Abstract
NAC genes, specific to plants, play important roles in plant development as well as in response to biotic and abiotic stresses. Here, a novel gene encoding a NAC domain, named as GhSNAC3, was isolated from upland cotton (Gossypium hirsutum L.). Sequence analyses showed that GhSNAC3 encodes a protein of 346 amino acids with an estimated molecular mass of 38.4 kDa and pI of 8.87. Transient localization assays in onion epidermal cells confirmed GhSNAC3 is a nuclear protein. Transactivation studies using a yeast system revealed that GhSNAC3 functions as a transcription activator. Quantitative real-time polymerase chain reaction analysis indicated that GhSNAC3 was induced by high salinity, drought and abscisic acid treatments. We overexpressed GhSNAC3 in tobacco by using Agrobacterium-mediated transformation. Transgenic lines produced longer primary roots and more fresh weight under salt and drought stresses as compared to wild-type plants. Collectively, our results indicated that overexpression of GhSNAC3 in tobacco can enhance drought and salt tolerances.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
NAC genes are plant-specific transcription factors that constitute one of the largest gene families in plants (Olsen et al. 2005). NAC proteins are characterized by a conserved NAC domain for DNA binding in their N-terminus, and a highly diverged C-terminus for regulation of transcription activity (Ooka et al. 2003). In 1996, the first NAC gene, NAM, which functions in boundary specification and shoot apical meristem formation was isolated and characterized in Petunia (Souer et al. 1996). Since then, thousands of NAC genes were identified or predicted from a number of plant species (Shen et al. 2009). For example, there are at least 117, 151, 205 and 145 NAC genes in Arabidopsis, rice and diploid cotton species, Gossypium raimondii, respectively (Mochida et al. 2010; Nuruzzaman et al. 2010; Shang et al. 2013).
NAC transcription factors function in a wide range of developmental processes in plants, such as lateral root development (He et al. 2005), leaf senescence (Takasaki et al. 2015), secondary cell formation (Nakano et al. 2015) and even plant nutrition (Uauy et al. 2006). Some NAC genes were reported as important regulators in response to biotic and abiotic stresses. In rice, OsNAC111 was induced by rice blast fungus inoculation. Overexpression of OsNAC111 in rice resulted in enhanced resistance to the rice blast fungus via regulating the expression of several defense-related genes (Yokotani et al. 2014). SNAC3 (rice ONAC003) was induced by drought, high temperature, salinity stresses and ABA treatment. Overexpression of SNAC3 in transgenic rice conferred increased tolerance to various environmental stresses (Fang et al. 2015). In Arabidopsis, the first identified NAC gene, ATAF1, was induced by drought stress and ABA treatment. Overexpression of ATAF1 in transgenic Arabidopsis plants resulted in enhanced drought tolerance and altered expression of several genes responsive to drought stress (Wu et al. 2009). Interestingly, ATAF1 was also found to be involved in biotic stress responses. Wang et al. (2009) reported that ATAF1 was downregulated after infection with fungal pathogen Botrytis cinerea, bacterial pathogen Pseudomonas syringae pv. tomato, or after treatment with several pathogen-associated hormones. Overexpression of ATAF1 in Arabidopsis resulted in increased susceptibility to P. syringae pv. tomato DC3000, B. cineea and Alternaria brassicicola, suggesting that ATAF1 acted as a negative regulator of defense response against necrotrophic fungal and bacterial pathogens (Wang et al. 2009).
