Abstract
The Transforming acidic coiled coil (TACC) proteins play a conserved role in normal development and tumorigenesis through interactions with multiple complexes involved in transcription, translation, and centrosomal dynamics. However, despite significant work on the function of TACC3 in the control of centrosomal mechanics, relatively little functional data is known about the family’s founding member, TACC1. From a continued analysis of clones isolated by an unbiased yeast two-hybrid assay, we now show direct physical interactions between the TACC1 and the FHL (Four and a Half LIM-only) family of proteins. The authenticity of these interactions was validated both in vitro and in cellular systems. The FHLs exhibit diverse biological roles such as the regulation of the actin cytoskeleton and are promiscuous coregulators for several transcription factors. The interaction of the endogenous TACC-FHL proteins is primarily localized to the nucleus. However, similar to FHL2, overexpression of TACC1A in HEK293 is able to sequester serum activated ERK to the cytoplasm. This has the effect of reducing the serum induced transcriptional response of the c-fos and c-jun genes. The observation that TACCs can interact with the FHLs and alter their serum induced activities raises the possibility that the TACCs participate in crosstalk between cell signaling pathways important for cancer development and tumor progression. The transforming acidic coiled coil genes are known to be important prognostic indicators for breast, ovarian and lung cancer. In this manuscript, we identify a novel interaction between the TACCs and the FHL protein family. This interaction has an affect on ERK and may in part explain the variable associations and changes in subcellular locations of each family with specific subtypes of malignancy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Previously, we described the cloning and genomic structure of the three human transforming acidic coiled-coil (TACC) genes (Lauffart et al. 2003; Still et al. 1999a,b). These genes encode proteins that are highly acidic and contain the evolutionary conserved 200 amino-acid TACC domain (Still et al. 2004). Based on a number of studies, TACCs are functionally linked to the processes of cell division and differentiation (Gergely et al. 2000b; Piekorz et al. 2002; Sadek et al. 2000). In addition, the exact role of the TACCs in tumorigenesis is currently under scrutiny, with clear links between loss of TACC protein expression in the pathogenesis of breast and ovarian tumors (Conte et al. 2002; Jacquemier et al. 2005; Lauffart et al. 2005), and upregulation or changes in splice variant expression in other cancers (Line et al. 2002; Stewart et al. 2004). Interestingly, loss of TACC2 and TACC3 may be of clinical prognostic value in breast tumors (Jacquemier et al. 2005). In contrast, microarray analysis has revealed that TACC3 mRNA increases during the transition of breast cancer from preinvasive ductal carcinoma in situ to invasive ductal carcinoma (Ma et al. 2003), suggesting that TACC3 imparts a proliferative advantage to a subset of breast cancers. A similar dichotomy was noted for ovarian cancer, where one study showed that the TACC3 protein is lost or mislocalized in the majority of ovarian tumors (Lauffart et al. 2005), while two smaller studies using Serial Analysis of Gene Expression suggested that some ovarian tumors exhibit increased TACC3 mRNA expression, and that this may be linked to chemoresistance (L’Esperance et al. 2006; Peters et al. 2005). Furthermore the retention of TACC1 observed in some ovarian tumors (Lauffart et al. 2005; Partheen et al. 2006) was recently linked to a more favorable prognosis (Partheen et al. 2006).
It is unclear whether the observed differences in TACC expression in these studies reflect the method of detection i.e. protein versus RNA, or more subtle tumor- or tissue-specific effects. In cell culture, the human TACCs are variably distributed within the interphase cell, although they primarily concentrate in the nucleus (Gergely et al. 2000a). Nuclear accumulation of TACC3 and TACC1 occurs in specific tissues during normal mouse development (Aitola et al. 2003; Lauffart et al. 2006; Sadek et al. 2003). Recently, we determined that the in vivo subcellular localization of TACC3 can be aberrant in ovarian tumors compared to normal ovarian surface epithelium (Lauffart et al. 2005). This also occurs during lung tumorigenesis (Jung et al. 2006). Although the mechanism underlying this nuclear to cytoplasmic relocation is currently unknown, it seems plausible that some of these observed differences may be dependent on the bioavailability of as yet uncharacterized TACC binding factors.
The list of known TACC interacting factors has been growing, and can be divided into two broad categories: (1) proteins with roles in centrosome/mitotic spindle dynamics, and (2) proteins involved in regulating gene expression (reviewed in Still et al. 2004). The subcellular distribution of the TACCs reflects these interactions, and the possibility that they may serve as signaling intermediates or scaffold/adaptor proteins in the cytosol and the nucleus. Indeed, TACC2 is known to migrate to the nucleus of microvascular endothelial cells upon stimulation with erythropoietin, implicating TACC2 in erythropoietin signaling (Pu et al. 2001). Furthermore, one of the first functional interactions defined for a TACC protein, mouse TACC3, was as a coregulator of arylhydrocarbon nuclear translocator 1 (ARNT1) in the transcriptional response to polyaromatic hydrocarbons and hypoxia (Sadek et al. 2000). We now present further evidence for the involvement of the TACCs in diverse protein complexes, by describing their direct physical association with members of the FHL (Four and a Half LIM-only) family of proteins. This raises the possibility that TACCs and FHLs participate in interconnected pathways that contribute to tumorigenesis.
Materials and methods
Yeast two-hybrid analysis and elucidation of protein binding domains of FHL and TACC proteins
The yeast two-hybrid methodology used to screen a Matchmaker human mammary epithelial cDNA library with a pASTACC1 bait plasmid, and controls to assess validity of initial positive clones was reported in Lauffart et al. (Lauffart et al. 2002). Construction of full length and subclones of TACC1, TACC2 and TACC3 in the glutathione-S-transferase expression vector pGEX5X2 were previously described (Gangisetty et al. 2004). Subclones of FHL3 and FHL2 were generated by the polymerase chain reaction (PCR) (primer sequences available on request) from full length I.M.A.G.E clones 2964682 and 5501154 and cloned into pET28c (Novagen, Madison WI, USA). In vitro expression and binding assays were performed as detailed in (Lauffart et al. 2003).
