Abstract
The definition of the number and nature of signal transduction pathways networking in the pathogenesis of osteosarcoma raised great interest. Intracellular calcium ions are important second messengers implicated in the control of cell death. The calcium concentration is regulated by signal transduction pathways, including the Phosphoinositides (PI) signaling. Phosphatydil inositol (4,5) bisphosphate (PIP2) is critical for many cellular activities. The levels of PIP2 are regulated by means of Phosphoinositide-specific Phospholipase C (PI-PLC) family of enzymes. We delineated the panel of expression of PI-PLC enzymes in four human osteosarcoma cell lines. In MG-63 cell line, PI-PLC β1, β2, β3, β4, γ1, γ2, δ1, δ3 and ε resulted expressed. In 143B cell line, PI-PLC β1, β2, β3, β4, γ1, γ2, δ1, δ3 and ε were expressed. In SaOS-2 cell line, PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3, ε and η1. In Hs888 cell line, PI-PLC β1, β3, β4, γ1, δ1, δ3, δ4, ε and η1 the administration of U-73122 to cultures briefly modifies the levels of PI-PLC transcripts. The obtained complete expression panel of PI-PLC isoforms will be a useful tool for further functional studies about the role of the PI signal transduction pathway in osteosarcoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma comprises less than 1 % of cancers diagnosed in the United States, occurring in less than 1000 patients per year (Mirabello et al. 2009a, b), and is the most common primary bone tumour in childhood and adolescence (Gatta et al. 2005).
The radiographic and histologic features, as well as the consistent spectrum of clinical presentations allow the diagnosis. The treatment includes surgery, radiotherapy and chemotherapy. Metastasizing tumours partially respond to current therapies and represent the primary cause of cancer related mortality (Meyers et al. 2005).
The complexity of osteosarcoma includes numerous abnormalities which were investigated (Gorlick et al. 2009). Progresses were achieved thanks to the use of epidemiologic features, genomic and molecular characterizations, and the study of osteosarcoma in animal models. A number of genetic abnormalities and environmental exposures were supposed to be involved in the development of osteosarcoma in laboratory models as well as in humans (Gorlick et al. 2009). Many efforts were made to bring multiple genetic risk factors together with epidemiologic and association data in order to identify a unifying recurrent event. However, despite the considerable improvement in the knowledge, no distinctive and specific dysregulation was identified and the pathogenesis of osteosarcoma is still largely unknown.
Osteosarcoma arises from a mesenchymal cell that has or can acquire the ability to produce osteoid (Gorlick et al. 2003; Marina et al. 2010). However, the cell of origin of osteosarcoma has been the subject of extensive discussions. A number of studies have also been performed as an attempt to decipher pathways associated with the pathogenesis of osteosarcoma. To define the number and the nature of the signal transduction pathways involved in the pathogenesis might improve the knowledge of the natural history of osteosarcoma. That also might help to refine the prognosis and to open the way to novel therapeutic strategies.
Intracellular calcium ions are important second messengers implicated in the control of cell death. Increases in calcium concentrations lead to necrosis or apoptosis following photodynamic therapy (Annunziato et al. 2003). Previous reports analyzed the role of calcium in osteosarcoma cells and the effect of concentration changes upon their viability. The antidepressant mirtazapine caused the death of MG-63 cells altering the cytosolic free calcium levels (Pan et al. 2006). In SaOS-2 cells, tissue factor VIIa and Xa induced transient intracellular calcium increase (Daubie et al. 2007). Calcium increase after treatment of UMR-106 cells with hematoporphyrin monomethyl ether and ultrasounds, followed by mitochondrial calcium reuptake, lead to cells death (Tian et al. 2010).
The calcium level is regulated by a complex network of signal transduction pathways, including the Phosphoinositides (PI) signaling. The regulation of PI signal transduction molecules is strictly related to a number of converting enzymes (Rhee et al. 1991). Phosphatydil inositol (4,5) bisphosphate (PIP2), one phosphorylated derivative of PI located mainly in the inner half of the plasma membrane lipid bilayer, is critical for many cellular activities, such as endo- and exocytosis, ion channel activity and cell motility. The levels of PIP2 are regulated by means of Phosphoinositide-specific Phospholipase C (PI-PLC) family of enzymes (Fukami et al. 2010).
