Abstract
Common bean (Phaseolus vulgaris L.) is a legume that is an important source of dietary protein in developing countries throughout the world. Utilizing the G19833 BAC library for P. vulgaris from Clemson University, 89,017 BAC-end sequences were generated giving 62,588,675 base pairs of genomic sequence covering approximately 9.54% of the genome. Analysis of these sequences in combination with 1,404 shotgun sequences from the cultivar Bat7 revealed that approximately 49.2% of the genome contains repetitive sequence and 29.3% is genic. Compared to other legume BAC-end sequencing projects, it appears that P. vulgaris has higher predicted levels of repetitive sequence, but this may be due to a more intense identification strategy combining both similarity-based matches as well as de novo identification of repeats. In addition, fingerprints for 41,717 BACs were obtained and assembled into a draft physical map consisting of 1,183 clone contigs and 6,385 singletons with ~9x coverage of the genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common bean (Phaseolus vulgaris L.) is a major legume crop with over 23 million metric tones produced per year [9] and is a major source of calories and dietary proteins in many developing countries throughout the world. In addition, common beans are high in fiber, a trait that is associated with hypocholesterolemic properties [13, 23]. The Phaseoleae, of which P. vulgaris is a member, contain 75% of the consumed legumes [9]. Although much of the breeding in common bean has focused on stable yields, many research programs are now working to increase yield as well as creating stress-tolerant, disease-resistant and high-quality beans.
DNA sequences are available for many crops, including full genome sequences of Arabidopsis [3], rice [20, 42], and most recently, poplar [38] and grapevine [19]. However, apart from the ongoing genome projects for Medicago truncatula, Lotus japonicus and Glycine max [24, 41], there is comparatively little sequence data from other legumes, including common bean. Recently, over 2,000 tentative consensus (TC)-contig sequences and more than 5,000 singleton sequences were described from ESTs generated for common bean by Ramirez et al. [35].
P. vulgaris is a member of the Phaseoloid/Millettioid clade of legumes. This clade is well adapted to tropical climates [14] and contains soybean, cowpea and pigeon pea. Among these, only soybean is being sequenced [24]. In contrast, among the Hologalegina clade or cool-season legume, M. truncatula and L. japonicus are currently being sequenced. Between the cool-season and warm-season legumes limited synteny has been observed [10]. However, the extent of synteny among the tropical legumes, P. vulgaris, Vigna radiata and soybean is quite extensive [8, 26, 32]. EST-based analyses and sequencing of homeologous regions in soybean have shown extensive retention of duplication within the soybean genome [36]. Most genetic markers in P. vulgaris are single-copy whereas, in contrast, most markers in soybean exist in 2–3 copies [21, 31, 39]. An examination of phylogenetic relationships between soybean and M. truncatula showed that all legumes likely shared an ancient duplication approximately 50 million years ago [14, 34], prior to the divergence of soybean and P. vulgaris approximately 19 million years [26]. The close relationship between soybean and common bean and the small size and relative simplicity of the P. vulgaris genome indicate that this species could be used as a reference model to study the dynamics of genome evolution and as an additional resource for gene discovery in soybean.
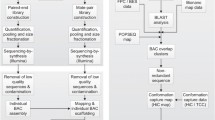
We report here the construction of a genome-wide, BAC-based physical map of the Phaseolus vulgaris genome. This map was developed using the HICF BAC-fingerprinting method [28] with BAC clones from G19833, a Peruvian landrace with low soil phosphate tolerance and high nutritional quality and has been used for genetic map development [7]. In addition, 89,017 BAC-end sequences were generated from the G19833 library and characterized for their repetitive DNA and gene content. These results were combined and compared with 1,404 shotgun sequences from BAT7, a cultivar from the Mesoamerican gene pool that was domesticated separately from the G19833 gene pool to give a first glimpse into the organization of the P. vulgaris genome.