Coimmunoprecipitation and immunoblotting
Commercial antibodies were obtained from the following companies, α-TACC antibodies were as previously described (Gangisetty et al. 2004), rabbit α-FHL2 (#BL455) from Bethyl laboratories Inc, Montgomery TX, USA, chicken IgY α-FHL3 (#A22332F) and rabbit α-IgY conjugated microbeads (RAY beads) #RAY-Microbeads from Genway, San Diego, CA, normal IgG and α-rabbit-horseradish peroxidase conjugates were purchased from Santa Cruz Biotechnology, Santa Cruz, CA, and normal chicken IgY and α-chicken horseradish peroxidase from Jackson laboratories, USA. Human embryonic kidney 293 (HEK293) cells were washed with ice-cold 1xPBS, and nuclear extract prepared according to the protocol of Schreiber et al. (1989), and diluted 1:2 in 50 mM Tris-HCl pH 7.4/0.5% v/v Nonidet-NP40 to reduce the salt concentration to 130 mM. Coimmunoprecipitation with FHL2 was carried out as described in (Gangisetty et al. 2004). For coimmunoprecipitation with FHL 3, 500 μg of the diluted nuclear extract was precleared with 10 μl of RAY beads for 30 min. The extract was centrifuged for 5 min at 4,000 g at 4°C, and the cleared supernatant incubated with primary antibody (5 μg) or matched IgY control (5 μg) overnight at 4°C. Ten microliters of RAY beads was then added and immunoprecipitation allowed to proceed for an additional hour at 4°C. Immune complexes were pelleted by centrifugation (1,000 g, 5 min at 4°C), washed three times with binding buffer, and immunoprecipitated proteins eluted by boiling with 2× Laemmli buffer (125 mM Tris-HCl (pH6.8 at 25°C), 4% w/v SDS, 20% v/v glycerol, 10%v/v β-mercaptoethanol, 0.004% w/v bromophenol blue). Cell lysates and eluted complexes were separated by 8% w/v SDS-PAGE and immunoblotted with respective antibodies as described in (Lauffart et al. 2002).
Indirect immunofluorescence
Cells were prepared as previously documented (Lauffart et al. 2002). Cells on coverslips were incubated with the α-TACC1 goat polyclonal antibody (Santa Cruz) together with either α-FHL2 or α-FHL3 antibody for 1 h at room temperature. The coverslips were washed with PBS (3 × 5 min) and incubated with a mixture of FITC-α-goat IgG (to detect TACC1) and rhodamine red-X-conjugated α-rabbit IgG (for FHL2) or rhodamine red-X-conjugated α-chicken IgY (for FHL3) for 30 min. Finally, the coverslips were washed first with PBS containing DAPI (4′,6′-diamidino-2-phenylindole hydrochloride) (1 μg/ml) and then twice with PBS, mounted with AquaPolyMount (Polysciences Inc., Warrington, PA) and examined at 60× magnification (oil). Secondary antibodies were obtained from Jackson Laboratories.
Transcriptional assay using the Secreted Alkaline Phosphatase (SEAP) assay system
The test plasmid or vector control (0.5 μg) was transfected simultaneously with the SEAP-reporter (0.5 μg) and pSV-βGAL (0.5 μg) plasmids into HEK293 using Lipofectamine 2000 (Invitrogen, Carlsbad, USA). After 48 h in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, media was collected and assayed for SEAP activity using the Great EscaPe SEAP chemiluminescence detection kit (Clontech, USA) and the chemiluminescent signal detected using a Zylux MPL2 microplate luminometer. Activity was normalized with respect to β-galactosidase and cellular protein. Fold activation was determined relative to the normalized vector control. Individual experiments were conducted in triplicate and verified for reproducibility in three independent experiments. Differences between the vector transfected and each test plasmid were analyzed using one way ANOVA followed by Dunnett’s Multiple Comparison Post Test (Graphpad Prism Version 3.0, Graphpad Prism Software Inc.).
Serum starvation experiments
HEK293 stably expressing full length TACC1, FHL2 and FHL3, fused to the C-terminus of EGFP were generated as previously described in (Lauffart et al. 2002). Serum starvation experiments were carried out essentially as described in (Purcell et al. 2004). Cytoplasmic and nuclear extracts were prepared according to the protocol of Schreiber et al. (Schreiber et al. 1989). Antibodies used for immunoblots were as follows: α-CREB (#238461, Calbiochem, San Diego, CA), α-pCREB (#9198, Cell Signaling Technology, Danvers, MA), α-ERK (#06-182 Upstate Biotechnology, Lake Placid, NY), α-pERK1/2 (#9101, Cell Signaling Technology), α-pp90RSK (#9341, Cell Signaling Technology), α-βtubulin (sc-9104, Santa Cruz) and α-Ku70 (#611893, BD Transduction Laboratories San Diego, CA). Immunoblots were performed using standard techniques (Gangisetty et al. 2004). RNA extraction and semi-quantitative rt-PCR was performed as noted in (Lauffart et al. 2006). Intron spanning primers were designed to specifically amplify c-fos and c-jun transcripts (primers available on request).
