Abstract
Oil palm (Elaeis guineensis Jacq.) is a perennial vegetable and a high oil-yielding crop (4–6 t/ha). There is a large scope for increasing the oil yield by selecting elite planting material for breeding programme in germplasm evaluation, characterization and utilization. In the present study, a diverse range of 150 oil palm genotypes were characterized using 12 quantitative variables with 54 genomic microsatellite markers. A wide variation was observed in the morphological traits among indigenous populations. Highly significant and positive correlations were observed between vegetative dry matter (VDM) and total dry matter (TDM) (0.862), and height and height increment (0.838). The first two principal component analyses explained 67.7% of total variation among morphological traits. The genotypes IC0610001-59 (Pune-2) and IC0610001-60 (Pune-2) were found highly promising based on less height increment, more TDM with high yield. For the mapping study, general linear model (GLM) approach, quantitative-trait loci (QTL) for annual height increment, number of bunches, bunch yield and bunch index were linked to simple-sequence repeat (SSR) loci mEgCIR3649 with phenotypic variance of 15.08, 10.43, 11.74, 15.39. TDM and VDM were linked to mEgCIR0192 (27.34 and 24.19%), mEgCIR3684 (16.84 and 18.30%), SPSC00163 (18.8 and 15.39%) and mEgCIR0555 (16.47 and 18.81%), with at a significant threshold (P) level of ≤0.001 and by mixed linear model (MLM) approach. TDM was linked to mEgCIR0555 with phenotypic variance of 20.72%, bunch yield and bunch index were linked to mEgCIR2813 at phenotypic variance of 17.11% and 12.88%, respectively, at a significant threshold (P) level of ≤0.01.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oil palm (Elaeis guineensis Jacq.) is one of the most important oil-bearing crops in the world. It is the highest edible oil-yielding crop among the vegetable oils up to 4.0–5.0 t/ha/year under good agricultural management practices. Portuguese introduced the oil palm to Brazil and other tropical countries in the 15th century, while Dutch imported oil palm seeds from Africa and planted four seedlings at Buitenzorg (Bogor) Botanical Gardens in Java and Indonesia during 1848. Commercial planting of oil palm started in Malaysia during 1917 onwards (Corley and Tinker 2003). It was first introduced in India during the year 1886 at National Royal Botanical Gardens, Kolkata. The Maharashtra Association for Cultivation of Sciences (MACS), Pune, later introduced African dura palms along canal bunds, home gardens and, to some extent, in forest lands near Pune during 1947 to 1959. The large-scale planting of oil palm was launched in Kerala from 1971 to 1984 by Plantation Corporation of Kerala (subsequently taken over by Oil Palm India) and in Little Andaman Islands of Andaman and Nicobar Islands during 1976 to 1985 by Andaman Forest and Plantation Development Corporation. Oil palm has been established as a successful crop in a number of states and the productivity levels of 4–6 t oil per ha could be achieved. The technology mission on oilseeds and pulses (TMOP) implemented by Government of India looks after the development of oil palm in the country through oil palm development programme (OPDP) along with other nine annual oilseed crops (Vision 2050 ICAR-IIOPR). At present, Andhra Pradesh, Karnataka, Tamil Nadu, Mizoram and Orissa are the major oil palm growing states in India which covers an area of about 331,476 lakh h with production of 1,638,759 mt fresh fruit bunches (FFBs) in 2017–2018 (Anon 2019a). Oil palm is being promoted by the government of India (Anon 2019b), realising the potential of crop for bridging the gap between demand and supply of edible oils in the country.
Oil palm, being a cross-pollinated crop has individual palm having variation in performance from each other and hence, assessment of individual palm is of utmost important. Evaluation of palm is needed for various purposes, including selection of superior palms (Mandal and Babu 2008). Accurate evaluation of oil palm is usually hindered by cumbersome data accumulated over the years (Oboh and Fakorede 1990). It is well known that some of these variables may not be adequate for germplasm evaluation, characterization and management. Both characterization and evaluation provide an effective source of information for genetic diversity and help in understanding the patterns of variation in crop species (Rao and Hodgkin 2002). There are a few reports on the phenotypic analysis (Mandal and Mathur 2015; Balakrishna et al. 2017) and association among the different phenotypic characters of oil palm (Balakrishna et al. 2017).