Materials and Methods
BAC Fingerprinting
A 55,296 clone BAC library constructed from HindIII digested and size selected Phaseolus vulgaris cultivar G19833 genomic DNA was obtained from the Clemson University Genomics Institute. The BAC clones were fingerprinted using the high information content fingerprinting (HICF) method and SNaPshot reagents as described by Luo et al. [28] and modified as described by Kim et al. [25]. Briefly, DNA from the first 125, 384-well BAC clone plates from the Pv_GBa P. vulgaris BAC library (48,000 clones) was extracted by a modified alkaline lysis protocol adapted for high throughput genome analysis [25]. BAC DNA was digested with BamHI, EcoRI, XbaI, XhoI and HaeIII. The four first enzymes produce cohesive termini that were labeled with fluorochrome tagged ddGTP, ddATP, ddCTP and ddTTP respectively; while the last enzyme produces blunt ends with a four-base cutter. Chromatograms were processed with GeneMapper v4.0 (Applied Biosystems, CA) to generate trace fingerprint size files. These files were submitted into an “in house” quality control pipeline that filtered and removed fingerprints based upon cross contamination (by close position in 96 and 384 well formats), empty clones, or clones with less than 25 or more than 179 bands.
BAC-end Sequencing
BAC-end sequences (BES) were generated using the identical BAC DNA preparations used for fingerprinting with the universal primers T7 (5’ TAA TAC GAC TCA CTA TAG GG 3’) and M13 reverse (5’ CAG GAA ACA GCT ATG ACC 3’) following the methods of Ammiraju et al. [2] and Kim et al. [25] at the Arizona Genomics Institute. Sequences were processed with Phred [16, 17] and LUCY software [11] and deposited in Genbank under the accession numbers EI415689–EI504705.
Shotgun Sequencing
Approximately five grams of frozen leaf was ground with liquid nitrogen, then extracted with CTAB at 65°C for 1h and further purified by chloroform:isoamylalcohol (24:1) extraction. Genomic DNA was precipitated with isopropylalcohol, looped-out with a glass hook and washed with 70% ethanol. Genomic DNA was then air dryed and dissolved in 500 µl TE with 1 µl of RNaseA (10 mg/ml). The sample was incubated at 37°C for 30 min and 65°C for 30 min. DNA was again extracted with phenol followed by phenol:chloroform. DNA was precipitated with one-tenth a volume of 3 M acetic acid and two volumes of 100% ethanol. The DNA was hooked-out and washed with 70% ethanol as above and redissolved in a suitable volume of TE buffer.
Genomic DNA was sheared (Genemachines Hydroshear) and treated with Mungbean nuclease (USB) at 30°C for 30 min. The sample was purified by QIAquick PCR purification Kit and the eluted DNA was treated with Shrimp Alkaline Phosphatase (USB) at 37°C for 1h and then 65°C for 10 min. DNA was blunted with dATP and GoTaq DNA polymerase (Promega) at 72°C for 30 min. DNA fragments (3–5 kb) were then size selected on an agarose gel and eluted using a QIAGEN Gel Extraction Kit. The DNA fragments were ligated into the pCR4-TOPO vector (Invitrogen) and transformed into TOP10 competent cell by electroporation. Sanger sequencing and sequence processing was performed using standard protocols at the Purdue Genomics Facility, Purdue University, West Lafayette, IN. All shotgun sequences were deposited in Genbank under accession numbers DX572374–DX573827.
FPC Assembly of HICF Fingerprints
Size files containing the fingerprint bands of each clone were assembled with Fingerprint Contig (FPC) software version 8.5.2 ([37], http://www.agcol.arizona.edu/). Initial assembly was performed using a tolerance value 4 and cutoff of 1e −50 and gel length of 17,000. Questionable clones (Q clones) were removed with the DQer function in three steps increasing the cutoff, allowing a maximum of 15% of Q clones per contig. The resulting contigs were auto-merged based on the comparison of all termini clones at a lower cutoff (1e −21) and 61 bands from the ends. The same parameters were used for the evaluation of clones (singletons) that did not overlap with any contigs with the initial settings. This same assembly pipeline was successfully used for the HICF physical map of maize [33, 40] and the wild rice species Oryza punctata [25].
Chloroplast contamination was tracked by BLAST of the P. vulgaris BES to the P. vulgaris organelle genome sequence (GenBank NC_009259) and using the FPC BSS function [15]. The result of these analyses is considered as a Phase I physical map of the Phaseolus vulgaris genome.