Results
We have previously documented the initial results from a yeast two hybrid based screening of an adult mammary epithelial cDNA library (Clontech, Delaware USA) with the full length TACC1 open reading frame (Lauffart et al. 2002). Thirty eight independent positive clones were isolated after the stringent two-stage screening process. In addition, to identifying YEATS4 (GAS41) (Lauffart et al. 2002), ch-TOG (Lauffart et al. 2002), and LSm-7 (confirming the report of Conte et al. 2002), sequence analysis revealed that three independent clones, M29, M42, and M56 corresponded to amino acids 6–280 of the Four and a Half LIM-only protein, FHL3 (Refseq: NP_004459). FHL3 belongs to a subfamily of LIM-only containing proteins with the characteristic four and one half repeats of the double zinc-finger LIM motif [CX2CX16–23HX2(C/H)X2CX2CX15–23CX2(C/H/D)]. To confirm and further refine the binding site of TACC1 on FHL3, smaller fragments of FHL3 were cloned from the full length FHL3 I.M.A.G.E clone 2964682 into pET28c. The choice of regions to clone and primers used were based on those previously reported (Li et al. 2001b; Samson et al. 2004; Turner et al. 2003), and therefore known to be able to form functional interactions with previously identified binding factors. Each in vitro translated subfragment was tested for its ability to bind to a full length glutathione-S-transferase (GST)-TACC1 fusion protein, as described in (Lauffart et al. 2003), and under conditions similar to those used to examine interactions of other FHL partners (Li et al. 2001b; Samson et al. 2004; Turner et al. 2003). TACC1 was found to specifically bind to FHL3 constructs containing LIM domain region 1, but not those constructs lacking this domain (Fig. 1b). The corresponding LIM domain 1 of FHL2 was similarly found to bind TACC1 (data not shown). This explains why the original FHL3 yeast two hybrid clones isolated by TACC1 were relatively full length (aa 6–280).
In vitro mapping of the TACC1-FHL3 binding domain. a Diagram and summary of deletion constructs for FHL3 binding domain mapping. Extents of the constructs are defined by the reference protein sequence NP_004459. The original clones (M29, M42 and M56) identified by yeast two-hybrid spanned amino acids 6–280 of the FHL3 protein. Subclones were obtained by PCR based cloning from full length I.M.A.G.E. clone 2964682. b Interaction of TACC1 with FHL3 constructs by GST pull down. Lane numbering corresponds to the numbered FHL3 construct in (a). Top panel: Autoradiograph of 12% SDS polyacrylamide gels with in vitro translated FHL3 construct pulled down with GST-TACC1 (T) or GST (G). I: 10% input of FHL3 protein; Bottom two panels represent Coomassie blue stained gels of the pull down experiment, verifying loading of the GST fusion proteins. c GST pull down assays between FHL and TACC family members: Top panel: Autoradiograph of 12% w/v SDS polyacrylamide gel with in vitro translated FHL1-3 constructs pulled down with GST-TACC1 aa596-ter (T1), GST-TACC2s (T2) aa2-ter, GST-TACC3 aa116-ter (T3) or GST (G). I: 10% input of in vitro translated FHL construct. Bottom two panels represent Coomassie blue stained gels of pull down experiment showing loading of GST-TACC proteins and GST. cDNAs corresponding to full length FHL1, FHL2, and FHL3 were subcloned into pcDNA3. Binding assays were then performed as described previously with stringent washing (23)
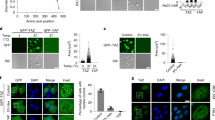
Together with FHL1 and 2, FHL3 forms a distinct subfamily of highly related and conserved LIM-domain proteins that are important regulators of cell determination, differentiation, remodeling of the cytoskeleton and pathological functions such as oncogenesis (Johannessen et al. 2006). Similarly, the TACCs form a distinct subfamily of coiled coil proteins (Still et al. 2004). Thus, we next determined whether the interaction between TACC1 and FHL3 was unique, or whether each of these proteins could interact with members of the other family through conserved domains within each protein. Figure 1c clearly establishes a conserved interaction between the TACC domain of TACC1 and each FHL member. Our data confirms a previously unsubstantiated report of an interaction between FHL2 and TACC1 quoted in a recent review on FHL2 (Table 2 of Johannessen et al. 2006). In addition, both TACC2 and TACC3 exhibited binding to FHL2 and 3, but showed no significant binding to FHL1. Under these in vitro conditions, FHL3 appears to interact more strongly with the TACCs compared to any other FHL. The interaction between TACC2 and FHL3 appears to be particularly strong, as indicated by the amount of FHL3 bound to the GST-TACC2. It should be noted however that this interaction may be multivalent, as FHL3 can also homodimerize (Fimia et al. 2000). Significantly, FHL1 and FHL2/3 exhibit differential binding to the TACCs in a similar manner to their interactions with proteins such as the α7β1 integrin receptor (Wixler et al. 2000), and the CREB (Fimia et al. 2000) and PLZF transcription factors (McLoughlin et al. 2002). Further evidence for in vivo interactions was indicated in coimmunoprecipitation experiments with commercial antibodies individually raised against native FHL2 and FHL3. In HEK293, endogenous TACC1 predominantly localizes to the nucleus with FHL2 and FHL3 (Fig. 2a), and both endogenous proteins can co-immunoprecipitate each endogenous TACC family member from this nuclear compartment (Fig. 2b). It is already known that the subcellular localization of each member of the FHL and TACC families is regulated in a temporal and tissue specific manner (Aitola et al. 2003; Lauffart et al. 2006; Li et al. 2001a; Martin et al. 2002; Muller et al. 2002; Sadek et al. 2003). Thus, this suggests a significant potential for selective and combinatorial interactions amongst FHL and TACC family members.
