Abstract
Two breakthrough techniques that have totally revolutionized biology in last 1 decade are the discovery of genome editing tools and growing the stem cells/primary tissue explants in defined 3D culture. In this regard the discovery of CRISPR-Cas9 as a specific gene editing tool and organoid culture from adult stem cell has provided easy handy tools to uncover the process of organ development and also modeling cancer. Genetically modified organoids have been developed by sequential knockout and knockin of driver mutations by genome editing followed by niche-based selection. The modified organoids when xenotransplanted in animal models faithfully recapitulate the neoplastic events of human tumors. The present review focuses on the merging of these two powerful technologies in understanding the complexities of colon and liver cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Innovating new biological tools are an integral part of scientific advances. The success of any biological tool is dependent on its high precision, reproducibility, ease of use, cost effectiveness, and utility in multiple areas of research. There are very few technologies that have made big impact in a very short time from its time of discovery, especially in the area of translational health. In this regard, the CRISPR-Cas9 genome editing tool, together with 3D organoid technology have revolutionized the science behind precision medicine. These next generation technologies have immensely helped in understanding the mechanisms involved in disease progression such as cancer.
The hallmark features of gastrointestinal cancer involve genetic and epigenetic alterations, in known oncogenes and tumor suppressors [1]. Heterogeneity is a mainstay in gastrointestinal cancers and understanding the molecular pathways contributing to specific pathologies is not only important in understanding the pathogenesis but also their prognosis. To study the various steps involved in carcinogenesis, appropriate animal and cell culture based models are often used. The advent of organoids as 3D cell culture system has not only helped in understanding the organization of organ development but also in modeling human disease in particular cancer. The first human and animal organoids were derived from the intestinal epithelium and later this technology has been utilized to derive patient derived tumoroids which can recapitulate very closely the tumor biology [2, 3]. Genetic engineering of normal organoids with overexpression or knockdown of genes by plasmid based or lentiviral based methods have been successfully tried. Additionally, when used together with genome editing tools the organoid technology not only model human disease, but also pave way for therapeutic corrections. There are basically three different types of genome editing viz., zinc-finger nucleases, transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9. All these three techniques vary in their efficiency and specific targeting. However, the ease of use of CRISPR/Cas9 has made it the most dominant and favored technique in genetically engineering organoid. In fact, the genome edited organoids closely mimic the human cancer and are described in later sections (Fig. 1).
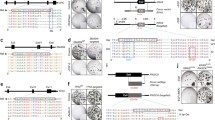
Combining organoid and genome editing CRISPR-Cas9 technology to develop modified organoids. Organoids are established using the progenitor cell (Lgr5+), by embedding in suitable matrix followed by culturing in defined medium consisting of cocktail of tissue specific growth factors (blue) and inhibitors (orange). The organoids can be genetically engineered using the CRISPR-Cas9 technology to knockout (KO) genes relevant to cancer such as APC, PTEN, and p53. The DNA damage repair pathways such as non-homologous end joining (NHEJ) or HDR (homologous DNA recombination) helps in indels or knockin to obtain genetically modified organoids. CRISPR-HOT is a new technology developed for knockin of genes in hard to transfect organoids like those derived from liver
The CRISPR-Cas9 system: from a bacterial adaptive immune system to a powerful genome editing tool