There are a very few reports on diversity studies of oil palm using molecular markers such as isozymes (Hayati et al. 2004), random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP) (Barcelos et al. 2000; Maizura et al. 2006). The analysis of DNA sequence variation is of major importance in genetic studies. Microsatellites are short-tandem repeat motifs and offer many advantages, such as high level of polymorphism (Smith et al. 1997), high accuracy and repeatability throughout the genome (Heckenberger et al. 2002), automated analysis (Sharon et al. 1997), rapidity, technical simplicity, low cost and requirement of only a small quantity of DNA (a few nanograms). Microsatellites or simple-sequence repeat (SSR) markers have been useful for integrating the genetic, physical and sequence-based physical maps in plant species and simultaneously have provided molecular breeders and geneticists with an efficient tool to link phenotypic and genotypic variation. There are only a few reports on oil palm using SSRs markers (Singh et al. 2008; Ting et al. 2010a, b; Claude et al. 2015), EST-SSRs (Ting et al. 2010b; Singh et al. 2008) and SNPs (Ong et al. 2015; Pootakham et al. 2015). However, very scarce information is available on genetic diversity of oil palm genotypes using molecular markers like SSRs. To bridge this gap, in the present study, the genetic diversity analysis of 150 oil palm genotypes was performed using genomic SSR markers.
The association mapping (AM) offers several advantages over linkage mapping (LM), where it involves natural population, high resolution mapping of the QTLs, more allele number, less time and broader reference population (Mott et al. 2000). Hence, the present study is aimed to use AM approaches for the identification of QTLs of important vegetative and oil yield related traits, which helps in identification of reliable, effective and major QTLs, and their further use in marker-assisted selection (MAS) of oil palm breeding in India. Hence, molecular marker technology can play an important role in identifying the markers linked to important traits used through AM. There are meagre reports available on comprehensive studies of oil palm genetic diversity and AM for important vegetative and yield related traits in oil palm germplasm. Hence, this study was taken up with the objectives of morphological and molecular characterization of oil palm.
Material and methods
The present research was carried out at ICAR-Indian Institute of Oil palm Research, Pedavegi, India. Experimental field located at (16o 81′ N latitude and 81o 13′ E longitudes) an elevation of 13.41 meter above the mean sea level. The soil is red sandy loam with good drainage and moderate water holding capacity. This experiment was conducted by using 52 accessions collected from different regions of India from more than 10-year-old plantations and these were maintained at field genebank at ICAR-IIOPR, Pedavegi with IC numbers from IC0610000 to IC0610051. These accessions were obtained from the source of Malaysia, Nigeria, Ivory coast, Papua New Guinea and Republic of Zaire and includes all dura, tenera and pisifera fruit forms. All germplasm lines (150) with source, IC number, germplasm block number, location of collection and fruit forms are provided in table 1 in electronic supplementary material at http://www.ias.ac.in/jgenet/.
Phenotypic analysis of PCA, correlation and phylogram
The observations of morphological traits were recorded as per standard protocols (Corley et al. 1971). Present study consists of 150 germplasm line collected from different parts of India, namely Karnataka, Andhra Pradesh, Maharashtra, Tamil Nadu, Andaman and Nicobar Islands which belongs to 52 accessions of different sources like Malaysia, Nigeria, Ivory coast, Papua New Guinea and Republic of Zaire. Morphological data were statistically analysed (PCA, correlation and phylogram) by using JMP software (JMP, v9.0.0. SAS, Cary, USA).
Genomic DNA extraction
For DNA isolation, young unopened spear leaves of oil palm were collected from each germplasm in aluminium foil and kept at –80°C. Total genomic DNA was isolated using a modified CTAB method (Murray and Thompson 1980), further purified by RNase treatment.
PCR analysis
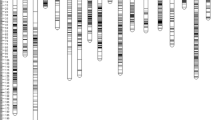
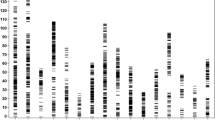
PCR was performed in 20 µL reaction volume containing 2 µL 10× buffer having 15 mM MgCl2, 0.2 µM of each forward and reverse primer, 2 µL of 2 mM dNTPs, 0.2 µL of 0.2 U of Taq DNA polymerase (Invitrogen, USA) and about 50 ng of template DNA. For genomic SSR loci, the amplifications were performed in a thermocycler (MJ Research, USA) programmed for an initial denaturation of 5 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at (different annealing temperatures for different primers), 1.0 min at 72°C, and a final extension of 10 min at 72°C and finally it will be hold at 4°C for completion of amplification. Scoring of gel picture was done by using 100 bp ladder (figure 1).
Molecular data analysis and AM
The dataset of 54 SSR loci on 150 accessions were used for statistical analysis using Power Marker v3.0 (Liu and Muse 2005). Association analysis was performed using morphological data of 150 oil palm germplasm and genotypic data of 54 SSR markers and Q matrix obtained from structure by using software TASSEL (Bradbury et al. 2007). The marker-trait association analysis was conducted using TASSEL 3.0 software along with the general linear model (GLM), mixed linear model (MLM) procedures (Pan et al. 2000). The significant threshold for the association was set at different levels P < 0.01 and P < 0.001.