Activated PI-PLC cleaves PIP2 into inositol trisphosphate (IP3) and diacylglycerol (DAG), both crucial molecules in signal transduction (Rhee et al. 1991). IP3, a small water-soluble molecule, diffuses rapidly to the cytoplasm, where it induces calcium release from the endoplasmic reticulum by binding to IP3-gated calcium-release channels located in the reticulum membrane. The initial increase induced by IP3 propagates as a wave through the cytoplasm often followed by a series of calcium spikes (Rhee et al. 1991). DAG remains bound to the membrane by its fatty acid tails and exerts two potential signalling roles. First, it can be further cleaved to release arachidonic acid, which either acts as a messenger or is used in the synthesis of eicosanoids (Tang et al. 2005). Second, DAG activates serine/threonine calcium-dependent protein kinase C enzymes (PKC). The calcium increase, induced by IP3, moves PKC to translocate from the cytoplasm to the cytoplasmic face of the plasma membrane. The translocation activates PKC, which subsequently phosphorylates specific serine or threonine residues on further target proteins (Rhee et al. 1991; Suh et al. 2008).
The eukaryote PI-PLC family comprises a related group of complex, modular, multi-domain enzymes which cover a broad spectrum of regulatory interactions, including direct binding to G protein subunits, small GTPases from Rho and Ras families, receptor and non-receptor tyrosine kinases and lipid components of cellular membranes (Rhee et al. 1991). PI-PLC enzymes were classified on the basis of amino acid sequence, domain structure and mechanism of recruitment. Thirteen mammalian PI-PLC enzymes were identified, divided into six subfamilies: β(1–4), γ(1–2), δ(1, 3, 4), ε(1), ζ(1), and η(1–2) (Suh et al. 2008).
Previous studies analyzed the expression of selected PI-PLC isoforms in human osteosarcoma. However, literature data lacked further information’s about others, such as ε and η subfamilies. The complete panel of expression of PI-PLC enzymes in tissues is critical for any kind of study. In fact, PI-PLC enzymes are strictly tissue specific (Suh et al. 2008; Bunney and Katan 2011), and our previous studies suggested that the expression panel is different in quiescent cells compared to the pathological counterpart (Lo Vasco et al. 2007, 2012). Therefore, knowledge of PI-PLC expression panel represents a preliminary tool in order to analyze the role of PI signal transduction pathway in the pathogenesis of osteosarcoma.
Materials and methods
Cell culture
We analyzed four cultured human osteosarcoma cell lines: MG-63, 143B, SaOS-2 and Hs888, obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured as previously described (Di Cristofano et al. 2010). Briefly, cells were grown under subconfluent or confluent conditions in medium, at 37 °C with 5 % of CO2. Cells were cultured in Dulbecco’s minimum essential medium (Sigma) supplemented with 10 % fetal bovine serum (GIBCO) with penicillin (100 μg/ml), streptomycin (100 U/ml) and sodium pyruvate. Cells were grown for 24 h, reaching a confluence of around 40–60 %. Experiments were independently repeated at least 3 times for each isoform. U-73122 (Sigma–Aldrich) was addicted to cultures at a concentration of 30 μmol in dimethyl sulfoxide (DMSO) (Takenouchi et al. 2005; Lo Vasco et al. 2010). Cultures were stopped 3 and 24 h after the administration of the molecule, and suspended in TRIzol reagent for molecular biology analysis. Experiments were independently repeated at least 3 times for each isoform. Cultures were stopped 3 and 6 h after the administration of U-73122 to be processed for western blot analyses. Experiments were independently repeated at least 2 times for each isoform.