The CRISPR-Cas9 component first discovered in eubacteria and arachea evolved as a potent innate defense mechanism against viral infection which helps in destruction of invading bacteriophage DNA/RNA. The CRISPR-Cas9 thus acts like molecular scissors and this finding has been efficiently harnessed as a tool for genome editing. The acronym CRISPR refers to clustered regularly interspaced short palindromic repeat sequences of 24–40 nt and was first recognized in E. coli and Haloferax mediterranei as a unique orderly repeat sequence [4, 5]. Later similar sequences were uncovered in other bacteria and arachea. The homologous genes accompanying the CRISPR loci are the Cas genes (for CRISPR-associated genes) which encode the endonuclease. The importance of CRISPR-Cas associated loci was realized when it was noted that the repeat sequence often flanked non-bacterial sequences which closely matched to bacteriophages/prophages. Thereby indicating that these external sequences were captured during invasion of the bacteria and serve as a memory to prevent future infections [6, 7]. The observation that bacteria carrying spacer sequences similar to phage were resistant to infection by bacteriophages harboring same sequence, in turn, led to the postulation that CRISPR-Cas system is analogous to mammalian RNAi [8, 9]. Subsequent work in Streptococcus thermophiles and E. coli showed that CRISPR locus is transcribed as mature CRISPR RNA (crRNAs). The maturation of crRNA in turn is mediated by trans-encoded small crRNA (tracrRNA), the endonuclease Cas9 and RNase III [10]. This functional CRISPR-Cas9 complex can cleave complementary viral sequences, depending on the type of CRISPR-Cas system. Additionally, the CRISPR-Cas system also utilizes protospacer adjacent motifs (PAMs) which are specific small nucleotide sequences required for effective and specific cleaving by the Cas nuclease. The recognition of the viral PAM sequence makes CRISPR-Cas9 effectively target the virus nucleic acid and not the host bacterial genome thereby acting as a potent antiviral defence system.
About 93 Cas genes are known till now, of which the most efficient endonuclease is the Cas9 which cleaves DNA. Other Cas proteins such as Cas13 can cleave RNA [11]. Additionally an enzymatically dead Cas9 (dCas9), instead of cutting the DNA, can modify the epigenome. While CRISPR-CAS system is an adaptive prokaryotic immune system, its potential power to precisely cut the nucleic acid sequence at the desired site has been harnessed as a tool for efficient genome editing for either knockout and knockin of genes [12]. The adaptation of bacterial originated CRISPR-Cas9 system as powerful genetic tool, together with its ease of its use has made it a very popular technology for a wide range of applications in life science, which has deserving won the Nobel prize for chemistry recently [13]. The genome editing tool of CRSIPR-Cas9 has been efficiently used in understanding the biology of cancer using the organoid model which is outlined below.
Organoid technology: from development biology to modeling cancer
From the two dimensional monolayer cell culture system, the development of three dimensional which can produce an organ-like in vitro has been a great achievement. Organoids are defined as self-organizing 3D structures grown in vitro and originate from either adult/pluripotent stem cell. This technology has been pioneered by the laboratory of Hans Clever and colleagues. The first successful organoids were established using leucine-rich repeat containing G protein-coupled receptor 5 (LGR5) + intestinal adult stem cells [14, 15]. Since then organoids have been established for almost all human organs. The organoids grow as multicellular structures closely mimicking not only the tissue architecture and pathology, but also perform tissue specific functions.
The growth of organoids for recapitulating organogenesis requires the presence of appropriate growth factors and extracellular matrix (3D). Basically there are three steps in establishment of organoid cultures (a) initiation, which involves the activation of key signaling events in sustaining the stem cell using defined media and matrix, (b) differentiation step, where stem cells in turn give rise to different cell lineage thereby inducing multicellularity to mimic tissue structure in the presence of cocktail of growth factors and small molecules and (c) finally expansion by leveraging self-renewal, so that they can be banked and propagated when required [16]. In contrast to the primary cell culture which stop replication as they undergo senescence with repeated cell division, organoids can be expanded for more than 1 year indicating that the stem cells from which they derive can self-renew besides undergoing lineage fate decision. The organoid technology was first pioneered using intestinal organoids and “mini-gut” thus obtained showed composite of paneth cells, enterocytes, goblet cells, and enteroendocrine cells [18]. The organoid technology has now been adapted for most of the organs.