Results and discussion
Phenotypic analysis
The mean and range of the 12 quantitative traits are given in table 1. The mean height of palm was 242.81 cm ranged from 95 to 645 cm. Height increment is an important dwarf related trait in oil palm. A wide variation was observed for annual height increment among the oil palm genotypes. The average annual height increment observed was 37.51 cm ranging from 19.00 to 71.20 cm. Variation in height of the palm and annual height increment was due to the genotypic character and also the age of the palm. Recently, Murugesan et al. (2015) reported the height of the palm from 300 to 1100 cm in African oil palm germplasm study in India, which comprised of 20 accessions of 50 individual palms. Here the height of the palm were more because the palm used were aged than in the present study. The results are supported by earlier studies in oil palm (Rance et al. 2001; Noh et al. 2012, 2014).
The mean girth of palm among oil palm genotype was 287.34 cm ranging from 199.50 to 377.35 cm, and the mean value of rachis length (RL) (cm) among the set of oil palm genotypes was 455.11 cm and the range obtained was 279.10 to 694 cm. The results are in congruence with Murugesan et al. (2015), who recorded 180 to 463 cm girth and RL from 139 to 692 cm in adult plantation of African germplasm at the age of 15 years. Similar findings were also supported by Noh et al. (2012) in oil palm and Noh et al. (2014) reported 4.8 m RL among different progenies. The range of leaf area (LA) varied from 2.92 to 13.82 m2 at a mean value of 6.89 cm2. These results were similar to the findings of Rance et al. (2001) and Noh et al. (2012, 2014).
The range of vegetative dry matter (VDM) varied from 60.7 to 356.44 kg at an average value of 145.79 kg/palm/year. A total of 58 genotypes had more than the mean value of VDM. The mean value of TDM was 198.22 kg at a range from lowest value of 62.05 to 457.15 kg/palm/year. In Malaysia, the average over the same age range was 20.7 (Corley and Gray 1976). In Indonesia, Gerritsma and Soebagyo (1999) recorded 22.5–23 leaves/year, and Breure (1994) found between 21 and 24.5 leaves/year in Papua New Guinea. TDM was also reported similarly by Corley and Tinker (2003) from different sources, namely Nigeria (129 kg/palm/year), Malaysia (249 kg/palm/year) and Ivory Coast (238 kg/pal/year). TDM varies between source and genotype.
The mean value of bunch number observed was 7.68 which varied from 1 to 18 among 150 genotypes. This variation in bunch number like 6.32 bunches in dura, 3.56 bunch in pisifera and 9.32 bunches in tenera was also reported in earlier studies (Mandal and Mathur 2015). Variation in bunch number was also reported in oil palm (Rance et al. 2001; Noh et al. 2012, 2014). More variation in yield may be due to the different fruit form of palms available in 150 genotypes (dura, tenera and pisifera). Usually, pisifera will bear less number of bunches, and dura and tenera will bear more fruit bunches per year. The FFB ranged from 8.25 to 274.89 kg with an average value of 100.05 kg. The average FFB ranged from 3.3 to 37.34 kg with a mean of 13.89 kg. The average bunch dry weight (BDW) was 52.78 kg ranged from lowest BDW of 4.35 kg to highest BDW of 145 kg. Mandal and Mathur (2015) reported an average FFB yield of 138.08 kg in dura, 62.56 kg in pisifera and 196.01 kg in tenera. The present results are similar to the findings of Noh et al. (2012, 2014) with respect to FFB yield BDW. The mean value of bunch index (BI) of oil palm genotype was 0.26 with a range from 0.03 to 0.52. The Tanzanian germplasm showed the highest mean BI and was significantly different than other germplasm with BI of 0.53 and 0.54 in duras and tenara, respectively (Fadila et al. 2011).
