Abstract
Cr(VI) is most toxic heavy metal and second most widespread hazardous metal compound worldwide. Present work focused on Cr(VI) reduction from synthetic solutions and polluted samples by Achromobacter xylosoxidans SHB 204. It could tolerate Cr(VI) up to 1600 ppm and reduce 500 ppm with 4.5 chromium reductase enzyme units (U) having protein size 30 kDa. Changes in morphology of cells on interaction with Cr(VI) metal ion was also studied using SEM–EDX and FTIR. Microcosm studies in pollutant samples for Cr(VI) reduction and adsorption isotherm with biomass of bacterium was best fitted with Langmuir model along with kinetic studies. This study focuses on significance of Cr reduction from synthetic solutions and polluted samples by A. xylosoxidans SHB 204 and its potential for bioremediation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Release of untreated effluents containing chromium (Cr) and other heavy metals in industrial effluents creates environmental pollution [1]. High concentrations of Cr in polluted samples enters human body through food chain creating carcinogenic, mutagenic and other tissue damaging effects and health hazards [2, 3]. Physico chemical methods practiced in heavy metal remediation fail to remove low concentrations of Cr in effluents [4]. Therefore biological methods such as biosorption, bioaccumulation, biomineralisation, bioleaching, enzymatic removal etc. are looked as options for removal of Cr in effluents and soil [5, 6]. These methods are advantageous due to low cost, insitu application, regeneration of biosorbents and easy waste disposal etc. Microbial reduction of toxic Cr(VI) to nontoxic Cr(III) is by extracellular and intracellular enzymes. Cr(VI) is used as terminal electron acceptor in membrane bound anaerobic respiration of bacteria. Uptake of metal ions by bacteria is due to complexation, ion exchange, adsorption and precipitation [7, 8]. Bacterial biomass is efficient in adsorption of heavy metal ions, consequently adsorption time and design of metals onto adsorbents are known from fitness of kinetics models [9]. Most of the literature supports Langmuir and Freundlich models as the best models for metal adsorption studies. Objective of the current study was to evaluate the ability of Cr(VI) tolerant Achromobacter xylosoxidans SHB 204 in reducing Cr from synthetic solutions and its possible removal from polluted environment.
Materials and Methods
Isolation and Identification of Heavy Metal Resistant Bacteria
Heavy metal contaminated samples like soil (Industrial area Balanagar, Hyderabad), sludge (paint and steel industry of Jeedimetla Hyderabad), drainage water (pharma industries, Patancheru, and Katedan industrial areas Hyderabad) were collected and subjected to liquid enrichment culturing by taking 10 gm or 10 ml in a total volume of 100 ml distilled water with addition of heavy metals like Cr, Ni, Pb 100 ppm each together in 250 ml conical flasks and kept for incubation at 37 °C, 200 rpm for 7 days. Samples from each flask were serially diluted and inoculated onto nutrient agar (NA) plates amended with Cr(VI) metal ion (100–2000 ppm). Morphologically similar colonies growing at highest concentration of Cr containing plate were picked and purified by re streaking on similar metal ion plates. About 50 colonies were selected randomly and sub cultured onto NA slants and preserved in refrigerator for further evaluation. Each colony was studied for its Cr resistance in broth culture by measuring its growth (turbidity) at different Cr concentrations. Isolate (SHB 204) growing at highest Cr concentration (1600 ppm) was selected for further detailed evaluation. Selected bacterium was identified by staining, morphological, cultural, microscopy, biochemical and 16srRNA sequencing. Biochemical tests such as indole, methyl red, Voges proskauer, and citrate utilization and other biochemical profile of bacterium was determined using Bergys manual of determinative bacteriology [10].