Subcellular and functional interactions between members of the TACC and FHL families in HEK293. a Indirect immunofluorescent detection of endogenous TACC1 and FHL2/3 in the nucleus of HEK293. Indirect immunofluorescence using the TACC1 antibody and a FITC-labeled secondary antibody (green) shows that TACC1 is predominantly localized to the nucleus of HEK293. FHL2 and FHL3 (red) show expression in the nucleus and cytoplasm of HEK293. Colocalization of TACC1 and FHL2/3 is revealed by the yellow/green signal in the nuclei of the cells, caused by the imposition of the green TACC1 signal on the red FHL2/3 signal. Nuclei were counterstained with DAPI. b HEK293 nuclear extracts were immunoprecipitated with antibodies to native FHL2, and FHL3, or normal species specific IgG, and immunoblots incubated with each α-TACC antibody. The α-FHL antibodies specifically immunoprecipitate the native TACC1, TACC2s and TACC3 proteins in HEK293. Nuc nuclear extract, IP immunoprecipitating antibody, WB western blot antibody. c TACCs and FHLs function as CREB coactivators in human cells. FHLs and TACCs coactivate the transcription from a cAMP response element attached to secreted alkaline phosphatase. Fold activation is supplied above the appropriate column compared to the SEAP reporter in the presence of the vector control plasmid. *p < 0.05, **p < 0.01. d Sustained activation of CREB in the absence of serum in TACC1, FHL2 and FHL3 overexpressing cell lines. HEK293 were stably transfected with TACC1A, FHL2 or FHL3 fused to the C-terminus of EGFP (EGT1, EGFHL2 and EGFHL3 respectively). These cell lines were grown for 24 h in the presence or absence of serum and whole cell protein extracts prepared in the presence of sodium orthovanadate. HEK293 cells transfected with EGT1 or EGFHL2 demonstrated a higher sustained level of CREB activation in the absence of serum, compared to GFP vector- or EGFHL3-transfected cells. β-Tubulin demonstrated equivalent loading of cell extracts
As presented above, the TACCs have the ability to bind to human members of the FHL family. Through the double zinc finger LIM motifs, FHLs engage in a wide range of protein–protein interactions, regulating the functions of cytoskeletal proteins (Coghill et al. 2003), enzymes (Lange et al. 2002) and transcription factors (Martin et al. 2002; McLoughlin et al. 2002; Turner et al. 2003). Similar to TACC3 (Garriga-Canut and Orkin 2004; Sadek et al. 2000), the first functions ascribed to the FHLs were as nuclear regulators through interactions with MCM7 (Chan et al. 2000), ZNF638 (Ng et al. 2002), and CREB (Fimia et al. 2000). In the latter case, the FHLs coactivate the basal activity of promoters containing cAMP response elements (CRE) by approximately two to fivefold, depending on the FHL family member, the promoter examined and the cell line used (Fimia et al. 2000). We have already shown that human TACCs can complex with nuclear chromatin remodeling enzymes and histone acetyltransferases required for CREB transcriptional activity, such as pCAF, and CBP (Gangisetty et al. 2004), interactions which have since been observed by other authors (Angrisano et al. 2006). As the endogenous FHL-TACC interaction was restricted to the nucleus of HEK293, we next determined whether the TACCs could act as independent CREB coregulators, in a similar manner to FHL2 and FHL3. Thus, we transiently transfected HEK293 with a CRE-responsive promoter attached to the secreted alkaline phosphatase (SEAP) reporter gene (CRE-SEAP from Clontech, Delaware USA). In this system, the basal activity of this promoter was activated between 1.7 and 2.2 fold for TACC1A, TACC2s and TACC3, significantly more than the 1.6 fold and 1.2 fold activation observed for FHL2 and FHL3 respectively (Fig. 2c). To our knowledge, this is the first time any stimulatory, transcriptional activity has been attributed to TACC1A and TACC2s.
Overexpression of the TACC1A isoform in mouse tissue and cell lines results in an increased activation of ERK, by an as yet uncharacterized mechanism (Cully et al. 2005). Similarly, FHL2 has been implicated in ERK signaling as overexpression of this protein inhibits ERK nuclear-cytoplasmic shuttling (Purcell et al. 2004). The interaction of TACC1 with FHL2 suggested that TACC1 and FHL2 could play an overlapping role in ERK regulation during tumorigenesis. To address this hypothesis, we established a series of HEK293 cell lines overexpressing TACC1A, FHL2 and FHL3 fused to the C-terminus of EGFP (EGT1, EGFHL2 and EGFHL3 respectively). Previously published data suggests that the EGFP moiety does not interfere with the functional interactions of either of the attached TACC1 or FHL protein (Garriga-Canut and Orkin 2004; Gergely et al. 2000a; Li et al. 2001b). These cell lines were grown for 24 h in the presence or absence of serum and whole cell protein extracts assayed to determine whether phosphorylation of serum activated proteins CREB and ERK was altered by the overexpressed proteins. In this system, both TACC1 and FHL2 demonstrated a higher sustained level of phosphorylation of CREB and ERK in the absence of serum, compared to vector- or FHL3-transfected cells (Figs. 2d and 3a respectively).
Effect of TACC1 and FHLs on pERK activation and nuclear sequestration. a The HEK293 stable transfectants were grown for 24 h in the presence or absence of serum and whole cell protein extracts prepared in the presence of sodium orthovanadate. HEK293 cells transfected with EGT1 or EGFHL2 demonstrated a higher sustained level of ERK activation in the absence of serum, compared to GFP vector- or EGFHL3-transfected cells. GFP, EGT1 and EGFHL3 demonstrated normal induction of pERK in response to 24 h serum stimulation. β-Tubulin demonstrated equivalent loading of cell extracts. b TACC1 and FHL2 have opposing effects on pERK activation and relocalization. Each stable cell line was transfected with increasing levels of TACC1 or FHL2 plasmid (from left to right per cell line: 0 μg, 0.5 μg, 1.0 μg). EGFP was used to control for effects of DNA concentration and any effect of the GFP moiety. Cytoplasmic and nuclear extracts were prepared as described in Schreiber et al. (1989). β-Tubulin and Ku70 were monitored to ensure equivalent loading of cytoplasmic and nuclear extracts respectively
Overexpression of FHL2 impacts ERK signaling by sequestration of phospho-ERK (pERK) to the cytoplasm of activated cells (Purcell et al. 2004). We next determined whether overexpression of TACC1A, similar to that observed in tumors, also acted to impede transport of pERK to the nucleus. This was investigated in two ways. First, we determined if overexpression of TACC1, FHL2 or FHL3 increased cytoplasmic sequestration of pERK (Fig. 3b). Each stable cell line was transfected with varying levels of TACC1 or FHL2 plasmid. EGFP was used to control for effects of DNA concentration and any effect of the GFP moiety. Noticeably, in EGT1-293, ERK phosphorylation increased as FHL2 levels were raised. Conversely, EGT1 inhibited FHL2-mediated activation and nuclear accumulation of ERK in the EGFHL2 cell line. Noticeably TACC1 increased phosphorylation and nuclear localization of ERK in FHL3 overexpressing HEK293 cells (Fig. 3b), suggesting that TACC1A function may be modified depending upon the relative levels of FHL2 to FHL3. Next, we determined the distribution of pERK in cells activated with serum, following the methodology of Morlon and Sassone-Corsi (Morlon and Sassone-Corsi 2003). Stable cell lines were starved of serum, treated with cycloheximide to block de novo protein synthesis and then stimulated with 20% serum for 4 h. As shown in Fig. 4a, normal activation and nuclear accumulation of ERK was noted in EGFP-293. However, pERK failed to accumulate in the nuclei of serum stimulated TACC1A overexpressing cells. This cytoplasmic accumulation appeared to be due to a defect in pERK shuttling into the nucleus as opposed to increased nuclear export because treatment with Ratjadone A, an inhibitor of the nuclear-cytoplasmic export system failed to increase nuclear accumulation of pERK. Notably, in EGFHL2-293, ERK activation was observed in the absence of serum, although little nuclear accumulation was noted. Serum stimulation subsequently increased nuclear localization (Fig. 4a). Interestingly, in the EGFHL3 cell line maximal nuclear accumulation of pERK was noted upon stimulation with UV light, or in the presence of Ratjadone A.