Principal component analysis (PCA), correlation and diversity analysis
Interrelationship among the morphological traits of oil palm genotypes was studied by PCA using JMP software (2009) (figure 2). The first two PCA components provided a reasonable summary of the data and explained 67.68% of the total variation and subsequent components contributed 9% or less. The first principal component (PC1) was the most important which explained 37.28% of the total variance. The traits bunch number and bunch index had largest negative loadings, which showed similar results in genetic variability study for yield, yield components and fatty acid traits in oil palm (E. guineensis Jacq.) germplasm using multivariate tools (Li-Hammeda et al. 2016). The second PC explained an additional 30.40% of total variance and was attributed to positive loadings of the traits FFB, BDW and TDM. Variables that have significant positive as well as negative impact on the PCs will contribute mostly to the genetic diversity, especially those on PC1 are likely to be the source of variation and can said to be the characters which differentiate the accessions in the oil palm germplasm (Iannucci et al. 2011; Hamza et al. 2014). Therefore, these traits can be used in the selection of diverse genotypes. The scatter plot of PCA1 and PCA2 showed overlapping of 150 genotypes and by these positive loadings, IC0610001-60, IC0610001-59, IC0610051-71, IC0610021-33 and IC0610027-206 were identified as promising genotypes among 150 genotypes. In oil palm, tenera was used only as commercial plantation, for this we need good parents like fertile dura and sterile pisifera. Except IC0610001-60, other selected palms are dura fruit form, so these can be used as mother palm in hybrid seed production programme. IC0610001-60 is tenera palm with good traits, this can be used in selfing programmes and we can get 1:2:1 (Dura:Tenera:Pisifera) palms. In this, we finally select sterile pisifera as male parent in hybrid seed production programme.
Correlation study was conducted to establish the relation between various growth parameters of oil palm germplasm (table 2). The height of plant was strongly correlated with height increment (0.838). VDM was strongly correlated with TDM (0.862). Highly significant correlation was observed between height with height increment (0.838) and VDM with TDM (0.862). The FFB was strongly correlated with BDW (1.00), whereas mesocarp to fruit ratio had significant negative correlation with shell to fruit ratio (–0.964). These results were supported by earlier reports in oil palm (Mathur and Kumar 2015). RL, TDM and VDM exhibited significant positive correlation with yield traits in oil palm (Li-Hammeda et al. 2015).
In the present study, 150 genotypes were evaluated for cluster analysis based on 12 morphological traits data, which separated genotypes as well as traits into two major groups. Group A consist of 56 genotypes and group B comprised of 94 genotypes. Group A was further divided into two subclusters, A1 and A2. The subcluster A1 consisted of all genotypes collected from Maharashtra and some genotypes from Andaman, Karnataka and Andhra Pradesh, while A2 cluster consisted of only Andaman originated genotypes. The subcluster B1 and B2 comprised of maximum of Andaman collected genotypes along with some genotypes of Karnataka, Andhra Pradesh and Tamil Nadu. The results showed that clustering pattern was not based on geographical origin. Hence, there was no association of genetic diversity with geographic distribution which was also reported by earlier workers (Kumar and Singh 2006; Khan et al. 2009).
Promising accessions with good agronomic traits were selected based on the quantitative data based on some cut-off with top five genotypes for different parameters. Annual height increment of <30 cm, girth of palm >300 cm, RL <400 cm, LA >10 m2, VDM >300 kg, TDM >350 kg, bunch number per year >15, FFB >170 kg, average bunch weight (ABW) >30 kg, BDW >100 kg, BI >0.38 have been taken into consideration for the selection of promising genotypes for particular traits based on 150 oil palm genotypes available data, with the aim of obtaining less height increment genotypes with more FFB. Based on the PCA analysis, TDM and FFB can be given more importance for selection of elite genotype, thus, IC0610001-59 (tenera), IC0610001-60 (Dura), IC0610027-206 (Dura) (figure 3), IC0610021-33 (Dura) (figure 4), IC0610051-71 (dura) were promising genotype selected for high FFB and IC0610001-59 (tenera), IC0610001-60 (Dura), IC0610005-96 (pisifera), IC0610005-97 (Dura), IC0610006-105 (tenera) were selected promising genotype for TDM. Based on all these morphological traits, genotypes IC0610001-59 (tenera) and IC0610001-60 (dura) were highly promising due to less height increment, more TDM with high FFB. The selected Tenera can be used in selfing programme to obtain a pisifera palms and further evaluation can be done in these pisifera to select female sterile pisifera as male parent in hybrid seed production and selected dura palms can be directly used as female parent in the hybrid seed production programme and these sprouts can be distributed to the farmers for commercial cultivation.
Marker-trait association by AM
Molecular analysis was performed in 150 oil palm genotypes using 54 SSR markers, the banding pattern is given in figure 1. The AM analysis was carried out by using GLM approaches of TASSEL software (Bradbury et al. 2007). The 150 oil palm genotypes mainly formed as two population structure (figure 5). In the present study, association of SSR allelic data with the morphological data revealed that 13 markers were linked to 12 traits at a significant threshold (P) level of ≤0.01 and ≤0.001 (table 3). The QTLs for height of palm was associated with locus, namely mEgCIR0192. Height of palm was strongly associated with the SSR marker mEgCIR0192. This locus is located on the 11th chromosome of oil palm. The SSR marker mEgCIR0192 linked to height of palm at a P value of 0.001 by explaining the phenotypic variance (R2) of 20.89%. This marker was found to be tightly linked, informative and significant, since it was linked at P value of 0.001 also 21% phenotypic variance. The QTL for annual height increment was linked to SSR locus mEgCIR3649, which was located on the 2nd chromosome. The SSR locus mEgCIR3649 was linked to annual height increment of palm at a P value of 0.001 by explaining the phenotypic variance (R2) of 15.08%. This marker is more informative and significant, since it was linked to P value of 0.001. The QTL for girth of palm was linked by SSR marker mEgCIR3286, which was located on 4th chromosome. In coconut, the QTLs were identified for height and girth using Ritter et al. (2000). In eucalyptus, the QTL for plant height was mapped using Byrne et al. (1997).