Cotton is the leading source of natural fibre being used in the textile industry and is cultivated worldwide. However, cotton plants suffer from periodic drought and salt stress, which severely constrain cotton growth and yield due to semiarid to subtropical growing environments (Huang et al. 2013). In 2009, six novel NAC genes were first isolated (GhNAC1–GhNAC6) from G. hirsutum cv Jinmian 19 using data mining and reverse transcription polymerase chain reaction (RT-PCR) methods. In addition, these six cotton NAC genes were identified to be involved in abiotic stress responses (Meng et al. 2009). Subsequently, 73 NAC genes were predicted from the G. hirstutum EST database and seven NAC genes (GhNAC7–GhNAC13) were isolated using the RT-PCR method. GhNAC7–GhNAC13 were preferentially expressed in roots and responsive to abiotic stresses (Huang et al. 2013). So far, hundreds of NAC genes are isolated or predicted from the diploid cotton species G. raimondii and tetraploid cotton species G. hirsutum (Kong et al. 2013; Shang et al. 2013; Shah et al. 2013, 2014). However, most of these NAC genes are not functionally characterized to date. In the present study, we report the identification of a putative NAC family transcription factor, denoted as GhSNAC3, from upland cotton (G. hirsutum L.). We show that GhSNAC3 is significantly induced by drought, high salinity and ABA treatments. Ectopic expression of this gene in tobacco confers drought and salt tolerance.
Materials and methods
Plant materials and treatments
Cotton (G. hirsutum L.) seeds (cultivar Lumian 418) were planted in clean sand and grown in a growth chamber with a photoperiod of 16-h light at \(28{^{\circ }}\hbox {C}\) and 8 h of darkness at \(25{^{\circ }}\hbox {C}\). A week after germination, the seedlings were transferred to full-strength Hoagland’s solution for three days to adapt to hydroponic system, and then transferred to Hoagland’s solutions supplemented with 20% polyethylene glycol (PEG molecular weight 6000), 200 mM NaCl and 100 \({\mu }\hbox {M}\) ABA for 2 h, respectively. We also subjected the seedlings to cold treatment at \(4{^{\circ }}\hbox {C}\) for 2 h. The cotyledon leaves of all treatments were collected and immediately frozen in liquid nitrogen and stored at \(-80{^{\circ }}\hbox {C}\) until use. Each stress treatment was replicated in three independent experiments.
Cloning of GhSNAC3
Total RNA was extracted from leaves of seedling using Trizol (Invitrogen), treated with RNase-free DNase I (Promega). The RNA samples were quantified by a spectrophotometer (NanoDrop 2000) and qualitatively examined by agarose gel electrophoresis. cDNA synthesis was performed using \(3 \, {\mu }\hbox {g}\) of total RNA, oligo-dT (18) and M-MLV reverse transcriptase (TaKaRa, Dalian, China) according to the manufacturer’s protocol. The primers GhSNAC3-FP and GhSNAC3-RP (table 1) were designed based on the Gorai. 002G073700.1 of G. raimondii. RT-PCR was performed under the following conditions: 4 min at \(95{^{\circ }}\hbox {C}\); 30 cycles of \(94{^{\circ }}\hbox {C}\) for 30 s, \(58{^{\circ }}\hbox {C}\) for 30 s, and \(72{^{\circ }}\hbox {C}\) for 80 s; and a final extension at \(72{^{\circ }}\hbox {C}\) for 10 min. The PCR product was purified, inserted into the pMD18-T vector (TaKaRa), and then transformed into Escherichia coli \(\hbox {DH5}\upalpha \) cells. At least three clones were sequenced. Genomic DNA was extracted from young leaves of cotton cv. Lumian 418 by using a modified CTAB method (Permingeat et al. 1998).
Sequence analysis of GhSNAC3
The NAC domain within the protein sequence was annotated according to Pfam (http://pfam.xfam.org/, Finn et al. 2015). Theoretical isoelectric point (pI) and molecular weight (Mw) were predicted by the Compute pI/Mw software (http://web.expasy.org/compute_pi/, Bjellqvist et al. 1993). The gene structure of GhSNAC3 was determined by comparing the cDNA sequence to its genomic sequence and visualized by GSDS2.0 (http://gsds.cbi.pku.edu.cn/, Hu et al. 2015). Motifs were identified by MotifScan (http://myhits.isb-sib.ch/cgi-bin/motif_scan, Sigrist et al. 2013) and NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/, Blom et al. 1999). The protein sequences of GhSNAC3 and ANACs from Arabidopsis were aligned by ClustalX program (ver. 2.0) using default parameters (Larkin et al. 2007). A phylogenetic tree was constructed using MEGA ver. 7.0 software (Kumar et al. 2016) by following the maximum likelihood (ML) method tuned to the parameters: 1000 bootstraps, Poisson model, uniform rates and complete deletion. Sequence identity between GhSNAC3 and other NACs was determined by BLASTP software (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Orthologous genes of GhSNAC3 were determined by searching the genome of G. raimondii and G. hirsutum (Paterson et al. 2012; Li et al. 2015; Zhang et al. 2015) using BLASTP software.