Molecular Characterization by 16S rRNA Sequencing
Selected isolate SHB 204 was characterized to species level by 16S rRNA sequencing. Pure bacterial colony was picked up with a sterile toothpick, and suspended in 0.5 ml of sterile saline in 1.5 ml eppendorf tube, centrifuged at 10,000 rpm for 10 min, supernatant was removed; the pellet was suspended in 0.5 ml of Insta Gene matrix (Bio-Rad, USA). This mixture was incubated at 56 °C for 30 min and then heated at 100 °C for 10 min. After heating, sample was centrifuged as above and supernatant was used for PCR. This sample was run on 0.8% agarose gel and band was observed.
PCR and Purification of PCR Products
1 ml of template DNA was added in 20 µl PCR reaction solution and 27F/1492R primers were used, and then 35 amplification cycles was performed at 94 °C for 45 s, 55 °C for 60 s and 72 °C for 60 s. Positive control (E. coli genomic DNA) and a negative control in the PCR was included. Unincorporated PCR primers and dNTPs from PCR products were removed by using Montage PCR clean up kit (Millipore).
Sequencing
Purified PCR products were sequenced using 2 primers (27F-AgA gTT TgA TCM TGG CTC Ag, 1492R-TAC ggY TAC CTT gTT ACg ACT T), (518F-CCA gCA gCC gCg gTA ATA Cg, 800R-TAC CAg ggT ATC TAA TCC) with Big Dye terminator cycle sequencing kit. Sequencing products were resolved on automated DNA sequencing system (Applied BioSystems, model 3730XL, USA). (This part of work was done with help of MACROGEN international corporation, Korea). Sequence procured was analysed using BLAST tool. Phylogenetic tree was constructed using Mega 4 software. 16S rRNA sequence was submitted to EMBL nucleotide sequence submission and accession number received.
Hexavalent Chromium Reduction by A. xylosoxidans SHB 204 Under Aerobic and Anaerobic Conditions
250 ml flasks containing 100 ml nutrient broth with Cr(VI) (100 ppm) were inoculated with 2% overnight grown SHB 204 bacterium (OD 0.6) and incubated at 37 °C, 200 rpm for 24–72 h. Similarly anaerobic culturing was done in sealed 180 ml serum vials without shaking. 1 ml samples were drawn at every 12 h interval, Cr(VI) concentration and extracellular chromium reductase enzyme activity were determined. Concentration of remaining Cr(VI) was determined by atomic absorption spectrophotometer (AAS) (A Analyst, 700, Perkin Elmer). Samples for AAS were processed by treating with 5% nitric acid, followed by filtration (Wattman No. 41) [11]. For enzyme activity 200 µl culture supernatant was added to 1.0 ml 1 mM K2Cr2O7(0.2 M phosphate buffer pH 7.4), incubated for 15 min at 37 °C, 1 ml TCA (30%) was added to stop enzyme reaction. Tubes were cemtrifuged and OD of supernatant was determined at 540 nm. K2Cr2O7 without enzyme served as control. One unit of Cr(VI) reductase activity is defined as decrease in 1.0 µM Cr(VI) ml−1 min−1 at 37 °C, protein content in enzyme solution was determined by Bradford method [12].
Chromium Reductase and Partial Purification
Overnight culture (100 ml) of A. xylosoxidans SHB 204 was centrifuged and supernatant was used for extracellular enzyme as described earlier. Cell pellet suspended in Tris buffer (40 ml, 10 mM, pH 7) lysed using French press (TS-series 0.75KW, Constants System UK) at 30 kV. Cell lysate was centrifuged (4000 rpm, 10 °C, 10 min) pellet and supernatant were separated; pellet was suspended in 10 ml Tris buffer. Each 10 ml (Extracellular/pellet/supernatant) was added with10 ppm Cr(VI) and incubated at 37 °C, 200 rpm for 6 h and analyzed for decrease in Cr(VI) content and enzyme activity [12]. Enzyme purification was done by ammonium sulfate (20–80%) precipitation, dialysis and gel filtration (Sephadex G-50) [13]. Molecular weight of enzyme was determined by 12% SDS-PAGE at a run time of one and half hour (Mini gel cassette, Biorad tetra pack, Coomassie briliant blue R-250) [14].