RT-PCR
After the confluent monolayer was obtained, 106 cells for each experiment were detached and suspended in TRIzol reagent (Invitrogen Corporation, Carlsbad, CA). Total RNA was isolated following the manufacturer’s instructions. The purity of the RNA was assessed using a UV/visible spectrophotometer (SmartSpec 3000, Bio-Rad Laboratories, Hercules, California, USA). 1 μg total RNA was reverse transcribed using a High Capacity cDNA Reverse Transcription (RT) kit (Applied Byosystems, Carlsbad, California, USA) according to manufacturer’s instructions. Briefly, RT buffer, dNTP mix, RT random primers, multiscribe reverse transcriptase, RNase inhibitor and DEPC-treated distilled water were added in RNase-free tubes on ice. The RNA sample was added. The thermal cycler was programmed as follows: 25 °C for 10 min; 37 °C for 120 min; the reaction was stopped at 85 °C for 5 min. The final volume was 20 μl. For PCR reactions we used the primer pairs (Bio Basic Inc, Amherst, New York, USA) listed in Table 1; for glyceraldehyde 3 phosphate dehydrogenase (GAPDH) (Bio Basic Inc, Amherst, New York, USA) forward 5′-CGAGATCCCTCCAAAATCAA-3′ reverse 5′-GTCTTCTGGGTGGCAGTGAT-3′. The specificity of the primers was verified by searching in the NCBI database for possible homology to cDNAs of unrelated proteins. Each PCR tube contained the following reagents: 0.2 μM of both sense and antisense primers, 1–3 μl (about 1 μg) template cDNA, 0.2 mM dNTP mix, 2.5 U REDTaq Genomic DNA polymerase (Sigma-Aldrich) and 1× reaction buffer. MgCl2 was added at a variable final concentration (empirical determination by setting the experiment). The final volume was 50 μl. The amplification was started with an initial denaturation step at 94 °C for 2 min and was followed by 35 cycles consisting of denaturation (30 s) at 94 °C, annealing (30 s) at the appropriate temperature for each primer pair and extension (1 min) at 72 °C. The PCR products were analysed by 1.5 % TAE ethidium bromide-stained agarose gel electrophoresis (Agarose Gel Unit, Bio-Rad Laboratories S.r.l., Segrate, IT). A PC-assisted CCD camera UVB lamp (Vilber Lourmat, Marne-la-Vallé France) was used for gel documentation. Gel electrophoresis of the amplification products revealed single DNA bands with nucleotide lengths as expected for each primer pair. RNA samples were also amplified by PCR without RT. No band was observed, excluding DNA contamination during the procedure (data not shown). The reaction products were further quantified with the Agilent 2100 bioanalyzer using the DNA 1000 LabChip kit (Agilent Technologies, Deutschland GmbH). The PCR products of cultures treated with U-73122 were analyzed by 1.5 % TBE agarose gel electrophoresis (Submarine Agarose Gel Unit, Hoefer, San Francisco, CA). Gels were previously stained with ethidium bromide (50 ng/100 ml). A PC-assisted CCD camera (GelDoc 2000 System/Quantity One Software; Bio-Rad) was used for gel documentation and quantification. Optical densities were normalized to the mRNA content of glyceraldehydes 3 phosphate dehydrogenase (GAPDH), a typical reference constitutive transcript. Gel electrophoresis of the amplification products revealed single DNA bands with nucleotide lengths as expected for all primer pairs. To exclude possible DNA contamination during the RT-PCR, RNA samples were amplified by PCR without reverse transcription. No band was observed, suggesting also in these experiments that there was no DNA contamination in the RNA preparation procedure. Experiments were independently repeated at least 3 times for each isoform.
Western blot
Whole-cell lysates (106 cells for each experiment) were prepared by lysing cells in RIPA buffer (50 mM Tris pH = 7.5, NP-40, 0.1 % SDS, 100 mM NaCl, 50 mM NaF, 1 mM EDTA) supplemented with a set of protease inhibitors: 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, 1 mM sodium benzamidine, and 1 mM phenylmethylsulfonyl fluoride. Proteins (50 μg) were separated on 12 % polyacrylamide, 0.1 % SDS gel. Then, incubation with a monoclonal antibody specific for each PI-PLC isoform (Santa Cruz, CA) followed. Immunoreactive bands were visualized using the enhanced chemiluminescence method. Experiments were independently repeated at least 2 times for each isoform.
For PI-PLC β2 and PI-PLC η2, positive controls were used to test the efficacy of primers in RT-PCR analyses and of antibodies in Western Blot analyses. For PI-PLC β2 human leucocytes were used as positive controls. For PI-PLC η2, nervous tissue was used as positive control.
Statistical analysis
The concentrations obtained for each isoform were compared in the different cell lines using the ANOVA test (http://www.physics.csbsju.edu/stats/anova_NGROUP_NMAX_form.html). The panels of expression, including dosages of all PI-PLC isoforms, were compared analyzing couple of cell lineages using the t-test (http://studentsttest.com/).