Subcellular localization of GhSNAC3 in onion epidermal cells
Subcellular localization analysis was performed in onion epidermal cells (Wang et al. 2016). The coding sequence (CDS) of GhSNAC3 without a terminator codon was amplified by PCR using primers GhSNAC3-SF and GhSNAC3-SR (table 1). The confirmed GhSNAC3 fragment was inserted between the BamHI and HindIII sites of the pBI221-GFP (green fluorescent protein) vector to create an intact in-frame fusion between GhSNAC3 and GFP. The recombinant (pBI221–GhSNAC3–GFP) and control (pBI221–GFP) constructs were introduced into onion epidermal cells. Transformations were performed by biolistic gene delivery method using the PDS-1000/He particle bombardment system (Biorad, Hercules, USA). Transformed onion cells were incubated on Murashige and Skoog (MS) mediums in darkness at \(25{^{\circ }}\hbox {C}\) for 18 h. Confocal imaging was performed using a LSM510 confocal laser microscope (Zeiss, Germany). Fluorescence of GFP was measured under the conditions: emission at 488 nm, beam splitter at 545 nm and excitation at 505–530 nm.
Transactivation analysis of GhSNAC3 in yeast
The full-length cDNA fragment of GhSNAC3 was amplified by PCR using primers GhSNAC3-TF and GhSNAC3-TR (table 1). The resulting PCR product was inserted between the NdeI and BamHI sites of the pGBKT7 vector (which contains the GAL4 DNA binding domain, Clontech, USA) to create a pGBKT7–GhSNAC3 construct. The recombinant and negative control (pGBKT7 empty vector) constructs were introduced into the yeast strain AH109. Transformed strains were cultured on SD/–Trp or SD/–Trp—His–Ade plates and cell growth was examined. For \(\upbeta \)-gal activity, plates were incubated at \(30{^{\circ }}\hbox {C}\) for 3 days and then assayed using 5-bromo-4-chloro-3-indolyl \(\beta \)-galactopyranoside (X-GAL) as substrate.
Expression analysis of GhSNAC3
Real-time quantification PCR was used to analyse GhSNAC3 expression under various stresses. The gene-specific primers, GhSNAC3-qF and GhSNAC3-qR and the internal reference gene Ubiquitin (AY189972, Ji et al. 2003) primers Ubi-qF and Ubi-qR were used (table 1). The relative \(2^{-\Delta \Delta {\mathrm{Ct}}}\) method was used to calculate relative expression of GhSNAC3 (Livak and Schmittgen 2001). Relative expression of GhSNAC3 was calculated from RT-PCR of four biological replicates.
Schematic diagram of the structure of GhSNAC3. The schematic diagram was visualized in Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/).
Functional analysis of GhSNAC3
The full coding sequence of GhSNAC3 was amplified from total RNA of the cotton cultivar, Lumian 418, by RT-PCR and inserted into binary vector pCAMBIA3301 between the XbaI and BamHI sites, under the control of the constitutive CaMV 35S promoter. The recombination constructs were introduced into Agrobacterium tumefaciens strain LBA4404 and then transformed into tobacco plants using the Agrobacterium-mediated leaf disc infiltration method (Horsch et al. 1985). Seeds of transgenic plant were screened on MS medium containing 10 mg \(\hbox {L}^{-1}\) BASTA. Seeds from the \(\hbox {T}_{3}\) homozygous lines and wild-type tobacco were grown on 1/2 MS plates. One week later, the seedlings were transferred vertically to 1/2 MS plates supplemented with 100 mM NaCl or 200 mM mannitol. Root lengths and weights of each seedling were measured 30 days after transplantation. All experiments were repeated at least three times.