Scanning Electron Microscopy (SEM)–Energy Dispersive X-ray Spectroscopy (EDS) and Fourier Transform Infra Red Spectroscopy Analysis
Cell pellet of 24 h grown SHB 204 was shaken with 100 ppm Cr(VI) for 72 h, centrifuged, pellet was ethanol washed (10–100%), observed under SEM (ZEISS, model EVO 18, dept physics, OU) and analyzed with EDS [15]. Functional groups and chemical nature of the extracted compounds was detected using FTIR. Lyophilized active dry samples were crushed with KBr into thin pellet with a pressure of 5–6 tons cm−2 with the help of hydraulic press and analyzed on Shimadzu UV FT-IR spectrometer in the wave range 4000–400 cm−1. Surface changes of treated and untreated cells were observed [16].
Microcosm Studies of the Sludge, Sewage and Tannery Samples by A. xylosoxidans SHB 204
Patancheru industrial area outlet, Jeedimetla pharma industry, Katedan area where textile, printing, dyeing wastes were dumped was selected for collection of sludge, sewage and tannery samples. Samples collected in sterile bottles were processed within 6 h. Chemical analysis was done at Institute of preventive medicine (IPM), water analysis center, Hyderabad. Processed samples were amended with additional Cr(VI) to make it 100 ppm. To these samples overnight grown 2% SHB 204 inoculum, pellet of same strain (1 g/100 ml) and 50% crude enzyme (v/v) (SHB 204) were added separately to 250 ml flasks containing 100 ml pollutant samples and incubated at 37 °C, 200 rpm for 72 h. Samples (2 ml) were drawn at regular intervals and observed for decrease in Cr(VI) content and Cr reductase enzyme activity.
Adsorption Isotherms and Kinetics by Modeling
Langmuir model and first and second order rate kinetics were used and found to be suitable to design the method [17, 18].
Statistical Analysis
All experiments were performed in triplicates, twice at different occasions. Mean values of different variables were calculated and compared using two way ANOVA without replication to indicate any significance between variables. The result was considered significant if p ≤ 0.05.
Results and Discussion
Isolation and Identification of Heavy Metal Resistant Bacteria
Toxic metal resistant bacteria were isolated by enrichment culturing from soil and water samples collected from varied polluted locations. One isolate labeled as SHB 204 was selected from 50 isolates which was able to grow on high concentrations (1600 ppm) of Cr(VI) (data not shown). It is reported that Pseudomonas aeruginosa and Serratia marcescens were tolerant to 200 ppm and 150 ppm Cr(VI) respectively [19]. Isolate from tannery effluent (Pseudomonas, aeromonas, micrococcus), Bacillus sphericus is reported to be tolerant to 800 ppm Cr(VI) [20, 21]. In comparison SHB 204 bacterial strain is found to be promising tolerance for Cr(VI). Isolate SHB 204 was Gram negative rod shaped bacterium, biochemically indole and methyl red negative and Voges proskauer and citrate utilization positive. The identity of SHB 204 bacterium was determined by 16S rRNA gene sequence analysis and gel image of the amplified sequence is shown in (Fig. S1). Strain SHB 204 showed 98% gene sequence similarity with Achromobacter xylosoxidans, which was analyzed using EZ-taxon database and also by phylogenic tree construction using MEGA version 4.0 (Fig. 1). Sequence of isolate had 1440 bp and deposited in the EMBL-Genbank database through BLAST (http://www.ncbi.nlm.nih.gov/BLAST) [22] with the accession no. LN610698.