Effects of TACC1, FHL2 and FHL3 on serum induced redistribution of pERK and transcriptional activity. a Stable cell lines were starved of serum, treated with cycloheximide (Chx) to block de novo protein synthesis and then stimulated with 20% serum for 4 h, following the methodology of Morlon and Sassone-Corsi (2003). Normal activation and nuclear accumulation of ERK was noted in EGFP-293, however, pERK failed to accumulate in the nuclei of serum stimulated TACC1A overexpressing cells. Inclusion of Ratjadone A (RatJ), an inhibitor of the nuclear-cytoplasmic export system failed to increase nuclear accumulation of pERK. The effect of UV light was also monitored as FHL2 has been implicated in responses to UV light (Morlon and Sassone-Corsi 2003). b TACC1A inhibits serum induced transcriptional activation of c-fos and c-jun genes. Stable cells expressing GFP, EGTACC1A (EGT1), EGFHL2 and EGFHL3 were treated as outlined in (a). Total RNA was then extracted and subject to rt-PCR (22) to determine expression from the endogenous c-fos and c-jun genes. Normal induction of c-fos and c-jun in response to brief serum stimulation was observed in all cell lines, except EGT1
Activation of ERK is known to impact several different downstream signaling molecules (Roux and Blenis 2004). p90RSK is a direct cytoplasmic substrate for pERK. Notably, an increase in p90RSK phosphorylation was noted in EGFHL2 and in FHL2 transfected EGT1-293 (Fig. 3b). However, we failed to observe any alteration in nuclear accumulation of pp90RSK in any HEK293 cell line (data not shown). Ultimately serum stimulation of ERK results in the activation of early serum response genes c-fos and c-jun, both of which are also targets for FHL2 (Reviewed in Johannessen et al. 2006). Stable cell lines overexpressing FHL2 and FHL3 showed comparable serum-induced transcription of the endogenous c-fos and c-jun genes. On the other hand, overexpression of TACC1A not only inhibited serum-induced transcription of c-fos, but also reduced the induction of c-jun that can occur in the presence of cycloheximide (Fig. 4b) (Rao and Mufson 1993). Thus altered expression of TACC1, depending on the level of other effectors, such as the FHLs, may determine the temporal and spatial response of tumor cells to ERK activation.
Discussion
In this report, we have identified a novel interaction between TACC1 and FHL3 during stringent screening of a yeast two-hybrid cDNA library. Using GST pull down of in vitro purified proteins we were able to map the interaction domains to the coiled coil of TACC1 and the region encompassing the first LIM domain of FHL3. We further expanded this analysis to analyze the other members of the TACC and FHL families and clearly demonstrated differential interactions between TACC and FHL family members.
A growing number of protein of diverse functions are known to interact with TACC proteins in Drosophila, Xenopus and humans (Still et al. 2004). Proteomic analysis has shown that the Caenorhabditis elegans ceTAC protein binds to the recently defined evolutionary conserved THAP zinc finger motif of lin36 and lin15A (Clouaire et al. 2005; Roussigne et al. 2003; Walhout et al. 2000). Furthermore, Simpson et al. (2004) recently reported that a small negatively charged C-terminal region of the TACC domain mediates the direct interaction of TACC3 with zinc finger domain 3 of FOG1. Thus, the LIM domain of the FHLs represents the third type of zinc finger to specifically interact with the TACC domain.
By interactions mediated via the LIM domains, members of the FHL family have been implicated in diverse cellular roles, including transcriptional regulation. FHL2 and FHL3 can act as transcriptional coactivators and corepressors for several transcription factors (Labalette et al. 2004; Turner et al. 2003). As the endogenous TACC-FHL interaction is localized to the nuclear compartment, we next examined whether, TACCs could also act as direct or indirect transcriptional coregulators of CREB. Indeed, we demonstrated that each TACC protein could upregulate transcription from a CRE-response element in a similar fashion to FHL2 and FHL3 (Fig. 2). Previously, TACC3 had been identified as a coregulator for three different transcription factors (ARNT1, KLF3 and FOG1) and is known to bind and recruit pCAF to promoter-bound MBD2 (Angrisano et al. 2006; Gangisetty et al. 2004; Garriga-Canut and Orkin 2004; Sadek et al. 2000; Simpson et al. 2004). Although TACC1 and TACC2 can bind components of theSWI/SNF complex and histone acetyltransferase complexes in the nucleus (Gangisetty et al. 2004; Lauffart et al. 2002), this is the first time that TACC1A and TACC2s have been shown to have any transcriptional stimulatory activity.