Besides growing organoids from healthy tissues, organoids can also be grown from diseased tissue and this has immensely helped in understanding biology of many diseases including cancer. Patient derived tumoroids show similar genomic alterations as the primary tumor implicating that they indeed faithfully recapitulate the tumor biology [2, 3].The missing components in the tumor organoid are the stromal cells and immune cell compartment, however, newer technologies using air liquid interface based systems can even mimic the immune tumor microenvironment [18]. The organoid technology holds great promises in understanding basic biology of embryonic development and lineage specification. In addition, organoid from normal tissue and tumoroids developed from corresponding cancer region have become useful in studying disease biology. Using this technology gastrointestinal organoids from colon, stomach, liver, and pancreas have been developed. Of these much progress in genetically modified organoids of colon and liver tissue which has been discussed in the present review.
Colon cancer organoids/tumoroids and advantages
Organoids were first described in colon from the LGR5 positive stem cells [12, 15]. Unlike other cancer the colorectal cancer especially the hereditary non-polyposis colorectal cancer (HNPCC) often is associated with autosomal inheritance of several mismatch repair genes viz., hMSH2, hMLH1, etc. In these patients the primary risk for cancer is the colon, however, these subjects are also at high risk of other cancers such as stomach, thyroid pancreas, etc. Other GI tract cancers, for colorectal cancer patients risk assessment and stratification becomes important. Compared to formaldehyde fixed tissues, generation of a renewable patient derived organoid bank of colon cancer will be extremely importance in predictive biology. Further, colorectal cancers are often heterogenous and many drugs effective in mice based cancer models often fail in clinical trials reiterating the genetic diversity of human cancer. Hence, in such conditions drug testing using patient derived cancer organoids/tumoroids with less turnaround time for drug testing will provide crucial information for clinical decisions. Recent work has shown utility of colon tumoroids in predicting metastatic disease. Tumoroids which did not respond to irinotecan-based palliative chemotherapy often show metastatic disease [19] thereby predicting the course of disease. Additionally genetic engineering of normal colon organoids has provided much needed information on the disease biology and described in later sections.
CRIPSR/CAS9 genome editing in human colon cancer organoids
Genetically modified organoids have become useful tools in understanding the biology of cancer. Genetic engineering of normal organoids with overexpression or knockdown of genes by plasmid or lentiviral based methods have been successfully tried. Additionally when used together with CRISPR-Cas9, organoid technology can not only model human disease, but also pave way for therapeutic corrections. In 2013, a seminar work by Schwank et al. [20] combined the two powerful technologies of organoid and CRISPR-Cas9 for correcting inherent genetic defects in cystic fibrosis using the intestinal organoid model. Much of the pioneering work on combining organoid with CRISPR-Cas9 has originated form the group of Dr. Hans Clever based in Netherland [21].
From an application perspective for an efficient knockin or knockout strategy of oncogene or tumor suppressor genes, the dual component of CRISPR-Cas system includes: (a) CRISPR RNA (crRNA) sequence which is specific to target gene together with a transactivating CRISPR RNA (tracrRNA) and (b) the endonuclease viz., Cas9. Together this constitutes the single guide RNA (sgRNA) entity. The specific RNA sequence guides the RNA–protein complex to corresponding DNA sequencing of host and the endonuclease cleavage creates a double-strand break (DSB). The damaged mammalian DNA is then repaired by either non-homologous end joining (NHEJ) or homology-directed repair (HDR) to create indels (insertion and deletions). The NHEJ pathway is mostly efficient in generating modified organoids, however, the method is error prone. On the other hand the HDR method is more specific but less efficient. [22,23,24]. The delivery method for the CRISPR-CAS9 complex is mostly extrachromosomal plasmid based or viral DNA delivery system. In addition, ribonucleoprotein (RNP) delivery of CRISPR-Cas9 is also preferred as it produces fewer off-target effects. The delivery of sgRNA mostly includes liposomal transfections, electroporation, lentiviral or in the form of exosomes [21]. All these various methods have been tried to successfully modify the organoids developed from normal epithelium for modeling cancer especially the intestinal cancer. The colorectal cancers are generally divided into four subtypes (a) Type 1 are microsatellite instable associated with hypermethylation and good prognosis, (b) Type 2 show the activation of WNT signaling and TP53 gene mutation, (c) Type 3 tumors have mutations in K ras gene and (d) Type 4 show the activation of TGFβ pathway with stromal infiltration [25]. Using the genome editing technology of CRISPR/Cas9 colorectal cancers Types 2, 3, and 4 have been modeled using organoids and described below.