Hexavalent Chromium Reduction by A. xylosoxidans SHB 204 Under Aerobic and Anaerobic Conditions
Achromobacter xylosoxidans SHB 204 reduced 100–500 ppm (total reduction) Cr(VI) with the production of 0.5–4.5 U Cr reductase enzyme in 24–72 h under aerobic conditions. In anaerobic conditions 48% of (10–100 ppm) Cr(VI) reduction was observed with 1.5 U Cr reductase (Fig. S2a and S2b). Previous studies showed that rhizobacterial strain 39 was able to reduce 15 ppm of Cr from synthetic solutions [23], P. aeruginosa could tolerate and reduce Cr(VI) up to 75 ppm [24]. Microbacterium sp. MP30 grew aerobically in the presence of 15 mM Cr but could not reduce it. However it reduced 100 µM Cr under anaerobic conditions [25]. A halophilic Nesterenkonia sp. MF2 was tolerant to 600 mM Cr and completely reduced 200 µM Cr(VI) in 24 h under aerobic condition [26]. There are reports on anaerobic reduction of Cr(VI) to Cr(III) by Achromobacter sp. [2] where as very fewer reports are observed for the reduction of Cr(VI) under both aerobic and anaerobic conditions. In comparison, our isolate SHB 204 showed better Cr(VI) reduction in both aerobic and anaerobic conditions.
Chromium Reductase and Partial Purification
Cr reductase activity of A. xylosoxidans SHB 204 in whole cell, cell free extract and cell wall was observed. Cr reduction and enzyme activity in extracellular medium was high (55%, 5.8 U) (Table 1). Purification profiling of chromium reductase process reveals that crude enzyme was 18.33 U/mg, as the purification steps such as dialysis and gel filtration were carried out enzyme activity was improved by 30 U/mg. Partially purified enzyme protein run on 12% SDS—PAGE was compared with controls as reported in literature and the isolate A. xylosoxidans SHB 204 showed protein expression on 30 kDa corresponding to Cr reductase enzyme [27] (Fig. S3).
SEM and EDS Study and FTIR Analysis
Ultra structure of 100 ppm Cr treated and untreated A. xylosoxidans SHB 204 cells were observed in SEM (Fig. S4a and S4b). These observations reveal that Cr untreated cells appeared regular in shape with smooth surfaces and Cr treated cells appeared slightly bulged and precipitated amorphous particles were found to be present adhered on the surface. These observations indicate effective binding of metal ions by cell wall component of this Gram negative bacterium. Similar observation is seen in a study, which showed that alteration in morphology of (Bacillus sp. ES 29) cells take place on exposure with Cr metal ion [28]. In FTIR analysis surface characterization of cells with and without chromium treatment revealed that mode of Cr(VI) uptake by cells is extracellular and cell wall consists of chemical functional groups which play important role in biosorption. Bands observed at 3736–3904 and 3416 cm−1 are indicative of hydroxyl and asymmetric stretching of amines of proteins on the bacterial surface. The peaks at 1200–900 cm−1 was observed due to stretching vibrations of C–C, C–O–C and CO characteristic of carbohydrates exopolysacharide (EPS), thus presence of EPS component could also be the reason for metal absorption [29] (Fig. S5a and S5b).