Targeted overexpression of the human TACC1A isoform to the mouse mammary gland promotes mammary tumorigenesis (Cully et al. 2005). This is linked to increased levels of phospho-AKT and pERK in whole cell extracts derived from the mammary tissue, suggesting that the oncogenic properties of TACC1A may be mediated through Ras and PI-3K pathways (Cully et al. 2005). The results presented in this manuscript have further elucidated one potential mechanism for this effect. We demonstrate that exogenous TACC1A can maintain higher levels of pERK during a 24 h period of serum starvation, relative to a normal cell control. However, a second and more potent effect was noted with the failure of pERK to accumulate in the nucleus of TACC1A overexpressing cells (Figs. 3b, 4a). When these cells were then stimulated with serum for 4 h, increased activation of pERK was noted in the cytosol, but pERK still failed to migrate to the nucleus. Intriguingly, FHL2 increased pERK activation and translocation to the nucleus in TACC1A overexpressing HEK293 cells. This appears contrary to the observation that FHL2 binds to pERK and inhibits its accumulation in the nucleus of cardiomyocytes (Purcell et al. 2004), and may suggest that these affects may be dependent on cellular origin. TACC1A overexpression also had opposing effects in HEK293 cells overexpressing FHL2 or FHL3 (Fig. 3). Increasing TACC1A in EGFHL2 cells resulted in a gradual decrease in ERK activation. Conversely, in EGFHL3-293, TACC1A increased ERK phosphorylation and translocation to the nucleus in a dose dependent manner. This could suggest that the FHLs differentially modify TACC1A behavior and its effects on the MAPK pathway.
Accumulation of pERK in the cytoplasm could be predicted to have multiple effects. First, it could potentially increase phosphorylation of ERK cytosolic substrates, such as p90RSK. Secondly, the decreased nuclear import of pERK could decrease phosphorylation of ERK nuclear targets with an associated alteration in downstream gene regulation. We only observed increased p90RSK phosphorylation when FHL2 induced activation of ERK, an effect that TACC1 inhibited in EGFHL2 cells. However, we did note that serum failed to activate the early serum response genes c-fos and c-jun within a 4 h window of serum stimulation in EGT1A-293 cells compared to the robust activation of these genes in control, EGFHL2- and EGFHL3-293 cells. This indicates that alterations in the expression and subcellular localization of TACC1A could affect cellular responses to external stimuli via perturbation of spatio-temporal dynamics of the MAPK pathway at the level of pERK. However, it should be noted that conclusions on the normal activity of endogenous TACCs must take into account the localization of the endogenous protein in the normal three-dimensional architecture of the tissue in the organism.
In summary, the findings presented here, together with those from previous studies, clearly indicate that the TACCs are a multifunctional family of proteins. Identifying interactions with the tissue-specific and developmentally regulated FHL proteins has now further increased the complexity of TACC function. This latter family has diverse biological roles in cell signaling pathways, actin cytoskeletal dynamics and transcriptional regulation in a variety of tissues. The potential combinatorial interactions between different TACCs and FHLs, together with hetero- and homodimerization within each family raise the possibility of dynamic associations that could have profound effects on cellular metabolism. Thus, one of the roles of the endogenous FHL-TACC interaction could be to temporally or spatially regulate specific, though currently unidentified, promoters during cell growth and differentiation. Indeed, both TACC1 and TACC3 are ideally suited to perform such regulatory roles as they are dynamically expressed during embryogenesis (Aitola et al. 2003; Lauffart et al. 2006; Sadek et al. 2003). For instance, besides the functional overlap between FHL2/3 and TACC1 in the ERK signaling pathway, both TACC3 and FHL3 have been implicated as modifiers of coregulators of the GATA1 transcription factor (Simpson et al. 2004; Turner et al. 2003). In fact, similar to the alteration in subcellular localization of pERK induced by overexpression of TACC1A, overexpression of TACC3 inhibits FOG1-mediated repression of GATA1-dependent transcription in a dose-dependent manner, by relocating FOG1 to the cytoplasm (Simpson et al. 2004). Thus, even minor changes in the expression or localization of these proteins may provide a mechanism of fine-tuning spatio-temporal responses to external stimuli. Our observations could also explain the variable associations of each family of proteins with specific subtypes of malignancy and the changes noted in their subcellular location in tumors compared to the normal tissue (Gabriel et al. 2004, 2006; Genini et al. 1997; Lauffart et al. 2005; Wei et al. 2003). This suggests that the relative protein expression levels of each TACC and FHL family member should also be considered when assessing their roles in tumor biology and cancer risk.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
BL performed the immunoprecipitation and immunolocalization studies. GVS confirmed the interactions between the TACC family and the FHL proteins by GST pull down. BL and GVS contributed equally to this manuscript. OG mapped the TACC1 binding domain on FHL2 and 3. MC performed the SEAP transcriptional assays. IHS performed the initial yeast two-hybrid assay, the serum starvation analysis, conceived the study and drafted the manuscript.