Genetically modified colon organoids derived by CRIPSR/CAS9 genome editing mimics adenocarcinoma of colon: single vs double hit approach
The proof of concept that CRISPR/CAS9 can modify driver genes in normal organoids thereby modeling cancer progression was first established by targeting the Adenomatous Polyposis Coli (APC) gene in the intestinal organoid model. Inactivation of APC due to its mutation is a hallmark feature of colon cancer. The tumor suppressor gene APC forms a destruction complex with kinases GSK3β and CK1α/ε (casein kinase 1α/ε) which helps in destruction of β‐catenin [26]. APC is a negative regulator of Wnt signaling. In fact, the actively dividing intestinal crypt cells secrete Wnt thereby stabilizing beta catenin and its nuclear entry leading to cellular proliferation. For growing the intestinal organoids Wnt activator R-spondin-1 is used in the growth factor cocktail as it is ligand for LGR5 intestinal stem cell and induces its proliferation. If APC is inactivated in the LGR5 cells, Wnt pathway will be activated and organoids can grow even in the absence of R-spondin and indeed this was proved by Schwank et al. in their classic experiment [20]. Inactivating APC gene in the intestinal LGR5 cells of organoids by the CRISPR/Cas9 resulted in mutant intestinal organoids with cystic morphology resembling colon cancer. In addition, this work also pioneered double knockout of RNF43 and Znrf3 which are also Wnt regulators like APC. This paved way for multiple loci targeting simultaneously in organoids using a sequential single transfection protocol in organoids derived from either normal or precursor lesions form adenomas (Table 1). In addition a hallmark feature of hereditary non-polyposis colorectal cancer (HNPCC) is an inherited mismatch repair (MMR) gene mutation viz., genes MLH1 or MSH2 which leads to microsatellite instability. The use of CRISPR-Cas9 molecular scissors to selectively delete mismatch repair gene MLH1 in human colon organoids resulted in the accumulation of genetic mutations which exactly match the mutation burden of mismatch repair–deficient colorectal cancers in human [26].
Genome editing of colon organoids by CRIPSR/CAS9: multiple hit approach
Colon cancers are usually considered to be clonal in nature and include sequential progressive genetic events of loss of tumor suppressors such as APC and p53 and gain of function in oncogenes such as K ras, SMAD4, etc. [1]. Unlike the existing mouse or cell culture models where at most one or two genetic mutants are obtained, in an elegant series of experiments Dorst et al. all using the CRISPR-Cas9 module performed genome editing for four important genes (APC, p53, K ras, SMAD4) and used this in combination with appropriate organoid culture medium specific for selection of mutant organoid [27]. Briefly, first the APC knockout mutant organoid was developed which could be selected in the absence of R-spondin culture media as explained in previous section. In the inactivated APC background, genome editing was done to inactivate p53 and cells were selected in the presence of p53 activator nutlin-3. Only those cells which have inactivated p53 could survive the genotoxic effects of Nutlin. This is because Nutlin is an activator of p53 and induces cytotoxicity of organoids with wild type p53. In the APC/p53 dual knockout background, an activating K rasG12 mutation was introduced. This gives a selective advantage to organoids to grow in the absence of epidermal growth factor. Lastly, TGFβ pathway was targeted by inactivating mutation of SMAD4 and selection of organoids in the absence of BMP pathway inhibitor noggin. This work elegantly generated a quadruple mutant intestinal organoids with activating mutation of K rasG12D and inactivation of APC/p53/ SMAD4 (Fig. 2). The quadruple mutant organoid when transplanted in mouse models showed invasive nature together with chromosomal instability thereby recapitulating the colorectal cancer as observed in humans. Yet another work used a quintuple strategy of targeting multiple genes viz., APC, SMAD4, TP53, K ras, and PIK3CA using human normal and adenoma derived organoids [28]. The engineered organoids could readily grow in minimal growth factor niche and also form tumors on xenotransplantation. However, macrometastasis could be seen only in the chromosomal instable intestinal adenoma derived organoids, indicating that quintuple driver mutations on their own are insufficient to drive metastasis and additional chromosomal aberrations are needed. This study supported the role of pre-existing human adenomas as precursor lesions which progress to neoplasia.