Results
RT-PCR
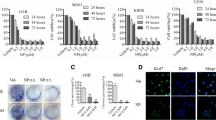
In MG-63 cell line, PI-PLC β1, β3, β4, γ1, γ2, δ1 and δ3 were expressed. Specific bands were obtained using gel electrophoresis analysis (Figs. 1, 2 and 3). Analysis with the bioanalyzer confirmed the data obtained with the gel electrophoresis. The concentrations obtained in 4 independent experiments measured with multiliquid analyzer are listed in Table 2. Analysis with the bioanalyzer detected a small peak for PI-PLC β2, although the quantification was not possible.
RT-PCR results in SaOS and Hs888. Left: SaOS-2 cell line. Visualization of specific bands for the transcripts of the genes codifying for PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3, ε and η1. Right: Hs888 cell line. Visualization of specific bands for the transcripts of the genes codifying for PI-PLC β1, β3, β4, γ1, δ1, δ3, δ4 and ε
Western Blot results of untreated cells (UT), after 3 and 6 h after U-73122 treatment in MG-63, 143B, SaOS, Hs888. Last line: β-actin loading controls. MW: antibody molecular weight. Visualization of specific bands indicate the presence of the PI-PLC isoforms and β-actin. The gel electrophoresis results did not show differences in the presence of PI-PLC enzymes after 3 and 6 h after addiction of U-73122 to the cultures with respect to untreated cells
In 143B cell line, PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3 and ε were expressed. Specific bands were obtained using gel electrophoresis analysis (Fig. 1). The concentrations obtained in 4 independent experiments measured with multiliquid analyzer are listed in Table 2. A weak signal for PI-PLC β2 was detected with gel electrophoresis analysis. Using the bioanalyzer, a small peak was detected, although the quantification was not possible.
In SaOS-2 cell line, PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3, ε and η1 were expressed. Specific bands were obtained using gel electrophoresis analysis (Fig. 2). Analysis with the bioanalyzer confirmed the data obtained with the gel electrophoresis. The concentrations obtained in 4 independent experiments measured with multiliquid analyzer are listed in Table 2.
In Hs888 cell line, PI-PLC β1, β3, β4, γ1, δ1, δ3, δ4 and ε were expressed. Specific bands were obtained using gel electrophoresis analysis (Fig. 2). Analysis with the bioanalyzer confirmed the data obtained with the gel electrophoresis. The concentrations obtained in 4 independent experiments measured with multiliquid analyzer are listed in Table 2.
For PI-PLC β2, in the human leucocytes used as positive controls, the corresponding specific band was obtained as expected. For PI-PLC η2, in the nervous tissue used as positive control, the corresponding specific band was obtained as expected.
After 3 h from administration of U-73122, the following results were detected: in MG-63, in143B, in SaOS-2 and Hs888 cell lines all the tested isoforms (PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3, ε, η1 and η2) resulted absent using the gel electrophoresis analysis (Tables 3, 4, 5 and 6). The GAPDH control specific band was visualized as a positive control with gel electrophoresis analysis.
After 24 h from administration of U-73122, RT-PCR identified specific bands were obtained using the gel electrophoresis analysis. In MG-63 cell line, PI-PLC β1, β3, β4, γ1, δ1, δ3, and ε resulted expressed (Table 3). In 143B cell line, PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3 and ε were expressed (Table 3). In SaOS-2 cell line, PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3, ε and η1 were expressed (Table 3). In Hs888 cell line, PI-PLC β1, β3, β4, γ1, δ1, δ3, δ4, ε and η1 were expressed (Table 3).
Western blot
The western blot analyses performed after administration of U-73122 did not show differences in the presence of PI-PLC enzymes after 3 and 6 h after addiction to the cultures with respect to untreated cells. For PI-PLC β2, in the human leucocytes used as positive controls, the corresponding specific band was obtained as expected. For PI-PLC η2, in the nervous tissue used as positive control, the corresponding specific band was obtained as expected.
Statistical analysis
The ANOVA test results were significant (p < 0.01) comparing the concentrations of each isoform in the 4 cell lines (Table 3, right column).
The t-test used to compare the panels of expression of the cell lines did not identify statistical significant results (Table 4).