Abbreviations
- ARNT:
-
arylhydrocarbon nuclear translocator
- KLF3:
-
kruppel like factor 3 (basic)
- ceTAC:
-
Caenorhabditis elegans TACC
- ch-TOG:
-
colonic and hepatic tumor over expressed
- CBP:
-
CREB binding protein
- CREB:
-
cAMP response element binding protein
- DAPI:
-
4′,6′-diamidino-2-phenylindole hydrochloride
- FHL:
-
four and a half LIM-only
- FOG1:
-
Friend of GATA 1
- GATA1:
-
GATA binding protein 1
- GST:
-
glutathione-S-transferase
- LIM:
-
Lin-11, Isl-1, Mec-3. Zinc-binding
- lin15A:
-
abnormal cell LINeage family member 15A
- lin36:
-
abnormal cell LINeage family member 36
- LSm-7:
-
U6 snRNA-associated Sm-like protein
- MBD2:
-
methyl-CpG binding protein 2
- MCM7:
-
minichromosome maintenance protein 7
- pCAF:
-
p300/CBP-associated factor
- PLZF:
-
promyelocytic leukemia zinc finger protein
- PCR:
-
polymerase chain reaction
- SEAP:
-
secreted alkaline phosphatase
- SWI/SNF:
-
switching/sucrose nonfermenting factor
- TACC:
-
transforming acidic coiled coil
- THAP:
-
Thanatos associated protein
- YEATS4:
-
YNK7/ENL/AF-9 and TFIIF small subunit domain containing 4
- ZNF638:
-
zinc finger protein 638
References
Aitola M, Sadek CM, Gustafsson JA, Pelto-Huikko M (2003) Aint/Tacc3 is highly expressed in proliferating mouse tissues during development, spermatogenesis, and oogenesis. J Histochem Cytochem 51(4):455–469
Angrisano T, Lembo F, Pero R, Natale F, Fusco A, Avvedimento VE, Bruni CB, Chiariotti L (2006) TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res 34(1):364–372
Chan KK, Tsui SK, Ngai SM, Lee SM, Kotaka M, Waye MM, Lee CY, Fung KP (2000) Protein–protein interaction of FHL2, a LIM domain protein preferentially expressed in human heart, with hCDC47. J Cell Biochem 76(3):499–508
Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP (2005) The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci USA 102(19):6907–6912
Coghill ID, Brown S, Cottle DL, McGrath MJ, Robinson PA, Nandurkar HH, Dyson JM, Mitchell CA (2003) FHL3 is an actin-binding protein that regulates alpha-actinin-mediated actin bundling: FHL3 localizes to actin stress fibers and enhances cell spreading and stress fiber disassembly. J Biol Chem 278(26):24139–24152
Conte N, Charafe-Jauffret E, Delaval B, Adelaide J, Ginestier C, Geneix J, Isnardon D, Jacquemier J, Birnbaum D (2002) Carcinogenesis and translational controls: TACC1 is down-regulated in human cancers and associates with mRNA regulators. Oncogene 21(36):5619–5630
Cully M, Shiu J, Piekorz RP, Muller WJ, Done SJ, Mak TW (2005) Transforming acidic coiled coil 1 promotes transformation and mammary tumorigenesis. Cancer Res 65(22):10363–10370
Fimia GM, De Cesare D, Sassone-Corsi P (2000) A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol Cell Biol 20(22):8613–8622
Gabriel B, Mildenberger S, Weisser CW, Metzger E, Gitsch G, Schule R, Muller JM (2004) Focal adhesion kinase interacts with the transcriptional coactivator FHL2 and both are overexpressed in epithelial ovarian cancer. Anticancer Res 24(2B):921–927
Gabriel B, Fischer DC, Orlowska-Volk M, zur HA, Schule R, Muller JM, Hasenburg A (2006) Expression of the transcriptional coregulator FHL2 in human breast cancer: a clinicopathologic study. J Soc Gynecol Investig 13(1):69–75
Gangisetty O, Lauffart B, Sondarva GV, Chelsea DM, Still IH (2004) The transforming acidic coiled coil proteins interact with nuclear histone acetyltransferases. Oncogene 23(14):2559–2563
Garriga-Canut M, Orkin SH (2004) Transforming acidic coiled-coil protein 3 (TACC3) controls friend of GATA-1 (FOG-1) subcellular localization and regulates the association between GATA-1 and FOG-1 during hematopoiesis. J Biol Chem 279(22):23597–23605
Genini M, Schwalbe P, Scholl FA, Remppis A, Mattei MG, Schafer BW (1997) Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol 16(4):433–442
Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW (2000a) The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA 97(26):14352–14357
Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW (2000b) D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J 19(2):241–252
Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, Adelaide J, Koki A, Houvenaeghel G, Hassoun J, Maraninchi D, Viens P, Birnbaum D, Bertucci F (2005) Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res 65(3):767–779
Johannessen M, Moller S, Hansen T, Moens U, Van Ghelue M (2006) The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci 63(3):268–284
Jung CK, Jung JH, Park GS, Lee A, Kang CS, Lee KY (2006) Expression of transforming acidic coiled-coil containing protein 3 is a novel independent prognostic marker in non-small cell lung cancer. Pathol Int 56(9):503–509
Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y (2004) Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and beta-catenin. Mol Cell Biol 24(24):10689–10702
Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Ehler E (2002) Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci 115(Pt 24):4925–4936
Lauffart B, Howell SJ, Tasch JE, Cowell JK, Still IH (2002) Interaction of the transforming acidic coiled-coil 1 (TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple TACC1-containing protein complexes in human cells. Biochem J 363(Pt 1):195–200
Lauffart B, Gangisetty O, Still IH (2003) Molecular cloning, genomic structure and interactions of the putative breast tumor suppressor TACC2. Genomics 81(2):192–201
Lauffart B, Vaughan MM, Eddy R, Chervinsky D, Dicioccio RA, Black JD, Still IH (2005) Aberrations of TACC1 and TACC3 are associated with ovarian cancer. BMC. Womens Health 5(1):8
Lauffart B, DiMatteo A, Vaughan MM, Cincotta MA, Black JD, Still IH (2006) Temporal and spatial expression of TACC1 in the mouse and human. Dev Dyn 235(6):1638–1647
L’Esperance S, Popa I, Bachvarova M, Plante M, Patten N, Wu L, Tetu B, Bachvarov D (2006) Gene expression profiling of paired ovarian tumors obtained prior to and following adjuvant chemotherapy: molecular signatures of chemoresistant tumors. Int J Oncol 29(1):5–24
Li HY, Kotaka M, Kostin S, Lee SM, Kok LD, Chan KK, Tsui SK, Schaper J, Zimmermann R, Lee CY, Fung KP, Waye MM (2001a) Translocation of a human focal adhesion LIM-only protein, FHL2, during myofibrillogenesis and identification of LIM2 as the principal determinants of FHL2 focal adhesion localization. Cell Motil Cytoskelet 48(1):11–23