Microcosm study of Sludge, Sewage and Tannery Samples Using A. xylosoxidans SHB 204
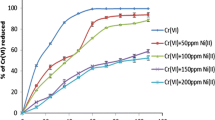
To evaluate Cr reduction by A. xylosoxidans SHB 204, microcosm studies were carried out to evaluate metal reduction/removal from sludge, sewage and tannery samples. Physicochemical examination and metal ion concentrations (Cr, Ni, Pb) of samples are shown in Table 2 of ESM. Reduction of Cr(VI) from sludge, sewage and tannery samples is shown in Fig. 2a, b, c. Cr(VI) concentrations in most of tanneries and industrial effluents were found to be 10–40 ppm. This study was carried out using 100 ppm of Cr to meet the tolerance and removal efficiency of the bacterium under study. Complete reduction of Cr(VI) was observed in the samples within 6 h of incubation with A. xylosoxidans SHB 204 and Cr amended samples showed variations in metal reduction. Cr reductase enzyme activity was relatively more in Cr amendments. Addition of actively growing culture resulted complete reduction in Cr(VI) and increase in Cr reductase to 5.5 U. Increased enzyme activity observed in above case could be due to presence of other metal ions in polluted samples. Biosorption studies also showed variation in sorption (86-100%), which could be due to chemical composition of the pollutant sample, presence of other metal ion (Ni and Pb) and high toxicity due to generation of reactive oxygen species during its reduction process. A consistent result was observed with A. denitrificans in 50% Cd removal from sediments [30], Enterobacter sp. in 85% Cd removal from industrial pollutants [31], similarly 85% of Zn and Pb biosorption was observed by Achromobacter xylosoxidans sp. [32]. With respect to above observations there are very few studies on complete removal of Cr(VI) from polluted industrial effluents by Achromobacter xylosoxidans sp. and our study focuses on ability of our bacterium in high efficient reduction of Cr(VI) from environmental samples. Extracellular enzyme was fairly able to reduce 80–95% of Cr(VI) and in a study partially purified Cr reductase enzyme produced by Bacillus methylotrophicus was able to reduce 91% of Cr(VI) from tannery samples [12]. This is found to be advantageous for environmental studies as it is non- toxic to other bacterial cells, lower cost and simpler production process involved. Significance of this study was biosorption coupled bio reduction considered to be conventional method in removal of metal ions from environmental samples.
a Cr(VI) reduction from sewage, sludge and Tannery samples by inoculating with 2% actively growing culture of A. xylosoxidans SHB 204, b Cr biosorption from sewage, sludge and Tannery samples by biomass of A. xylosoxidans SHB 204, c Cr(VI) bio reduction from Sewage, Sludge and Tannery samples by partially purified Cr reductase enzyme of A. xylosoxidans SHB 204
Adsorption Isotherms and Kinetics Study
Adsorption isotherms and kinetics of adsorption using modeling studies was carried out and in Langmuir adsorption isotherm study biosorption isotherm was plotted for Cr uptake (Q) against remaining metal concentration Cf (final concentration) and result was fitted and mathematically expressed using A. xylosoxidans SHB 204. The coefficient of determination (R2) was 0.96253 and close to 1, indicates higher affinity of biomass in biosorption of Cr and favourable adsorption of Cr(VI) on the cell surface. Rate kinetics was calculated at specific time intervals of biosorption of Cr onto biomass and data obtained and plotted against time to determine a suitable kinetic model. The adsorption data was fitted into first order and second order kinetics (Fig. S6a, b and c). The correlation coefficient R2 was 0.99347 for first order, 0.95907 for second order states that pseudo first order best fitted with experimental values and it is found closer to 1 [33]. According to Ghani et al. kinetics study for adsorption of metal ions Cd, Cu and Pb etc. on varied adsorbents was carried out [34] and our study is considered as first kinetic study with A. xylosoxidans biomass as adsorbent in Cr(VI) removal from industrial pollutant sites.
Conclusion
This study shows that A. xylosoxidans SHB 204 can tolerate high concentration of heavy metal and reduced Cr(VI) both aerobically and anaerobically. The biomass of bacterium and chromium reductase was able to reduce Cr(VI) ions from synthetic as well as contaminated environmental samples. Adsorption process is best fitted into Langmuir model and it follows first order kinetics. Further research and exploration is needed to use A. xylosoxidans SHB 204 as a bioremediation tool both at lab scale and industrial sludge treatment plants.
