics assay showed the potential binding sites of LINC00313 and miR-135a. Moreover, STAT6 may be a target of miR-135a. Luciferase reporter assay and Ago2-RIP proved that LINC00313 could bind to miR-135a, and STAT6 was a direct target of miR-135a. It was further revealed by qRT-PCR that overexpression of LINC00313 decreased the miR-135a level in the THP-1 cells. STAT6 is an important transcriptional factor during the process of M2 macrophage differentiation [42]. Then Western blotting was performed to determine the effect of LINC00313 and miR-135a on the STAT6 expression. It was found that LINC00313 knockdown decreased the expression of STAT6, leading to reduced M2 marker expression, while overexpression of STAT6 could reverse the effect of LINC00313 knockdown. In vivo studies showed that overexpression of LINC00313 promoted tumor progression and M2 macrophage differentiation. These results demonstrated that exosomal LINC00313 promotes tumor growth and M2 macrophage differentiation.
Studies have shown that macrophages play important roles in tumor progression and metastasis [43]. In this study, the specific regulatory mechanism of macrophage differentiation was explored. However, there are still many limitations in this study. For instance, the biological functions of macrophages in NSCLC cell migration or angiogenesis, the specific function of macrophages such as phagocytosis, and the subsequent cytokine production and cytotoxicity remain unknown. Therefore, the role of macrophages in NSCLC will be further described in future studies.
Conclusions
In summary, cancer cell-derived exosomal LINC00313 promotes M2 macrophage differentiation in NSCLC by up-regulating STAT6 as miR-135a-3p sponge. This study provides a new mechanism and direction for prevention of M2 macrophage differentiation in NSCLC.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–94.
Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–55.
Zhang J, Li H, Wu Q, Chen Y, Deng Y, Yang Z, Zhang L, Liu B. Tumoral NOX4 recruits M2 tumor-associated macrophages via ROS/PI3K signaling-dependent various cytokine production to promote NSCLC growth. Redox Biol. 2019;22: 101116.
Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC, Chen YY, Liu YC, Hong TH, Yu SL, Chen JJ, et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. 2015;5:14273.
Chen Z, Bu N, Qiao X, Zuo Z, Shu Y, Liu Z, Qian Z, Chen J, Hou Y. Forkhead box M1 transcriptionally regulates the expression of long noncoding RNAs Snhg8 and Gm26917 to promote proliferation and survival of muscle satellite cells. Stem Cells. 2018;36(7):1097–108.
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–45.
Chen X, Zhou X, Lu D, Yang X, Zhou Z, Chen X, Chen Y, He W, Feng X. Aberrantly expressed long noncoding RNAs are involved in sevoflurane-induced developing hippocampal neuronal apoptosis: a microarray related study. Metab Brain Dis. 2016;31(5):1031–40.
Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. Chem Med Chem. 2014;9(9):1932–56.
Wang J, Zhang L, Wang C, Chen Y, Sui X. LINC00313/miR-4429 axis provides novel biomarkers for the diagnosis and prognosis of non-small cell lung cancer. Acta Biochim Pol. 2022;69(2):343–8.
Zhai Y, Liu Y, Wang Z, Wang W, Zhou J, Lu J. Long non-coding RNA LINC00313 accelerates cervical carcinoma progression by miR-4677-3p/CDK6 axis. Onco Targets Ther. 2021;14:2213–26.
Chen H, Wahafu P, Wang L, Chen X. LncRNA LINC00313 knockdown inhibits tumorigenesis and metastasis in human osteosarcoma by upregulating FOSL2 through sponging miR-342-3p. Yonsei Med J. 2020;61(5):359–70.
Shao L, He Q, Liu Y, Liu X, Zheng J, Ma J, Liu L, Li H, Li Z, Xue Y. UPF1 regulates the malignant biological behaviors of glioblastoma cells via enhancing the stability of Linc-00313. Cell Death Dis. 2019;10(9):629.
Chen L, Yang W, Guo Y, Chen W, Zheng P, Zeng J, Tong W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE. 2017;12(9): e0185406.
Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 2018;17(21–22):2411–22.
Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, Gurley EC, Liang G, Chen W, Lai G, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes hepatic stellate cell activation and cholestatic liver fibrosis. Hepatology. 2019;70(4):1317–35.
Du C, Xie H, Zang R, Shen Z, Li H, Chen P, Xu X, Xia Y, Tang W. Apoptotic neuron-secreted HN12 inhibits cell apoptosis in Hirschsprung’s disease. Int J Nanomed. 2016;11:5871–81.
Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A, Sjostrand M, Gabrielsson S, Lotvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9.
Hu W, Ru Z, Xiao W, Xiong Z, Wang C, Yuan C, Zhang X, Yang H. Adipose tissue browning in cancer-associated cachexia can be attenuated by inhibition of exosome generation. Biochem Biophys Res Commun. 2018;506(1):122–9.