Analysis of BAC-end and Shotgun Sequences
Analysis of repetitive sequences in the BESs was done using a repeat database constructed from the P. vulgaris shotgun sequences. This database was constructed using a de novo approach with RECON [5] and annotated using the All Plant repeat database at TIGR (http://www.tigr.org/tdb/e2k1/plant.repeats/). Two rounds of annotation were done with BLASTN [1]; the first round at e = 10−4 and the second at e = 10−2. A vast majority of the repeats remained unannotated using these criteria. As such, they are labeled as ‘Phaseolus novel repeats. A custom legume specific repeat library for Repeat Masker (http://www.repeatmasker.org/) was generated using the annotated P. vulgaris repeat database described above a database of Medicago truncatula full-length LTR retrotransposons, constructed using LTR STRUCT [30], Glycine max (http://www.tigr.org/tigr-scripts/e2k1/rpStat.cgi?DB=Glycine), Medicago truncatula (http://www.tigr.org/tigr-scripts/e2k1/rpStat.cgi?DB=Medicago) and Lotus japonicus (http://www.tigr.org/tigr-scripts/e2k1/rpStat.cgi?DB=Lotus) repeat databases at TIGR. This library was then used to mask repetitive sequences found in the BAC-end sequences using RepeatMasker version open-3.1.5, default mode, with BLASTP version 2.0MP-WashU as the search engine. The masked sequences were then categorized into different classes of repetitive elements. All of this data is available at http://phaseolus.genomics.purdue.edu/data/index.shtml.
Annotation of potential genic sequences from the BAC-end sequences was accomplished by using the RepeatMasker masked BAC-end sequences as a query to the NCBI non-redundant database with BLASTX [1]. An e-value cutoff of 1e−6 was initially used. The BLAST output was parsed in an XML format and loaded into Blast2Go for mapping each blast-based high-identity match to an associated GO annotation term [12]. Pie-charts were generated based upon the biological process class of an associate GO term with an alpha score of at least 0.6 and an ontology depth level of 3.
Results and Discussion
Analysis of BAC-end and Shotgun Sequences
The Phaseolus vulgaris cv. G19833 BAC library PV_GBa was obtained from CUGI (http://www.genome.clemson.edu) and contains 55,296 clones with an average insert size of 145 kb. Taking into consideration the estimated genome size of P. vulgaris is approximately 637 to 675 Mb [4, 6] for an average of 656 Mb, the library has an estimated coverage of 12.2 haploid genome equivalents, thereby providing an excellent substrate for the generation of a whole genome physical map.
A total of 96,000 BAC-end sequence (BES) reads were attempted and resulted in a total of 89,071 (96%) successful sequences with an average high-quality read length of 619 bp. This figure represents almost 62.6 Mb, approximately ~9.54% of the estimated size of the P. vulgaris genome. Distribution of forward and reverse BES displayed a 1:1 ratio (44,236 F: 44,781 R). From those clones, 42,553 have both forward and reverse reads available (89% of the sequenced clones). 40,527 BAC-end sequenced clones have fingerprints assembled in the physical map.
Analysis of the BAC-end sequences using a custom Repeat Masker library found that ~30.8 Mb or 49.3% of sequences contained repetitive elements (Table 1). The P. vulgaris novel class is the most abundant type of element that masked 36.65% of the total BESs. LTR retrotransposons were the second most abundant class of repetitive elements masking 9.58% of BESs; Within this class, Ty-3 gypsy were the most frequent (2.72%), followed by Ty-1 copia, calypso and other unidentified LTRs (Table 2). Of the total repetitive fraction, 74.03% were considered novel P. vulgaris repeats which could not be annotated using our criteria (see methods and materials). When the repeat masker library was comprised of only the G. max, M. truncatula and L. japonicus repeats and excluded the novel P. vulgaris repeats, only 15.31% of the bases in the BAC-end sequences were masked (as compared to 47.05%). Similarly, when the G. max, M. truncatula and L. japonicus repeats were used to mask the P. vulgaris Bat7 shot-gun sequences, only 16.63% were masked. Thus, a high proportion of repetitive sequences in P. vulgaris have little or no similarity to other legume sequences in the Fabaceae family, suggesting the presence of a large number of P. vulgaris-specific repetitive sequences.