Discussion
The tight regulation of calcium concentration is critical to many physiological events. Unregulated elevation alters protein functions, apoptosis regulation, secretion mechanisms and cell contractile activity (Annunziato et al. 2003). The metabolism of calcium is regulated by a complex network of signalling pathways, including the PI transduction system (Rhee et al. 1991). Great attention was paid to PI-PLC enzymes, effector molecules in PI signalling, due to their involvement in a growing number of diseases (Rhee et al. 1991; Suh et al. 2008).
Calcium metabolism was also indicated to be critical for cells viability in human osteosarcoma cell lines (Barry 2000). Previous literature data reported the expression of PI-PLC β1, β3, γ1, γ2 and δ1 in a number of osteosarcoma cell lines, including MG-63 and SaOS-2 cell lines (Hoberg et al. 2005). We analyzed the expression of PI-PLC enzymes in four cultured human osteosarcoma cell lines: MG-63, 143B, SaOS-2 and Hs888.
MG-63 cells, used as experimental models for human osteoblasts, have low levels of alkaline phosphatase activity, and PTH unresponsive adenylate cyclase (Fukayama and Tashjian 1990). The present results (Tables 2 and 3; Fig. 1) accord to literature data with regard to the expression of PI-PLC β1, β3, γ1, γ2 and δ1. Our results partially contrast to previous literature data about PI-PLC β2. Previous reports demonstrated a specific role of PI-PLC β2 in the mechanotransduction and cell attachment, although the basal concentration of the enzyme in unstimulated cells was not reported (Hoberg et al. 2005). Further reports highlighted that in osteoblasts PI-PLC β2 transduces signals from prostaglandin E2 and other prostanoids (Kondo and Togari 2004). In the present study, PI-PLC β2 resulted weakly expressed, and the concentration was not measurable. The expression of PI-PLC β2 seems usually to be correlated to specific stimuli, and probably our results differ in that we analyzed unstimulated MG-63 cells. The basal expression of PI-PLC β2 in osteoblast-like cells might be low. Increase might follow to different and specific stimuli, both mechanic and biochemical. No further data were available about expression of the remaining enzymes in MG-63 line. In the present study, PI-PLC β4 and PI-PLC δ3 were expressed. PI-PLC ε known to be a downstream effector of Ras superfamily GTPases and an upstream activator of small GTPases, both Ras and Rap (Seifert et al. 2008), was weakly expressed. The expression of PI-PLC β1 is significantly higher. The expression PI-PLC β3, β4, γ2 and ε were significantly lower with respect to the other cell lines. These observations require further studies in order to evaluate the role of PI-PLC β2 and PI-PLC ε in MG-63 cells.
143B, thymidine kinase negative cells, are known to develop osteolytic tumours (Kaminski et al. 2003). No literature data were available for PI-PLC expression in 143B cell line. In the present study, PI-PLC β1, β3, β4, γ1, γ2, δ1, δ3 and ε were expressed (Table 2; Fig. 1). PI-PLC β2 was weakly expressed and the concentration not measurable. The expression of PI-PLC ε is significantly higher with respect to the other cell lines. Further studies are required to evaluate the role of PI-PLC β2 and PI-PLC ε in 143B cells.
SaOS-2 is an established epithelial-like human osteosarcoma cell line (Fogh et al. 1975) used as experimental model for human osteoblasts (McQuillan et al. 1995; Rodan et al. 1987). The present results (Table 2; Fig. 1) accord to previous literature data with regard to the expression of PI-PLC β1, β3, γ1, γ2 and δ1. No further data were available about the expression of the remaining isoforms. In the present study, PI-PLC β4, δ3, ε and η1 were also expressed. Moreover, the expression of PI-PLC δ3 was significantly higher with respect to the other cell lines. These data suggest a possible role of the PI-PLC induced calcium osteosynthesis even in osteosarcoma.
Hs888 cells were derived from lung metastasis of osteosarcoma. No literature data were currently available for PI-PLC expression in Hs888 cell line. In the present study, PI-PLC β1, β3, β4, γ1, δ1, δ3, δ4 and ε were expressed. Surprisingly, PI-PLC γ2 resulted unexpressed, whereas it was detected in the other three cell lines we analyzed. Therefore, in the present study, PI-PLC γ2 resulted absent exclusively in the metastatic counterpart of the human osteosarcoma. Previous literature data suggested that PI-PLC γ2 isoform is required for the early phase in osteoclast differentiation (Kertèsz et al. 2012). PI-PLC δ4 was expressed exclusively in Hs888. Noteworthy, studies of breast cancer MCF-7 cells suggested that the abnormal expression of PI-PLC δ4 contributes to oncogenesis through up-regulation of ErbB expression and activation of ERK pathway (Leung et al. 2004). The expression of PI-PLC β and PI-PLC δ1 was significantly higher with respect to the other cell lines.