Chu D, Huttenlocher PR, Levin DN, Towle VL. Reorganization of the hand somatosensory cortex following perinatal unilateral brain injury. Neuropediatrics. 2000;31(2):63–9.
Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95.
Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ. Enhancer-mediated control of macrophage-specific arginase I expression. J Immunol. 2004;172(12):7565–73.
Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56(3):347–59.
Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51.
Chen JJ, Yao PL, Yuan A, Hong TM, Shun CT, Kuo ML, Lee YC, Yang PC. Up-regulation of tumor interleukin-8 expression by infiltrating macrophages: its correlation with tumor angiogenesis and patient survival in non-small cell lung cancer. Clin Cancer Res. 2003;9(2):729–37.
Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42(6):717–27.
Ma J, Liu L, Che G, Yu N, Dai F, You Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer. 2010;10:112.
Spalinger MR, Sayoc-Becerra A, Santos AN, Shawki A, Canale V, Krishnan M, Niechcial A, Obialo N, Scharl M, Li J, et al. PTPN2 regulates interactions between macrophages and intestinal epithelial cells to promote intestinal barrier function. Gastroenterology. 2020;159(5):1763-1777.e1714.
Zong Z, Zou J, Mao R, Ma C, Li N, Wang J, Wang X, Zhou H, Zhang L, Shi Y. M1 macrophages induce PD-L1 expression in hepatocellular carcinoma cells through IL-1β signaling. Front Immunol. 2019;10:1643.
Koelink PJ, Bloemendaal FM, Li B, Westera L, Vogels EWM, van Roest M, Gloudemans AK, van’t Wout AB, Korf H, Vermeire S, et al. Anti-TNF therapy in IBD exerts its therapeutic effect through macrophage IL-10 signalling. Gut. 2020;69(6):1053–63.
Jaguin M, Houlbert N, Fardel O, Lecureur V. Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin. Cell Immunol. 2013;281(1):51–61.
Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20.
Buchacher T, Ohradanova-Repic A, Stockinger H, Fischer MB, Weber V. M2 Polarization of human macrophages favors survival of the intracellular pathogen Chlamydia pneumoniae. PLoS ONE. 2015;10(11): e0143593.
Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–79.
Zhao H, Shang Q, Pan Z, Bai Y, Li Z, Zhang H, Zhang Q, Guo C, Zhang L, Wang Q. Exosomes from adipose-derived stem cells attenuate adipose inflammation and obesity through polarizing M2 macrophages and beiging in white adipose tissue. Diabetes. 2018;67(2):235–47.
Lankford KL, Arroyo EJ, Nazimek K, Bryniarski K, Askenase PW, Kocsis JD. Intravenously delivered mesenchymal stem cell-derived exosomes target M2-type macrophages in the injured spinal cord. PLoS ONE. 2018;13(1): e0190358.
Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, Chen K, Zhang Y, Liu H, Gan L, et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37(28):3822–38.
Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci. 2018;19(10):2958.
Li J, Zhang Q, Jiao H. LncRNA NRON promotes M2 macrophage polarization and alleviates atrial fibrosis through suppressing exosomal miR-23a derived from atrial myocytes. J Formos Med Assoc. 2021;120(7):1512–9.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Wu XS, Wang F, Li HF, Hu YP, Jiang L, Zhang F, Li ML, Wang XA, Jin YP, Zhang YJ, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18(10):1837–53.
Binnemars-Postma K, Bansal R, Storm G, Prakash J. Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer. FASEB J. 2018;32(2):969–78.
Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99(Pt B):180–5.
Acknowledgements
We would like to thank all the researchers and subjects for their contributions.
Author information
Authors and Affiliations
Contributions
WK conceived and designed the study, and drafted the manuscript. LZ and YC collected, analyzed and interpreted the experimental data. ZY and ZZ revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of 900TH Hospital of Joint Logistics Support Force and conducted in accordance with the ethical standards.
Informed consent
Subjects signed the informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Figure S1. LINC01116 was related to the prognosis of NSCLC and up-regulated in exosomes from NSCLC cells.
(A) TCGA database showed that the five lncRNAs (LINC01116, LINC00968, LINC00313, lncRNA C1orf132 and lncRNA TMPO-AS1) were related to the prognosis of NSCLC. (B) qRT-PCR showed an increased LINC00313 level in NSCLC cells (H1299, A549, H1975) and MRC-5. n=3, data were described by mean ± SEM. * (*p<0.05; **p<0.01; ***p<0.001) was considered different from control group. (JPG 2198 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, W., Zhang, L., Chen, Y. et al. Cancer cell-derived exosomal LINC00313 induces M2 macrophage differentiation in non-small cell lung cancer. Clin Transl Oncol 24, 2395–2408 (2022). https://doi.org/10.1007/s12094-022-02907-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02907-7