Repeat-masked BAC-end sequences and masked shotgun sequences were putatively annotated with BLASTX searches against the NCBI non-redundant database and functionally classified using Blast2Go. Of the 89,071 total masked BAC-end sequences analyzed, 21,302 had blast hits and 10,321 were assigned to GO categories. Similarly, of the 1,404 total masked shotgun sequences, 324 had blast hits and 233 were assigned to GO categories. All of the classifications are shown in Fig. 1. The largest categories in both the BAC-end sequence and shotgun sequence analysis were cellular process, metabolic process, response to stimulus and localization. This is not surprising since these classes comprise both housekeeping genes as well as gene families generally involved in transcription and cellular communication, two of the largest functional gene categories found in Arabidopsis [3].
GO classification of shotgun (a) and BAC-end Phaseolus (b) sequences. GO classifications were based upon the biological process class of an associate GO term with an alpha score of at least 0.6 and an ontology level of 3 from Blast2Go [12]
When compared to the overall sequence composition of other legume BAC-end sequencing analyses, P. vulgaris appears to have a much higher percentage of repetitive sequence (49.3%) than the 8.5% identified for Trifolium repens L., white clover [18], or the estimated 33.5% for soybean [29]. The striking disparity between P. vulgaris and white clover is almost certainly because the white clover BES analysis used only sequence-similarity based methods instead of a combination of de novo repetitive sequence prediction and sequence-similarity based identification.
BAC Fingerprinting and Assembly of the Physical Map
We obtained a total of 41,717 SNaPshot fingerprints [25, 28] out of 48,000 attempted reactions from the G19883 P. vulgaris BAC library for a success rate of 86%. This figure satisfies the quality standard for the technique [25]. Fingerprinting failures were primarily due to either empty clones (8%) or cross-contaminations (4%). The remaining failures were mainly due to the low quality of the labeling reaction. On average, the P. vulgaris BAC clones displayed 101 bands, and clones at the extremes of the distribution were eliminated from analyses. Clones with more than 180 bands were eliminated because of the possibility of chimeras, and clones with fewer than 25 bands were eliminated because they had very small inserts.
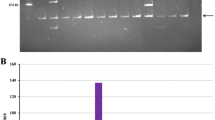
Assembly of the Phase I P. vulgaris physical map was done using FPC version 8.5.2 [37]. This initial assembly contained 41,717 clones where 34,264 (83% of the total) assembled into 1,183 contigs with the remaining 6,385 clones existing as singletons. Figure 2 depicts the frequency of the distribution of the number clones per contig. These results show a highly skewed distribution in favor of less populated contigs. In fact, 90% of the contigs contained less than 80 clones, with ~44% of the contigs comprised of fewer than 10 clones. This distribution is consistent with a stringent build that provides fewer clone misassembles and clone conflicts that require further manual editing. FPC clone contig 730 (Fig. 3) contains 14 BACs and is shown as a representative image of the fingerprints in an average BAC contig.
The average genome size of P.vulgaris is ~656 Mb and with an insert size of ~145 kb and 40,749 good clones were included in the assembly, then the haploid genome equivalent is estimated to be ~9X. The average number of consensus band units (CB units) per clone was 101, where one consensus band is estimated to be approximately 1200 bp. Given that there are 374,410 bands total for all contigs, then the estimated length of the map is 449,292 kb, quite smaller than the estimated genome size. This suggests that the CB unit estimate is underestimated given that it has not been experimentally validated. Traditionally, CB units are determined by dividing the average size of sequenced BACs by the average number of fingerprinted bands (for those BACs). However, since no BACs from P. vulgaris G19833 have been sequenced, the CB unit is an estimate and may be skewed.
Contig 1 in the current FPC build (Fig. 4a) contains the largest number of clones, 968, and is highly stacked with 555 buried clones indicating a high level of overlapping among the clones. This pattern is indicative of clone grouping as the result of BACs containing the chloroplast genome. BAC-end sequences from these clones show high BLAST-based sequence similarity to the organelle genome sequence (DQ886273; [22]), an observation that is in agreement with the alignment of the consensus Cap3 sequence contigs to the P. vulgaris chloroplast genome (Fig. 4b). This FPC contig covers 285Cb units (an estimated 342,000 bp) and was marked as a “dead contig” in FPC to avoid possible confusion in future editions of the physical map.