RL was linked to mEgCIR3672 which was located on the 12th chromosome. Rance et al. (2001) reported that RL was closely linked to marker SP1016 with phenotypic variance of 23.8% and located at 14th chromosome. In E. oleifera x E. guineensis population, RL was located in chromosome number 13 reported by Osorio-Guarín et al. (2019). LA was linked to four SSR loci: mEgCIR1773, mEgCIR0192, mEgCIR2518 and mEgCIR3653, which were located at 12, 11, 3 and 11, respectively. In eucalyptus, QTL for LA was mapped by Vaillancourt et al. (1995) and Byrne et al. (1997). The QTLs of VDM and TDM were linked to four SSR loci, namely mEgCIR0192, mEgCIR3684, SPSC00163 and mEgCIR0555. VDM and TDM were highly positively correlated in the present study and also these TDM was positively correlated with height, LA, VDM and TDM was positively correlated with average bunch weight (ABW). Due to the positive correlation, same markers were linked to TDM and VDM. Bunch number (BN) was linked to two SSR loci, mEgCIR0782 and mEgCIR3649. FFB, BI and BDW was linked with mEgCIR3649. ABW was linked with three SSR loci mEgCIR1773, mEgCIR3653 and SPSC00163. BI was positively correlated with FFB, in turn it was positively correlated with BN and BDW, all are interrelated traits, finding common marker-associated QTLs effects for these traits is not surprising. BN was positively correlated with FFB (Ooi et al. 1973; Vander-vossen 1974). Rance et al. (2001) located a putative QTL on LG13 that explained 5.5% of the variance in FFB and 19.7% for BN, while a QTL detected for ABW on LG4 explained 19.7% of the variance for that trait. Jeennor and Volkaert (2014) reported that QTLs of FFB and ABW were accounted for ~12 and 54% of the variation, respectively.
Association of SSR allelic data with the morphological data revealed that eight markers were linked to nine traits at a significant threshold (P) level of ≤0.01 (table 4). The association analysis was performed using MLM approaches of TASSEL software (Bradbury et al. 2007). The QTLs for height of palm was associated with SSR marker, namely mEgCIR3328. This locus is located on the 8th chromosome of oil palm. The SSR marker mEgCIR3328 linked to height of palm at a P value of 0.01 by explaining the phenotypic variance (R2) of 11.43%. The QTL for girth of palm was linked by SSR marker mEgCIR0243, which was located on the 10th chromosome. Another SSR locus mEgCIR0243 linked to girth of palm at a P value of 0.01 by explaining the phenotypic variance (R2) of 10.38%. In coconut, QTLs were reported for height and girth (Ritter et al. 2000). In eucalyptus, QTL for plant height was mapped by Vaillancourt et al. (1995) and Byrne et al. (1997). The QTL for RL was linked by SSR locus mEgCIR3672, which was located on the 12th chromosome at P value of 0.01 by explaining the phenotypic variance (R2) of 12.87%. Rance et al. (2001) reported that RL is closely linked to marker SP1016 with phenotypic variance of 23.8% and located at 14th chromosome.
LA was linked to two SSR loci mEgCIR3672 and mEgCIR2518 at P value 0.01, which were located on the 12th and 3rd chromosomes, respectively. The SSR locus mEgCIR0555 was linked to TDM at P value of 0.01 with phenotypic variance of 20.72% and this locus was located on chromosome 8. In oil palm, LA was linked to SP1029 marker and it was located at chromosome 5 (Rance et al. 2001). In eucalyptus, QTL for LA was mapped by Vaillancourt et al. (1995) and Byrne et al. (1997). The FFB, BDW and BI were linked with SSR marker mEgCIR2813 which were located on the 5th chromosome at P value of 0.01 by explaining the phenotypic variance (R2) of 17.11%, 7.11% and 12.88 %, respectively. BI is a product of FFB and BDW. Hence, the common markers were found to be linked to the above three traits, as they were correlated each other. The QTL for ABW (kg) was linked by SSR marker mEgCIR0246 at 0.01 P value with phenotypic variance of 15.39% which was located on chromosome 8. In coconut, yield related QTLs were mapped by Lebrun et al. (2001). Rance et al. (2001) located a putative QTL on LG13 that explains 5.5% variance in FFB, while a QTL detected for ABW on LG4 explained 19.7% of the variance for that trait. The QTLs detected by Jeennor and Volkaert (2014) accounted for ~12 and 54% of the variation for FFB and ABW, respectively.