References
Gibb HJ, Lees PS, Pinsky PF, Rooney BC (2000) Lung cancer among workers in chromium chemical production. Am J Ind Med 38:115–126. doi:10.1002/1097-0274(200008)38:23.3
Zhu W, Chai L, Ma Z, Wang Y, Xiao H, Zhao K (2008) Anaerobic reduction of hexavalent chromium by bacterial cells of Achromobacter sp. strain Ch1. Microbiol Res 163:616–623
Talapatra S, Banerjee S (2007) Detection of micronucleus and abnormal nucleus in erythrocytes from the gill and kidney of Labeo bata cultivated in sewage-fed fish farms. FCT 45:210–215. doi:10.1016/j.fct.2006.07.022
Zouboulis A, Loukidou M, Matis K (2004) Biosorption of toxic metals from aqueous solutions by bacteria strains isolated from metal-polluted soils. Process Biochem 39:909–916. doi:10.1016/S0032-9592(03)00200-0
Khan MY, Swapna TH, Hameeda B, Reddy G (2015) Bioremediation of heavy metals using biosurfactants. Adv Biodegrad Bioremediat Ind Waste. doi:10.3389/fmicb.2015.01555
Poguberović SS, Krčmar DM, Maletić SP, Kónya Z, Pilipović DDT, Kerkez DV, Rončević SD (2016) Removal of As (III) and Cr(VI) from aqueous solutions using “green” zero-valent iron nanoparticles produced by oak, mulberry and cherry leaf extracts. Ecol Eng 90:42–49. doi:10.1016/jecoleng2016.01.083
Knauer K, Behra R, Sigg L (1997) Adsorption and uptake of copper by the green alga scenedesmus subspicatus (chlorophyta) 1. J Phycol 33:596–601. doi:10.1111/j.0022-3646.1997.00596
Swapna TH, Narendra Kumar P, YahyaKhan M, Gopal reddy M, Hameeda B (2016) Bioreduction of Cr(VI) by biosurfactant producing marine bacterium Bacillus subtilis SHB 13. jsir 75:432–438. http://nopr.niscair.res.in/handle/123456789/34708
Hossain MA, Ngo HH, Guo W (2013) Introductory of Microsoft Excel SOLVER function-spreadsheet method for isotherm and kinetics modelling of metals biosorption in water and wastewater. J Water Sustain 3:223–237. doi:10.11912/jws.3.4.223-237
Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins, Baltimore. doi:10.1002/9781118960608
Jayabarath J, Shyam S, Arulmurugan R, Giridhar R (2009) Bioremediation of heavy metals using biosurfactants. Int J Biotechnol Appl 1:50–54. doi:10.9735/0975-2943.1.2.50-54
Mala JGS, Sujatha D, Rose C (2015) Inducible chromate reductase exhibiting extracellular activity in Bacillus methylotrophicus for chromium bioremediation. Microbiol Res 170:235–241. doi:10.1016/j.micres.2014.06.001
Sallau AB, Inuwa HM, Ibrahim S, Nok AJ (2014) Isolation and properties of chromate reductase from Aspergillus niger. Int J Mod Cell Mol Biol 3:10–21
Laemmli VK (1970) Cleavage of structural proteins cooling water of an electricity generating station. J Nat 277:680–685
Damodaran D, Raj MB, Vidya KS (2013) The uptake mechanism of Cd (II), Cr(VI), Cu (II), Pb(II), and Zn (II) by mycelia and fruiting bodies of Galerina vittiformis. Biomed Res Int. doi:10.1155/2013/149120
Maria Antonieta G, Maria Cristina M (2012) Purification of peptides from Bacillus strains with biological activity, chromatography and its applications. ISBN 978-953- 51- 0357-8
Langmuir I (1918) The adsorption of gases on plane surfaces of glass, mica and platinum. J Am Chem Soc 40:1361–1403. doi:10.1021/ja02242a004
Rathinam A, Maharshi B, Janardhanan SK, Jonnalagadda RR, Nair BU (2010) Biosorption of cadmium metal ion from simulated wastewaters using Hypnea valentiae biomass: a kinetic and thermodynamic study. Bioresour Technol 101:1466–1470. doi:10.1021/ja02242a004
Kafilzadeh F, Saberifard S (2016) Isolation and identification of chromium (VI)-resistant bacteria From Soltan Abad river sediments (Shiraz-Iran). Jundishapur J Health Sci. doi:10.17795/jjhs-33576