Remarkably, PI-PLC η1 was detected exclusively in SaOS-2 and Hs888 cells. PI-PLC η1, the most calcium sensitive isoform, is essential for downstream ERK1/2 phosphorylation (Kim et al. 2011). Although the role of PI-PLC η1 is not fully elucidated, the activation of PI-PLC η1 is known to enhance the GPCR mediated calcium pathway (Kim et al. 2011).
Further investigations, performed using RT-PCR, demonstrated that the transcription of PLC genes was absent after 3 h from the administration of U-73122, a specific inhibitor for PI-PLC isoforms. These observations accord to previous literature data described in other tumours (Lo Vasco et al. 2011) indicating that U-73122 also acts upon the genes’ expression. After 24 h from the administration of U-73122, the expression returns similar to untreated cells, accordingly to previous data (Lo Vasco et al. 2011).
Given the tissue specificity of PI-PLC enzymes, the panel of expression presently delineated slightly differed in the MG-63, 143B, SaOS and Hs888 osteosarcoma cell lines. Selected enzymes, such as PI-PLC β1, β3, β4, γ1, δ1 and δ3, were commonly expressed, although at variable concentrations, in all four cell lines. PI-PLC η2 was commonly unexpressed in all the analyzed cell lines. Beside the results regarding PI-PLC β2 and PI-PLC ε, already commented, the remaining enzymes were differently expressed. In our previous report, we had recently analyzed the expression of PI-PLC isoforms in cultured human fibroblasts (Lo Vasco et al. 2013). PI-PLC β2, η1 and η2 had resulted unexpressed and PI-PLC δ4 inconstantly expressed (Lo Vasco et al. 2013). Interestingly, also in the osteosarcoma cell lines we presently analyzed, PI-PLC β2 and η2 were not expressed and PI-PLC δ4 inconstantly expressed. The present results suggest that human cultured fibroblasts and human osteosarcoma cell lines share similar expression panel for PI-PLC isoforms. This observation will require further studies in order to investigate the possible relationship.
Moreover, although it is not possible to formulate conclusions about these findings, the differences in the panel of expression of PI-PLC enzymes might be due to the different commitment of the cell lines. In fact, PI-PLC γ2 was expressed in MG-63, 143B and SaOS-2, whereas it was absent in Hs888 cells. The concentration measured in 143B and SaOS-2 cells was about 50 % higher than in MG-63 cells. Notably, recent literature data indicated that PI-PLC γ2 is involved in osteoclast differentiation and bone resorption (Kertèsz et al. 2012). Therefore, unique among the analyzed cell lines, Hs888 did not express PI-PLC γ2 and expressed PI-PLC δ4. The peculiar expression of these two isoforms in Hs888 cells might be related to the metastasizing ability of the lineage.
PI-PLC η1 was expressed in SaOS-2 epithelial-like lineage and in Hs888 cells. PI-PLC η1 is considered a sensor activated by very small calcium increases, and, therefore, involved in the finest regulation of signalling.
The statistical comparison of the results resulted significant. However, further studies are required in order to confirm these observations and the possible use of this finding as diagnostic or prognostic tool. Moreover, the peculiar expression of specific enzymes in distinct cell lines deserves further studies in order to analyze the specific role and crosstalk of each expressed enzyme in specific osteosarcoma lineages.
Little is known about the events leading to osteosarcoma, with special regard to the complex network of molecular pathways which regulate the cells survival and metabolism. The present work offers a first contribute to further studies addressed to analyze the role of PI signal transduction pathway in osteosarcoma cells, representing a tool to improve our knowledge of the metabolic activity during the disease.