A WebFPC version of this initial assembly is available at http://phaseolus.genomics.purdue.edu/WebAGCoL/Phaseolus/WebFPC/. In addition, through the Phaseolus vulgaris genomics website (http://phaseolus.genomics.purdue.edu), users may submit sequences for overgo design and hybridization to the PV_GBa BAC library filters. All of these markers will be added to the physical map as new builds are assembled. Collaborative efforts are currently underway to map EST-based genetic markers to the physical map as an integrated genetic and physical map. The construction of a physical map in P. vulgaris presents an excellent genomic tool for positional gene cloning. In addition, the coverage presented by this initial build provides the P. vulgaris research community with an excellent resource for eventual sequencing of the genome, either by a clone-by-clone approach or whole genome shotgun.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ammiraju JSS, Luo M, Goicoechea JL, Wang W, Kudrna D, Mueller C, Talag J, Kim HR, Sisneros NB, Blackmon B, Fang E, Tomkins JB, Brar D, MacKill D, McCouch S, Kurata N, Lambert G, Galbraith D, Arumuganathan K, Rao K, Walling JG, Gill N, Yu Y, SanMiguel P, Soderlund C, Jackson S, Wing RA (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res 16:140–147
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Arumuganathan K, Earl ED (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol 9:229–233
Bao Z, Eddy SR (2002) Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 12:1269–1276
Bennett MD, Leitch IJ (1995) Nuclear DNA amount in angiosperms. Ann Bot 76:113–176
Blair MW, Pedraza F, Buendia HF, Gaitán-Solís E, Beebe SE, Gepts P, Tohme J (2003) Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor Appl Genet 107:1362–1374
Boutin SR, Young ND, Olson TC, Yu Z-H, Shoemaker RC, Vallejos CE (1995) Genome conservation among three legume genera detected with DNA markers. Genome 38:928–937
Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.)—model food legumes. Dev Plant Soil Sci 99:55–128
Choi HK, Mun JH, Kin DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, Young ND, Cook DR (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101:15289–15294
Chou HH, Holmes MH (2001) DNA sequence quality trimming and vector removal. Bioinformatics 17:1093–1094
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Costa NM, Low AG, Walker AF, Owen RW, Englyst HN (1994) Effect of baked beans (Phaseolus vulgaris) on steroid metabolism and non-starch polysaccharide output of hypercoholesterolaemic pigs with or without an ileo-rectal anastomosis. Br J Nutr 71:871–886
Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Phys 131:900–910
Engler FW, Hatfield J, Nelson W, Soderlund CA (2003) Locating sequence on FPC maps and selecting a minimum tiling path. Genome Res 13:2152–2163
Ewing B, Hillier M, Wendle MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Febrer M, Cheung F, Town CD, Cannon SB, Young ND, Abberton MT, Jenkins G, Milbourne D (2007) Construction, characterization, and preliminary BAC-end sequencing analysis of a bacterial artificial chromosome library of white clover (Trifolium repens L.). Genome 50:412–421
The French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature DOI 10.1038/nature06148
Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun W-L, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. spp. Japonica). Science 296:92–100
Freyre R, Skroch PW, Geffroy V, Adam-Blondon AF, Shirmohamadali A, Johnson WC, Llaca V, Nodari RO, Pereira PA, Tsai SM, Thome J, Dron M, Nienhuis J, Vallejos CE, Gepts P (1998) Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theor Appl Genet 97:847–856
Guo X, Castill-Ramirez S, Gonzalez V, Bustos P, Fernandez-Vazgueq JL, Santamaria RI, Arellano J, Cevallos MA, Davila G (2007) Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics 8:228
Han K-H, Sekikawa M, Shimada K-I, Sasaki K, Ohba K, Fukushima M (2004) Resistant starch fraction prepared from kintoki bean affects gene expression of genes associated with cholesterol metabolism in rats. Exp Biol and Med 229:787–792
Jackson SA, Rokshar D, Stacey G, Shoemaker RC, Schmutz J, Grimwood J (2006) Toward a reference sequence of the soybean genome: a multiagency effort. Crop Sci 46:S-55–S-61
Kim HR, San Miguel P, Nelson W, Collura K, Wissotski M, Walling JG, Kim JP, Jackson SA, Soderlund C, Wing RA (2007) Comparative physical mapping between Oryza sativa (AA genome type) and O. punctata (BB genome type). Genetics 176:379–390