Sinha SN, Biswas M, Paul D, Rahaman S (2011) Biodegradation potential of bacterial isolates from tannery effluent with special reference to hexavalent chromium. Biotechnol Bioinform Bioeng 1:381–386. doi:10.6084/m9.figshare.1279416
Pal A, Paul A (2004) Aerobic chromate reduction by chromium-resistant bacteria isolated from serpentine soil. Microbiol Res 159:347–354. doi:10.1016/j.micres.2004.08.001
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr1
Pramono A, Retno Rosariastuti MMA, Ngadiman N, Prijambada ID (2013) Bacterial Cr(VI) reduction and its impact in bioremediation. J Ilmu Lingkung 11:120–131. doi:10.13140/2.1.3961.4086
Munawaroh HSH, Gumilar GG, Nandiyanto ABD, Kartikasari S, Kusumawaty D, Hasanah L (2017) Microbial reduction of Cr(VI) into Cr(III) by locally isolated Pseudomonas aeruginosa. Mater Sci Eng. doi:10.1088/1757-899X/180/1/012296
Pattanapipitpaisal P, Brown NL, Macaskie LE (2001) Chromate reduction and 16S rRNA identification of bacteria isolated from a Cr(VI) contaminated site. Appl Microbiol Biotechnol 57:257–261
Amoozegar MA, Ghasemi A, Razavi MR, Naddaf S (2007) Evaluation of Hexavalent chromium reduction by chromate resistant moderate halophile Nesterenkonia sp. strain MF2. Process Biochem 42:1475–1479. doi:10.1016/j.procbio.2007.07.001
Thacker U, Madamwar D (2005) Reduction of toxic chromium and partial localization of chromium reductase activity in bacterial isolate DM1. W J Microbiol Biotechnol 21:891–899. doi:10.1007/s11274-004-6557-7
Camargo F, Okeke B, Bento F, Frankenberger W (2003) In vitro reduction of hexavalent chromium by a cell-free extract of Bacillus sp. ES 29 stimulated by Cu2+. Appl Microbiol Biotechnol 62:569–573. doi:10.1007/s00253-003-1291-x
Deng K, Tian H, Zhang P, Ren X, Zhong H (2009) Synthesis and characterization of a novel temperature-pH responsive copolymer of 2-hydroxypropyl acrylate and aminoethyl methacrylate hydrochloric salt. Express Polym Lett 3:97–104. doi:10.3144/expresspolymlett.2009.13
Abyar H, Safahieh A, Zolgharnein H, Zamani I (2012) Isolation and identification of achromobacter denitrificans and evaluation of its capacity in cadmium removal. Pol J Environ Stud 21:1523–1527
Abbas SZ, Rafatullah M, Ismail N, Lalung J (2014) Isolation, identification, characterization, and evaluation of cadmium removal capacity of Enterobacter species. J Basic Microbiol 54:1279–1287. doi:10.1002/jobm.201400157
Bhattacharya A, Gupta A (2013) Evaluation of Acinetobacter sp. B9 for Cr(VI) resistance and detoxification with potential application in bioremediation of heavy-metals-rich industrial wastewater. Environ Sci Pollut Res 20:6628–6637. doi:10.1007/s11356-013-1728-4
Jain M, Garg VK, Kadirvelu K (2013) Chromium removal from aqueous system and industrial wastewater by agricultural wastes. Bioremediat J 17:30–39. doi:10.1080/10889868.2012.731450
Abdel-Ghani NT, El-Chaghaby GA (2014) Biosorption for metal ions removal from aqueous solutions: a review of recent studies. Int J Latest Res Sci Technol 3:24–42
Acknowledgements
Authors acknowledge the financial support and fellowship to THS by OU-DST PURSE and SERB Govt. of India (SR/FT/LS-17/2011) to carry out this research work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tadishetty Hanumanth Rao, S., Papathoti, N.K., Gundeboina, R. et al. Hexavalent Chromium Reduction from Pollutant Samples by Achromobacter xylosoxidans SHB 204 and its Kinetics Study. Indian J Microbiol 57, 292–298 (2017). https://doi.org/10.1007/s12088-017-0654-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-017-0654-4