Abbreviations
- PI:
-
Phosphoinositides
- PIP2:
-
Phosphatydil inositol (4,5) bisphosphate
- PI-PLC:
-
Phosphoinositide-specific Phospholipase C
- IP3:
-
Inositol trisphosphate
- DAG:
-
Diacylglycerol
- PKC:
-
Protein kinase C enzymes
- GAPDH:
-
Glyceraldehyde 3 phosphate dehydrogenase
- ANOVA:
-
Analysis of variance
References
Annunziato L, Amoroso S, Pannaccione A, Cataldi M, Pignataro G, D’Alessio A, Sirabella R, Secondo A, Sibaud L, Di Renzo GF (2003) Apoptosis induced in neuronal cells by oxidative stress: role played by caspases and intracellular calcium ions. Toxicol Lett 139(2–3):125–133
Barry EL (2000) Expression of mRNAs for the alpha 1 subunit of voltage-gated calcium channels in human osteoblast-like cell lines and in normal human osteoblasts. Calcif Tissue Int 66(2):145–150
Bunney TD, Katan M (2011) PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem Sci 36(2):88–96
Daubie V, De Decker R, Nicaise C, Pochet R (2007) Osteosarcoma cell-calcium signaling through tissue factor-factor VIIa complex and factor Xa. FEBS Lett 581(14):2611–2615
Di Cristofano C, Leopizzi M, Miraglia A, Sardella B, Moretti V, Ferrara A, Petrozza V, Della Rocca C (2010) Phosphorylated ezrin is located in the nucleus of the osteosarcoma cell. Mod Pathol 23(7):1012–1020
Fogh J et al (1975) In: Fogh J (ed) Human tumor cells in vitro. Plenum Press, NY, pp 115–159
Fukami K, Inanobe S, Kanemaru K, Nakamura Y (2010) Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res 49(4):429–437
Fukayama S, Tashjian AH Jr (1990) Stimulation by parathyroid hormone of 45Ca2+ uptake in osteoblast-like cells: possible involvement of alkaline phosphatase. Endocrinology 126(4):1941–1949
Gatta G, Capocaccia R, Stiller C, Kaatsch P, Berrino F, Terenziani M (2005) EUROCARE Working Group. Childhood cancer survival trends in Europe: a EUROCARE Working Group study. J Clin Oncol 3(16):3742–3751
Gorlick R et al (2009) Osteosarcoma. J Bone Miner Res 25(4):683–691
Gorlick R, Toretsky J, Marina N (2003) Bone tumors. In: Kufe D, Pollock R, Weichselbaum R et al (eds) Cancer medicine vol. 2, 6th edn. BC Decker, Hamilton, pp 2383–2406
Hoberg M, Gratz HH, Noll M, Jones DB (2005) Mechanosensitivity of human osteosarcoma cells and phospholipase C beta2 expression. Biochem Biophys Res Commun 333(1):142–149
Kaminski M, Masaoka M, Karbowski M, Kedzior J, Nishizawa Y, Usukura J, Wakabayashi T (2003) Ultrastructural basis for the transition of cell death mode from apoptosis to necrosis in menadione-treated osteosarcoma 143B cells. J Electron Microsc 52:313–325
Kertész Z, Gyori D, Körmendi S, Fekete T, Kis-Tóth K, Jakus Z, Schett G, Rajnavölgyi E, Dobó-Nagy C, Mócsai A (2012) Phospholipase Cγ2 is required for basal but not oestrogen deficiency-induced bone resorption. Eur J Clin Invest 42(1):49–60
Kim JK, Choi JW, Lim S, Kwon O, Seo JK, Ryu SH, Suh PG (2011) Phospholipase C-η1 is activated by intracellular Ca(2+) mobilization and enhances GPCRs/PLC/Ca(2+) signaling. Cell Signal 23(6):1022–1029
Kondo A, Togari A (2004) Activation of osteoblastic functions by a mediator of pain, bradykinin. Biochem Pharmacol 68(7):1423–1431
Leung DW, Tompkins C, Brewer J, Ball A, Coon M, Morris V, Waggoner D, Singer JW (2004) Phospholipase C delta-4 overexpression upregulates ErbB1/2 expression, Erk signaling pathway, and proliferation in MCF-7 cells. Mol Cancer 3:15
Lo Vasco VR, Fabrizi C, Artico M, Cocco L, Billi AM, Fumagalli L, Manzoli FA (2007) Expression of phosphoinositide-specific phospholipase C isoenzymes in cultured astrocytes. J Cell Biochem 100(4):952–959