Various genome editing approaches for modeling cancer using organoids. Genome editing by a CRISPR-Cas9 knockout followed by niche-based selection in defined growth medium, b chromosomal engineering and introduction of R-spondin fusion gene and c CRISPR-Cas9 library pool for discovering novel genes. KO, knockout; Overexp overexpression, + plus, − minus
Genome editing of colon organoids by chromosomal engineering and gene fusion
Colorectal cancers are heterogenous in nature and one such type is the serrated colorectal type which has now been modeled in BRAFV600E mutant organoids followed with TGFβ treatment (Fig. 2). In general, the tumor adenoma (TA) derived organoids from human adenoma colon polyps undergo apoptosis when treated with TGFβ. However, adenoma derived organoids modified with CRISPR/CAS9 for activating mutation of BRafV600E resisted cell death in the presence of TGFβ [29]. Intriguingly the mutant BRAF modified adenoma in the presence of TGFβ showed features of serrated colorectal cancer neoplasia which closely mimicked the Type 4 type of colorectal cancer with poor prognosis. Recently the traditional serrate adenomas (TSA) of colon were engineered using the human intestinal derived organoids using a newer approach of CRISPR/CAS9 chromosomal engineering [30]. The traditional serrate adenomas often show fusions in the R-spondin gene with either EIF3 or PTPRK and rare fusion of BRAF with DLGB1 gene is also seen [31,32,33]. To develop R-spondin fusion, Sato’s group in Japan has developed a novel approach of chromosomal engineering [34]. This involved genetic reconstruction whereby CRISPR-Cas9 targeted fusion breakpoints to fuse R-spondin to either EIF3E or PTPRK genes by chromosomal deletion. The R-spondin fusion was done in a modified intestinal organoid having activating BRAF and mutant GREM1. In this mutant organoid background R-spondin promoted tumor formation with pathology similar to traditional serrate adenomas when transplanted in xenograft mouse model. Further, unlike the other colon cancer organoid models described earlier, the gene fusion organoid model showed the methylator phenotype which is commonly seen in human colorectal cancers.
CRISPR/CAS9 genetic screen in colon cancer organoids
While the above strategies utilized sequential genome editing, in a recent development organoids were subjected to high throughput CRISPR-Cas9-based screens to find novel candidate genes in CRC progression [35]. The CRISPR/CAS9 screen utilized lentiviral transfection of guide RNAs pool targeting 9–10 genes in the intestinal organoids with an APC/K ras mutant background. The transformed organoids were xenotransplanted in mouse model and monitored for tumor formation. This CRISPR/CAS9 screen has identified novel tumor suppressors, viz, Activins (Acvr1b, Acvr2a) and chromatin remodeller, Arid2, in colorectal cancer, while TP53 was identified as candidate gene responsible for CRC metastasis.