Li HY, Ng EK, Lee SM, Kotaka M, Tsui SK, Lee CY, Fung KP, Waye MM (2001b) Protein–protein interaction of FHL3 with FHL2 and visualization of their interaction by green fluorescent proteins (GFP) two-fusion fluorescence resonance energy transfer (FRET). J Cell Biochem 80(3):293–303
Line A, Slucka Z, Stengrevics A, Li G, Rees RC (2002) Altered splicing pattern of TACC1 mRNA in gastric cancer. Cancer Genet Cytogenet 139(1):78–83
Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC (2003) Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA 100(10):5974–5979
Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kuhl M, Behrens J, von der MK, Starzinski-Powitz A, Wixler V (2002) The LIM-only protein FHL2 interacts with beta-catenin and promotes differentiation of mouse myoblasts. J Cell Biol 159(1):113–122
McLoughlin P, Ehler E, Carlile G, Licht JD, Schafer BW (2002) The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein. J Biol Chem 277(40):37045–37053
Morlon A, Sassone-Corsi P (2003) The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc Natl Acad Sci USA 100(7):3977–3982
Muller JM, Metzger E, Greschik H, Bosserhoff AK, Mercep L, Buettner R, Schule R (2002) The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J 21(4):736–748
Ng EK, Chan KK, Wong CH, Tsui SK, Ngai SM, Lee SM, Kotaka M, Lee CY, Waye MM, Fung KP (2002) Interaction of the heart-specific LIM domain protein, FHL2, with DNA-binding nuclear protein, hNP220. J Cell Biochem 84(3):556–566
Partheen K, Levan K, Osterberg L, Horvath G (2006) Expression analysis of stage III serous ovarian adenocarcinoma distinguishes a sub-group of survivors. Eur J Cancer 42(16):2846–2854
Peters DG, Kudla DM, Deloia JA, Chu TJ, Fairfull L, Edwards RP, Ferrell RE (2005) Comparative gene expression analysis of ovarian carcinoma and normal ovarian epithelium by serial analysis of gene expression. Cancer Epidemiol Biomark Prev 14(7):1717–1723
Piekorz RP, Hoffmeyer A, Duntsch CD, McKay C, Nakajima H, Sexl V, Snyder L, Rehg J, Ihle JN (2002) The centrosomal protein TACC3 is essential for hematopoietic stem cell function and genetically interfaces with p53-regulated apoptosis. EMBO J 21(4):653–664
Pu JJ, Li C, Rodriguez M, Banerjee D (2001) Cloning and structural characterization of ectacc, a new member of the Transforming Acidic Coiled Coil (TACC) gene family: cDNA sequence and expression analysis in human microvascular endothelial cells. Cytokine 13(3):129–137
Purcell NH, Darwis D, Bueno OF, Muller JM, Schule R, Molkentin JD (2004) Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol Cell Biol 24(3):1081–1095
Rao P, Mufson RA (1993) Interleukin-3 inhibits cycloheximide induction of C-jun mRNA in human monocytes: possible role for a serine/threonine phosphatase. J Cell Physiol 156(3):560–566
Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP (2003) The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci 28(2):66–69
Roux PP, Blenis J (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68(2):320–344
Sadek CM, Jalaguier S, Feeney EP, Aitola M, Damdimopoulos AE, Pelto-Huikko M, Gustafsson J (2000) Isolation and characterization of AINT: a novel ARNT interacting protein expressed during murine embryonic development. Mech Dev 97(1–2):13–26
Sadek CM, Pelto-Huikko M, Tujague M, Steffensen KR, Wennerholm M, Gustafsson JA (2003) TACC3 expression is tightly regulated during early differentiation. Gene Expression Patterns 3(2):203–211
Samson T, Smyth N, Janetzky S, Wendler O, Muller JM, Schule R, von der MH, von der MK, Wixler V (2004) The LIM-only proteins FHL2 and FHL3 interact with alpha- and beta-subunits of the muscle alpha7beta1 integrin receptor. J Biol Chem 279(27):28641–28652
Schreiber E, Matthias P, Muller MM, Schaffner W (1989) Rapid detection of octamer binding proteins with ‘mini-extracts,’ prepared from a small number of cells. Nucleic Acids Res 17(15):6419
Simpson RJ, Yi Lee SH, Bartle N, Sum EY, Visvader JE, Matthews JM, Mackay JP, Crossley M (2004) A classic zinc finger from friend of GATA mediates an interaction with the coiled-coil of transforming acidic coiled-coil 3. J Biol Chem 279(38):39789–39797
Stewart JP, Thompson A, Santra M, Barlogie B, Lappin TR, Shaughnessy Jr J (2004) Correlation of TACC3, FGFR3, MMSET and p21 expression with the t(4;14)(p16.3;q32) in multiple myeloma. Br. J. Haematol. 126(1):72–76
Still IH, Hamilton M, Vince P, Wolfman A, Cowell JK (1999a) Cloning of TACC1, an embryonically expressed, potentially transforming coiled coil containing gene, from the 8p11 breast cancer amplicon. Oncogene 18(27):4032–4038
Still IH, Vince P, Cowell JK (1999b) The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics 58(2):165–170
Still IH, Vettaikkorumakankauv AK, DiMatteo A, Liang P (2004) Structure-function evolution of the transforming acidic coiled coil genes revealed by analysis of phylogenetically diverse organisms. BMC Evol Biol 4(1):16
Turner J, Nicholas H, Bishop D, Matthews JM, Crossley M (2003) The LIM Protein FHL3 Binds Basic Kruppel-like Factor/Kruppel-like Factor 3 and Its Co-repressor C-terminal-binding Protein 2. J Biol Chem 278(15):12786–12795
Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M (2000) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287(5450):116–122
Wei Y, Renard CA, Labalette C, Wu Y, Levy L, Neuveut C, Prieur X, Flajolet M, Prigent S, Buendia MA (2003) Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J Biol Chem 278(7):5188–5194
Wixler V, Geerts D, Laplantine E, Westhoff D, Smyth N, Aumailley M, Sonnenberg A, Paulsson M (2000) The LIM-only protein DRAL/FHL2 binds to the cytoplasmic domain of several alpha and beta integrin chains and is recruited to adhesion complexes. J Biol Chem 275(43):33669–33678
Acknowledgements
This work was supported in part by developmental funds support from Arkansas Tech University and the Roswell Park Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lauffart, B., Sondarva, G.V., Gangisetty, O. et al. Interaction of TACC proteins with the FHL family: implications for ERK signaling. J. Cell Commun. Signal. 1, 5–15 (2007). https://doi.org/10.1007/s12079-007-0001-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-007-0001-3