In conclusion, the present study revealed that a wide variation was observed in height, yield and other vegetative traits among oil palm genotypes. In the mapping study based on GLM approach, 13 SSR markers were linked to 12 QTLs. The QTLs for annual height increment, number of bunches, FFB yield and BI were linked to SSR loci mEgCIR3649; TDM and VDM were linked to mEgCIR0192, mEgCIR3684, SPSC00163 and mEgCIR0555. By MLM approach, there were eight SSR markers linked to nine QTLs, TDM was linked to mEgCIR0555; FFB and BI were linked to mEgCIR2813. These QTLs can be used in oil palm breeding programme. IC0610001-59 (tenera), IC0610001-60 (dura), IC0610027-206 (dura), IC0610021-33 (dura), IC0610051-71 (dura) were promising genotype selected for high FFB. Based on all these morphological traits, genotypes IC0610001-59 (tenera) and IC0610001-60 (dura) were highly promising due to less height increment, more TDM with high FFB. Selected tenera can be used in selfing programme to obtain a pisifera palms as male parent in hybrid seed production and selected dura palms can be used as female parent in the hybrid seed production programme. Selected palms can be used for further breeding programme in seedling production and that can be evaluated for next breeding cycle. Further evaluation can be done in germplasm block for selection of high yielding mother palms (dura) and female sterile pisifera as male parent. Tenera x tenera crosses can be made for production of pisifera male parent. Dura × dura crosses can be made for production of good mother parent (high yielding and dwarf) and further evaluation can be done. Dura × pisifera crosses can be made in selected parents for commercial production of seedlings. The identified QTLs, polymorphic SSR markers and promising genotypes can be further used in the oil palm crop improvement programmes.
References
Anonymous 2019a Oil palm hybrid seed production, pp. 5. ICAR-IIOPR, Pedavegi.
Anonymous 2019b Annual report 2018-19, ICAR-Indian Institute of Oil palm Research, Pedavegi- 534 450, Andhra Pradesh, India, pp.1–3 (http://dopr.gov.in/Reports/IIOPR%20AR%202018-09%20Final_1.pdf).
Balakrishna P., Pinnamaneni R., Pavani K. V. and Mathur R. K. 2017 Genetic diversity in oil palm genotypes by multivariate analysis. Int. J. Curr. Microbiol. Appl. Sci. 6, 1180–1189.
Barcelos E., Amblard P., Berthaud J. and Seguim M. 2000 The genetic diversity of the American oil palm (Elaeis oleifera Kunth.) Corte’s revealed by nuclear RFLP markers. In International symposium on oil palm genetic resources and utilization, pp. 173–192. The Malaysian Palm oil Board, Kuala Lumpur.
Bradbury P., Zhang Z., Kroon D., Casstevens T. and Ramdoss Y. 2007 TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635.
Breure C. J. 1994 Development of leaves in oil palm (Elaeis guineensis Jacq.) and the determination of leaf opening rates. Exp. Agric. 30, 467–472.
Byrne M., Murrell J. C., Owen J. V., Kriedemann P., Williams E. R. and Moran G. F. 1997 Identification and mode of action of quantitative trait loci affecting seedling height and leaf area in Eucalyptus nitens. Appl. Genet. 94, 674–681.
Claude B., Wickneswari R., Siju S., Rajanaidu N., Kushairi A. and Billotte N. 2015 Genetic diversity of the world’s largest oil palm (Elaeis guineensis Jacq.) field genebank accessions using microsatellite markers. Genet. Resour. Crop Evol. 62, 349–360.
Corley R. H. V. and Gray B. S. 1976 Growth and morphology. In Oil palm research (ed. R. H. V. Corley, J. J. Hardon and B. J. Wood), pp. 7–21. Wiley-Blackwell, Hoboken.
Corley R. H. V. and Tinker P. B. 2003 Dry matter production (4. 1. 2). In The oil palm, pp. 91. Wiley-Blackwell, Hoboken.
Corley R. H. V., Hardon J. J. and Tan G. V. 1971 Analysis of growth of the oil palm (Elaeis guineensis Jacq.) and estimation of growth parameters and application in breeding. Euphytica 20, 307–315.