Genetically modified hepatic organoids mimicking liver cancer
Unlike the intestinal organoid models which are easily amenable to genome editing tools, the hard to transfect liver organoids are a real challenge. Only recently, genome editing have been achieved in liver organoid model. Artegiani et al., [36] have pioneered a new tool called CRISPR—Cas9-mediated homology-independent organoid transgenesis (CRISPR–HOT), which allows insertion of gene (knockin) in organoids with a reporter tag for easy visualization of cells and tracing their division. The CRISPR-HOT makes use of NHEJ for genomic integration. This technology was tried in liver organoids for knockin of cadherin and beta-tubulin genes to label hepatocyte membrane and mitotic spindle, respectively, for monitoring of hepatocyte division. Recent evidence suggests that increased ploidy state is a feature of hepatocellular carcinoma [37]. Combining CRISPR-HOT knockin model with p53 knockout liver organoid, the authors showed aberrant mitotic spindle formation with increased aneuploidy and polyploidy. This work suggested that loss of p53 is related to mitotic spindle aberrations which can contribute to hepatocarcinogenesis.
Genetically modified organoids from biliary tract cancer (cholangiocarcinoma, gall bladder, and pancreas)
The biliary tract organs such as bile duct, gall bladder, and pancreas have all been successfully utilized to develop patient derived organoids. Normal human organoids have been transformed to tumoroid by introducing specific oncogenic mutations for cholangiocarcinoma, pancreatic adenocarcinoma, and gall bladder cancer. Ductal derived cholangiocyte organoids with the deletion of p53 and BAP1 genes using the CRISPR/Cas9 vectors resulted in organoids with the loss of epithelial polarity, reduced cell-to-cell adhesion and increased motility mirroring the effects of cholangiocarcinoma with increased cell proliferation [38]. In yet another report, liver organoids with an activated K ras background were subjected to a genome editing using CRISPR-Cas9 to inactivate the tumor suppressors PTEN and p53 [39]. When these modified liver organoids were transplanted to mice model they gave rise to tumors resembling intrahepatic cholangiocarcinomas.
There are very few reports on organoids derived from gallbladder cancer. Erlangga et al. [40] derived murine gallbladder organoids expressing mutant K ras or overexpression of ERBB. These organoids when subjected to CRISPR-Cas9 gene editing technique for deletion of p53 or PTEN and transplanted in immunocompromised mice resulted in gallbladder cancer (GBC) that resembles the human disease.
Using normal pancreatic organoids, sequential genome editing was performed with knockin of mutant K ras together with deletion of tumor suppressors, viz., TP53, CDKN2A, and SMAD4. The resulting genome edited organoids when transplanted to immunodeficient mice, resulted in the development of pancreatic intraepithelial neoplasia [41].
Newer advancements in organoid system in personalized medicine
As the organoids are stable and keep renewing, a major advancement in this field is biobanking the patient derived organoids (PDO) from various organs. The large scale production of organoids/tumoroids have proved useful in capturing and preserving the cancer heterogeneity in terms of various histological types of cancer. The generation paired normal and cancer originated PDOs have proved to be extremely useful for not only high throughput testing for drug testing but also understanding the genomic alterations. However, one limitation of using organoids is lack of immune cells and vasculature. Tumor microenvironment consists of a variety of nonparenchymal cells such as the infiltrating immune cells, the endothelial cells, pericytes, and tumor associated fibroblasts. In this regard, the latest development includes coculturing the organoids with tumor associated fibroblasts and with patient derived immune cells to resemble the tumor. Accordingly, pancreatic ductal adenocarcinomas have been co-cultured with cancer associated fibroblasts to mimic desmoplastic reaction and inflammatory reaction [42]. These newer advancements of coculturing PDOs with patient derived T cells will prove advantageous in future for predicting the response to immunotherapy. Interestingly, one report has shown that patient derived tumoroids may contain the infiltrating lymphocytes which when treated with anti-PD-1 and/or anti-PD-L1 resulted in the activation of T cells residing in the organoid cultures and caused tumor cell elimination [43]. Hence, the current technology of PDOs will take us one step closer to precision medicine. As cancer often involves multiple genetic lesions, effective strategies are needed for multitargeting. As changes in epigenome is a hallmark feature of cancer, CRISPR-based dCas9 epigenome editing is required for epigenetic manipulations. While the genome editing studies in organoids have been well studied for intestine, the studies are far too less in organoids from other organs as they are difficult to transfect. Hence better delivery systems based on next generation based viral vector, nanoparticles or exosomes are needed. Most of the CRISPR-Cas9 vectors make use of homologous DNA repair which is less efficient compared to NHEJ, however, the NHEJ based recombination method is usually error prone. The most frequent challenge in CRISPR/CAS9 is off-target effects and missing out the specific target. To understand the cancer signaling networks CRISPR/CAS9 large screening strategies similar to RNAi libraries are needed.