Lo Vasco VR, Fabrizi C, Panetta B, Fumagalli L, Cocco L (2010) Expression pattern and sub cellular distribution of Phosphoinositide specific Phospholipase C enzymes after treatment with U-73122 in rat astrocytoma cells. J Cell Biochem 110(4):1005–1012
Lo Vasco VR, Leopizzi M, Chiappetta C, Businaro R, Polonia P, Della Rocca C, Litta P (2012) Expression of Phosphoinositide-specific Phospholipase C enzymes in normal endometrium and in endometriosis. Fertil Steril 98(2):410–414
Lo Vasco VR, Leopizzi M, Chiappetta C, Puggioni C, Di Cristofano C, Della Rocca C (2013) Expression of Phosphoinositide-specific phospholipase C enzymes in human skin fibroblasts. Connective Tissue Res 54(1):1–4
Marina N, Gorlick R, Bielack S (2010) Pediatric osteosarcoma. In: Carroll W, Finlay J (eds) Cancer in children and adolecents. Jones and Bartlett, Sudbury, pp 383–394
McQuillan DJ, Richardson MD, Bateman JF (1995) Matrix deposition by a calcifying human osteogenic sarcoma cell line (SAOS-2). Bone 16(4):415–426
Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W, Gebhardt M, Goorin AM, Harris MB, Healey J, Huvos A, Link M, Montebello J, Nadel H, Nieder M, Sato J, Siegal G, Weiner M, Wells R, Wold L, Womer R, Grier H (2005) Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol 23(9):2004–2011
Mirabello L, Troisi RJ, Savage SA (2009a) International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer 125:229–234
Mirabello L, Troisi RJ, Savage SA (2009b) Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 115:1531–1543
Pan CC, Cheng HH, Huang CJ, Lu YC, Chen IS, Liu SI, Hsu SS, Chang HT, Huang JK, Chen JS, Jan CR (2006) The antidepressant mirtazapine-induced cytosolic Ca2+ elevation and cytotoxicity in human osteosarcoma cells. Chin J Physiol 49(6):290–297, Erratum in: Chin J Physiol. 2007 Feb 28;50(1):41
Rhee SG, Kim H, Suh PG, Choi WC (1991) Multiple forms of phosphoinositide-specific phospholipase C and different modes of activation. Biochem Soc Trans 19(2):337–341
Rodan SB, Imai Y, Thiede MA, Wesolowski G, Thompson D, Bar-Shavit Z, Shull S, Mann K, Rodan GA (1987) Characterization of a human osteosarcoma cell line (Saos-2) with osteoblastic properties. Cancer Res 47(18):4961–4966
Seifert JP, Zhou Y, Hicks SN, Sondek J, Harden TK (2008) Dual activation of phospholipase C-epsilon by Rho and Ras GTPases. J Biol Chem 283(44):29690–29698
Suh PG, Park J, Manzoli L, Cocco L, Peak JC, Katan M, Fukami K, Kataoka T, Yuk S, Ryu SH (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep 41:415–434
Takenouchi T, Ogihara K, Sato M, Kitani H (2005) Inhibitory effects of U73122 and U73343 on Ca2+ influx and pore formation induced by the activation of P2X7 nucleotide receptors in mouse microglial cell line. Biochim Biophys Acta 1726(2):177–86
Tang CH, Yang RS, Fu WM (2005) Prostaglandin E2 stimulates fibronectin expression through EP1 receptor, phospholipase C, protein kinase Calpha, and c-Src pathway in primary cultured rat osteoblasts. J Biol Chem 280(24):22907–22916
Tian Z, Quan X, Leung AW, Xiang J, Xu C (2010) Hematoporphyrin monomethyl ether enhances the killing of ultrasound on osteosarcoma cells involving intracellular reactive oxygen species and calcium ion elevation. Integr Cancer Ther 9(4):365–369
Acknowledgments
The authors thank the ‘Serena Talarico Association’ for the support to this research and encouragement, Dr Liselotte Setter for technical support and Dr Stefano Bussolon for constructive discussion.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lo Vasco, V.R., Leopizzi, M., Chiappetta, C. et al. Expression of Phosphoinositide-specific phospholipase C enzymes in human osteosarcoma cell lines. J. Cell Commun. Signal. 7, 141–150 (2013). https://doi.org/10.1007/s12079-013-0194-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-013-0194-6