Fadila A. M., Norziha A., Mohd Din A. and Rajanaidu N. 2011 Evaluation of bunch index in the mpob germplasm collections. Paper presented at the International Seminar on Breeding for Sustainability in Oil Palm, held on 18 November 2011 in Kuala Lumpur, Malaysia, pp. 59–70. Jointly organised by the International Society for Oil Palm Breeders (ISOPB) and Malaysian Palm Oil Board (MPOB).
Gerritsma W. and Soebagyo F. X. 1999 An analysis of the growth of leaf area of oil palms in Indonesia. Exp. Agric. 35, 293–308.
Hamza A. M., Collins A., Ado S. G., Ikuenobe C. E., Ataga C. D. and Odewale J. O. 2014 Proximate compositions evaluation and variability among cultivars of date palm (Phoenix dactylifera L.) in Nigeria. Int. J. Plant Soil Sci. 3, 248–259.
Hayati A., Wickneswari R., Maizura I. and Rajanaidu N. 2004 Genetic diversity of oil palm (Elaeis guineensis Jacq.) germplasm collections from Africa: implications for improvement and conservation of genetic resources. Theor. Appl. Genet. 108, 1274–1284.
Heckenberger M., Bohn M., Ziegle J. S., Joe L. K., Hauser J. D., Hutton M. et al. 2002 Variation of DNA fingerprints among accessions within maize inbred lines and implications for identification of essentially derived varieties. Mol. Breed. 10, 181–191.
Iannucci A., Codianni P. and Cattivelli L. 2011 Evaluation of genotype diversity in oat germplasm and definition of ideotypes adapted to the mediterranean environment. Int. J. Agron. 8, 1–8.
Jeennor S. and Volkaert H. 2014 Mapping of quantitative trait loci (QTLs) for oil yield using SSRs and gene-based markers in African oil palm (Elaeis guineensis Jacq.). Tree Genet. Genom. 10, 1–14.
Khan M. A., VonWitze-Ehbrecht S., Maass S. L. and Becker H. C. 2009 Relationships among different groups, agromorphology, fatty acid compositiom and RAPD marker diversity in safflower (Carthanus tinctorius). Genet. Resour. Crop Evol. 56, 19–30.
Kumar R. R. and Singh S. P. 2006 Multivariate analysis and clustering of Cuphea procubens inbred lines. Genetika 38, 23–30.
Lebrun P., Baudouin L., Bourdeix R., Konan J. L. and Barker J. H. A. 2001 Construction of a linkage map of the Rennell Island Tall coconut type (Cocos nucifera L.) and QTL analysis for yield characters. Genome 44, 962–970.
Li-Hammeda M. A., Kushairib A., Rajanaidub N., Mohd Sukria H., Che Wan Zanariah C. W. N. and Jalania B. S. 2015 Multivariate analysis of vegetative and physiological traits in oil palm (Elaies guineensis Jacq) Germplasm. Expert Opin. Environ. Biol. 4, 219–226.
Liu K. and Muse M. 2005 Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129.
Maizura I., Rajanaidu N., Zakri A. and Cheah S. 2006 Assessment of genetic diversity in oil palm (Elaeis guineensis Jacq.) using restriction fragment length polymorphism (RFLP). Genet. Resour. Crop Evol. 53, 187–195.
Mandal G. and Mathur R. K. 2015 Performance of segregating Tenera x Tenera population in oil palm. Int. J. Bio-res. Env. Agric. Sci. 1, 108–113.
Mandal P. K. and Babu K. M. 2008 Bunch analysis of oil palm. In Technical bulletin, No. 8. (ed. M. Kochu Babu). National Research Centre on Oil Palm, Pedavegi.
Mott R., Talbot C. J., Turri M. G., Collins A. C. and Flint J. A. 2000 A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97, 12649–12654.
Murray M. G. and Thompson W. F. 1980 Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326.
Murugesan P., Mary R. K. L., Ramajayam D., Sunil K. K., Mathur R. K., Ravichandran G. et al. 2015 Genetic diversity of vegetative and bunch traits of African oil palm (Elaeis guineensis) germplasm in India. Indian J. Agric. Sci. 85, 32–35.
Noh A., Rafii M. Y., Saleh G., Kushairi A. and Latif M. A. 2012 Genetic performance and general combining ability of oil palm deli dura × AVROS pisifera tested on inland soils. Sci World J. article ID792601.
Noh A., Rafii M. Y., Mohd Din A., Kushairi A., Norziha A., Rajanaidu N. et al. 2014 Variability and performance evaluation of introgressed Nigerian dura x deli dura oil palm progenies. Genet. Mol. Res. 13, 2426–2437.
Oboh B. O. and Fakorede M. A. B. 1990 Interrrelations among vegetative, yield and bunch quality traits in short term oil palm progenies. Euphytica 46, 7–14.