Prospects
Patient derived organoids are proving extremely useful in precision medicine as a tool for drug testing and making clinical decisions. Most of the studies describing cancer modeling have utilized CRISPR/CAS9 technology by introducing driver mutations in normal organoids to generate tumoroids to model cancer. However, there are hardly any papers where the driver mutations were inactivated using genome editing scissors in tumoroid models to reverse the process towards normalcy. The genome editing of organoids and generation of modified clones require niche-based selection and there is always a possibility that the organoids become resistant to growth factor withdrawal due to spontaneous epimutations introduced during continuous culture conditions. While currently much of the genome editing using organoids are restricted to in vitro settings, a greater challenge will be genetically organoids as effective tools for treatment of solid cancers in the in vivo setting. Can the edited tumoroid with cancer reversing phenotype be used for precision medicine is still a long awaited question? Genome editing holds great promise for immunotherapy for cancer. In a recent study Lu et al., [44] have used CRISPR/Cas9 to ablate PD1 on T cells and the autologous cells were infused back to lung cancer patients. This study reports minimal off-target effects and one patient showed stable disease after treatment as the edited T cells were retained for till 72 weeks. More studies are required for harnessing the new technologies of organoids and genome editing for clinical use. In spite of some of the current limitations, combining these two powerful technologies has taken us closer towards reality of precision medicine.
References
Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70.
Weeber F, Wetering M, Hoogstraact M, Dijkstra KK, Krijgsman O, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. PNAS. 2015;112:13308–11.
Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–45.
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–33.
Mojica FJM, Juez G, Rodriguez FV. Transcription at different salinities of Haloferax mediterranei sequence adjacent to partially modifies Pstl sites. Mol Microbiol. 1993;9:613–21.
Mojica FJM, Villasenor ChcD, Martinez JG, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82.
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindromic repeats (CRISPRs) have spacers of extra chromosomal origin. Microbiology. 2005;151:2551–61.
Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanism of action. Biol Direct. 2006;7:1–26.
Morange MJ. What history tells us XXXIX. CRISPER-Cas: from a prokaryotic immune system to a universal genome editing tool. J Biosci. 2015;40:829–32.
Rodriguez DRR, Solis RR, Elizondo MAG, Rodriguez MLG, Saldana HAB. Genome editing: a perspective on the application of CRISPER/Cas 9 to study human diseases (review). Int J Mol Med. 2019;43:1559–74.
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucelic acid detection with CRISPER-Cas13a/C2c2. Science. 2017;356:438–42.
Doudna JA, Charpentier E. The new frontier of genom engineering with CRISPER-Cas9. Science. 2014;346:1258096–1–1258096–2.
Cohen J. A cut above: pair that developed CRISPER earns historic award. Science. 2020;370:271–2.
Sato T, Vries RG, Snippert HJ, Wetering M, Barker N, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5.
Sato T, Clevers H. Growing self organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–4.
Clinton J, Koeppen PW. Initiation, expansion, and cryopreservation of human primary tissue-derived normal and disease organoids in embedded three dimensional culture. Curr Protoc Cell Biol. 2019;82:1–20.
Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol Biol. 2013;945:319–28.
Neal JT, Li X, Zhu J, Giangarra V, Grezeskwaik CL, et al. Organoids modeling of the tumor immune microenvironment. Cell. 2018;175:1972–88.
Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11(513):eaay2574.
Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–8.
Fujii M, Clevers H, Sato T. Modeling human digestive diseases with CRISPER-Cas9-modified organoids. Gasteroenterology. 2019;156:562–76.
Yang Q, Oost KC, Liberali P. Engineering human knock-in organoids. Nat Cell Biol. 2020;22:261–3.
Hendrinks D, Clevers H, Artegiani B. CRISPER-Cas tool and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell. 2020;27:705–31.
Yip BH. Recent advances in CRISPR/Cas9 delivery strategies. Biomolecule. 2020;10:839.
Wang W, Kandimalla R, Hung H, Zhu L, Li Y, et al. Molecular subtyping of colorectal cancer. Recent progress, new challenges and emerging opportunities. Cancer Biol. 2019;55:37–52.
Schneikert J, Behrens J. The canonical Wnt signaling pathway and its APC partner in colon cancer development. Gut. 2007;56:417–25.
Drost J, Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7.
Matano M, Date S, Shimokawa M, Takano A, Fujii M, et al. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–62.
Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med. 2016;8:745–60.
Kawasaki K, Fujii M, Sugimoto S, Ishikawa K. Chromosome engineering of human colon derived organoids to develop a model of traditional serrated adenoma. Gastroenterology. 2019;158:638–51.
Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–4.
Han T, Schatoff EM, Murphy C, Zafra MP, Wilkinson JE, et al. Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun. 2017;8:15945.
Bettington ML, Chetty R. Traditional serrated adenoma: an update. Hum Pathol. 2015;46:933–8.
Kawasaki K, Fujii M, Sugimoto S, Ishikawa K, Matano M, et al. Chromosome engineering of human colon derived organoids to develop a model of traditional serrated Adenome. Gastroenterology. 2020;158:638–51.
Takeda H, Kataoka S, Ali MAE, Oshima H, Yamamoto D, et al. CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc Natl Acad Sci USA. 2019;116(31):15635–44.
Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, et al. Fast and efficient generation of knock-in human organoids using homology independent CRISPER-Cas9 precision genome editing. Nat Cell Biol. 2020;22:321–31.
Bou-Nader M, Caruso S, Donne R, Celton-Morizur S, Calderaro J, et al. Polyploidy spectrum: a new marker in HCC classification. Gut. 2020;69:355–64.
Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell. 2019;24:927–43.
Saborowski A, Wolff K, Spielberg S, Beer B, Hartleben B, et al. Murine liver organoids as a genetically flexible system to study liver cancer In vivo an In vitro. Hepatol Commun. 2019;3:423–36.
Erlangga Z, Wolff K, Poth T, Peltzer A, Nahnsen S, Spielberg S, et al. Potent antitumor activity of liposomal irinotecan in an organoid- and CRISPR-Cas9-based murine model of gallbladder cancer. Cancers. 2019;11:1904.
Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–67.
Tsai S, McOlash L, Palen K, Johnson B, Duris C, Yang Q, et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancers. 2018;18:335.
Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–88.
Lu Y, Xue J, Deng T, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26:732–40.
Acknowledgements
Work in laboratories of GR and NT are funded by Dept. of Science and Technology (DST-FIST) and Department of Biotechnology, respectively. Part of the figures were drawn using Biorender. We thank the anonymous reviewers for their expert comments in improving the manuscript.
Author information
Authors and Affiliations
Contributions
GR wrote the manuscript. PBE and GR drew the figures. PBE, RS, and NT edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Gayatri Ramakrishna, Preedia E. Babu, Ravinder Singh and Nirupma Trehanpati declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ramakrishna, G., Babu, P.E., Singh, R. et al. Application of CRISPR-Cas9 based gene editing to study the pathogenesis of colon and liver cancer using organoids. Hepatol Int 15, 1309–1317 (2021). https://doi.org/10.1007/s12072-021-10237-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-021-10237-z