Lavin M, Herendenn PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. System Biol 54:530–549
Lee JM, Grant D, Vallejos CE, Shoemaker RC (2001) Genome organization in dicots. II. Arabidopsis as a ‘bridging species’ to resolve genome evolution events among legumes. Theor Appl Genet 103:765–773
Luo MC, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OE, Dvorak J (2003) High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing restriction fragments by capillary electrophoresis. Genomics 82:378–389
Marek LF, Mudge J, Darnielle L, Grant D, Hamson N, Paz M, Huihuang Y, Denny R, Larson K, Foster-Hartnett D, Cooper A, Danesh D, Larsen D, Schmidt T, Staggs R, Crow JA, Retzel E, Young ND, Shoemaker RC (2001) Soybean genomic survey: BAC-end sequences near RFLP and SSR markers. Genome 44:572–581
McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19:362–367
McClean PE, Lee RK, Otto C, Gepts P, Bassett MJ (2002) Molecular and phenotypic mapping of genes controlling seed coat pattern and color in common bean (Phaseolus vulgaris L.). J Heredity 93:148–152
Menancio-Hautea D, Fatokun CA, Kumar L, Danesh D, Young ND (1993) Comparative genome analysis of mungbean (Vigna radiata L. Wilczek) and cowpea (V. unguiculata L. Walpers) using RFLP mapping data. Theor. Appl. Genet. 86:797–810
Nelson WM, Bharti AK, Butler E, Wei F, Fuks H, Kim HR, Wing RA, Messing J, Soderlund C (2005) Whole-genome validation of high-information-content fingerprinting. Plant Physiol 139:27–38
Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54:441–454
Ramirez M, Graham MA, Blanco-Lopez L, Silvente S, Medrano-Soto A, Blair MW, Hernandez G, Vance CP, Lara M (2005) Sequencing and analysis of common bean ESTs. Building a foundation for functional genomics. Plant Phys 137:1211–1227
Schlueter JA, Lin J-Y, Schlueter SD, Vasylendo-Sanders IF, Deshpande S, Yi J, O’Bleness M, Roe BA, Nelson RT, Scheffler BE, Jackson SA, Shoemaker RC (2007) Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8:330
Soderlund C, Humphray S, Dunham A, French L (2000) Contigs built with fingerprints, markers, and FPC v4.7. Genome Res 10:1772–1787
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen G-L, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, de Pamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehiting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Peple J-C, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson V, Pereda G, Peter R, Philippe G, Pilate G, Poliakov P, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai C-J, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van de Peer Y, Rokhsar D (2006) The genome of black cottonwood, Populus trichocarpa (Torr & Gray). Science 313:1596–1604
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Wei F, Coe E, Nelson W, Bharti A, Engler F, Butler E, Kim H, Goicoechea J, Chen M, Lee S, Fuks G, Sanchez-Villeda H, Schroeder S, Fang, Z, McMullen M, Davis G, Bowers J, Paterson A, Schaeffer M, Gardiner J, Cone K, Messing J, Soderlund C, Wing R (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genetics 3:1254–1263
Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the gene spaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137:1174–1181
Yu J, Hu S, Wang J, Wong GKS, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H (2002) A draft sequence of the rice genome (Oryza sativa L. spp. indica). Science 296:79–92
Acknowledgments
The authors would like to thank all of the collaborators in this project for their work and additions to this manuscript. We would also like to acknowledge the technical staff at the AGI BAC/EST Resource, Sequencing and Bioinformatics Centers for their contributions to this project. The project was supported by the USDA Cooperative State Research, Education and Extension Service—National Research Initiative—Plant Genome Program Grant number 06-35300-17266.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jessica A. Schlueter, Jose Luis Goicoechea, and Kristi Collura, are the Co-first authors.
Rights and permissions
About this article
Cite this article
Schlueter, J.A., Goicoechea, J.L., Collura, K. et al. BAC-end Sequence Analysis and a Draft Physical Map of the Common Bean (Phaseolus vulgaris L.) Genome. Tropical Plant Biol. 1, 40–48 (2008). https://doi.org/10.1007/s12042-007-9003-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-007-9003-9