Ong P. W., Maizura I., Abdullah N. A. P., Rafii M Y., Ooi L. C. L., Low E. T. L. et al. 2015 Development of SNP markers and their application for genetic diversity analysis in the oil palm (Elaeis guineensis). Genet. Mol. Res. 14, 12205–12216.
Ooi S. C., Hardon J. J. and Phang S. 1973 Variability in the Deli dura breeding population of the oil palm (Elaeis guineensis Jacq.) components of bunch yield. Malay. Agric. J. 49, 112–121.
Osorio-Guarín J. A, Garzón-Martínez G. A., Delgadillo-Duran P., Bastidas S., Moreno L. P., Enciso-Rodríguez F. E. et al. 2019 Genome-wide association study (GWAS) for morphological and yield-related traits in an oil palm hybrid (Elaeis oleifera x Elaeis guineensis) population. BMC Plant Biol. 19, 533.
Pan Q., Wendel J. and Fluhr R. 2000 Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50, 203–213.
Pootakham W., Jomchai N., Ruang-areerate P. Shearman J. R., Sonthirod C., Sangsrakru D. et al. 2015 Genome-wide SNP discovery and identification of QTL associated with agronomic traits in oil palm using genotyping-by-sequencing (GBS). Genomics 105, 288–295.
Li-Hammeda M. A., Kushairib A., Rajanaidub N., Mohd Sukria H., Che Wan Zanariah C. W. N. and Jalania B. S. 2016 Genetic variability for yield components and fatty acid traits in oil palm (Elaeis guineensis Jacq.) germplasm using multivariate tools. Int. J. Agric. For. Plantation 2, 219–226.
Rance K. A., Mayes S., Price Z., Jack P. L. and Corley R. H. V. 2001 Quantitative trait loci for yield components in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 103, 1302–1310.
Rao V. R. and Hodgkin T. 2002 Genetic diversity, conservation and utilization of plant genetic resources. Plant Cell, Tiss. Org. 68, 1–19.
Ritter E., Rodriguez M. J. B., Herran A., Estioko L., Becker D. and Rohde W. 2000 Analysis of quantitative trait loci (QTL) based on linkage maps in coconut (Cocos nucifera L.). In Plant genetic engineering towards the third millennium (ed. A. Arencibia), pp. 42–48. Elsevier, Amsterdam.
Sharon E. M., Kresovich S., Jester C. A., Hernandez C. J. and Szewc-McFadden A. K. 1997 Application of multiplex PCR and fluorescence-based, semi-automated allele sizing technology for genotyping, plant genetic resources. Crop Sci. 37, 617–624.
Singh R., Zaki N. M., Ting N. C., Rosli R., Tan S. G., Low E. T. L. et al. 2008 Exploiting an oil palm EST database for the development of gene-derived SSR markers and their expoitation for assessment of genetic diversity. Biologia 63, 227–235.
Smith J. S. C., Chin E. C. L., Shu H., Smith O. S., Well S. J., Senior M. L. et al. 1997 An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor. Appl. Genet. 95, 163–173.
Ting N. C., Noorhariza M. Z., Rozana R., Low E. T. L., Ithnin M., Cheah S. C. et al. 2010a SSR mining in oil palm EST database: application in oil palm germplasm diversity studies. J. Genet. 89, 135–145.
Ting N. C., Zaki N. M., Rozana R. and Singh R. 2010b SSR mining in oil palm EST database: Application in oil palm germplasm diversity studies. J. Genet. 89, 135–145.
Vander-Vossen H. A. M. 1974 Towards more efficient selection for oil yield in the oil palm (Elaeis guineensis Jacquin). M. Sc. Thesis, Wageningen University, Wageningen.
Acknowledgment
First author is thankful to the Director Dr R. K. Mathur and Scientists P. Murugesan, K. Sunil Kumar, K. Suresh, Goutham Mandal, A. G. K. Reddy, D. Ramajayam, G. Ravichandran, S. Rahana, B. Kalyana Babu, P. Naveen Kumar, P. Anitha and Somasundaram and other staff including RA, SRFs and technical persons etc. who worked for collection, conservation, cataloguing of oil palm germplasm project at ICAR-Indian Institute of Oil Palm Research, Pedavegi and University of Horticultural Sciences, Bagalkot, for providing the facilities to conduct my research in the Institute as a part of my Ph.D. work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Manoj Prasad
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bhagya, H.P., Kalyana Babu, B., Gangadharappa, P.M. et al. Identification of QTLs in oil palm (Elaeis guineensis Jacq.) using SSR markers through association mapping. J Genet 99, 19 (2020). https://doi.org/10.1007/s12041-020-1180-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-020-1180-4