Results
Cloning of GhSNAC3 gene
RT-PCR was performed using cDNA from cotton leaves. The product was about 1000 bp in size, determined by agarose gel electrophoresis. PCR products were purified and then sequenced. The sequencing results indicated GhSNAC3 consists of 1089-bp nucleotides, including 1041 bp of an intact open reading frame (ORF), encoding for 346 amino acids (figure 1 in electronic supplementary material at http://www.ias.ac.in/jgenet/). To identify the intron–exon structure of GhSNAC3, the genomic sequence of GhSNAC3, 1273 bp, was amplified from genomic DNA using primers GhSNAC3-FP and GhSNAC-RP. A comparison between cDNA and genomic DNA sequences indicated GhSNAC3 consists of three exons and two introns (figure 1; figure 2 in electronic supplementary material). The first two exons encode the highly conserved NAC domain at the N-terminal region, whereas the third exon encodes the highly variable transcriptional domain at the C-terminus (figure 1).
Subcellular localization of the GhSNAC3 protein in onion epidermal cells. Both GhSNAC3–GFP fusion protein and control GFP protein were transiently expressed in onion epidermal cells and visualized by fluorescent microscopy (Zeiss LSM510, wavelength 488 nm). (i–iii) The control GFP protein. (iv–vi) The GhSNAC3–GFP fusion protein.
Sequence analysis of GhSNAC3 protein
GhSNAC3 was predicted to encode a putative protein of 346 amino acids with an estimated molecular mass of 38.4 kDa and pI of 8.87. The deduced GhSNAC3 protein contains a conserved NAC domain from 10 to 168 amino acid determined by using Pfam software (Finn et al. 2015). The NAC domain can be divided into five subdomains (I–V) based on Arabidopsis and rice NAC proteins (Ooka et al. 2003) (figure 3 in electronic supplementary material). Motif analysis indicated GhSNAC3 contains 28 phosphorylation sites, seven myristoylation sites, four glycosylation sites and one amidation site. The protein sequence of GhSNAC3 was used to search against the genome of G. hirsutum and 307 GhNACs were identified \((E\,\underline{\le } \underline{\mathrm{e}}^{-10})\). Sequence analysis showed GhSNAC3 is the same as Gorai.002G073700.1 and Gh_D01G0514, and has a high similarity with NAC proteins from other plant species. GhSNAC3 has the highest sequence identity to GhNAC72 (97%), TcNAC1 (87%), GaNAC72 (84%) and GrNAC72 (83%) (table 2).
Transactivation analysis of the GhSNAC3 in yeast. The pGBKT7–GhSNAC3 fusion protein and control pGBKT7 vector were expressed in yeast strain AH109. The transformed yeast cells were cultured on SD medium without tryptophan (i), tryptophan, histidine and adenine (ii) and subjected to \(\beta \)-galactosidase assay (iii). GN3, GhSNAC3; CK, pGBKT7.
Phylogenetic analysis of GhSNAC3 protein
Firstly, we constructed a phylogenetic tree from the protein sequences of GhSNAC3, 105 Arabidopsis ANACs and 307 GhNACs. Phylogenetic analysis revealed that GhSNAC3 is clustered together with ANACs belonging to the ATAF subfamily, namely ANAC002 (ATAF1), ANAC019, ANAC032, ANAC055, ANAC072, ANAC081 (ATAF2) and ANAC102. We then reconstructed a simplified phylogenetic tree from the protein sequences of GhSNAC3 and 12 known ANACs including the above seven ANACs from the ATAF subfamily (figure 2). GhSNAC3 has close relationship to ANAC019, ANC055 and ANAC072 with an overall sequence identity of 68.97, 66.44 and 60.96%, respectively. ANAC019, ANAC055 and ANAC072 were reported to be stress-responsive transcription factors (Takasaki et al. 2015) and belong to the ATAF subfamily (Ooka et al. 2003). Therefore, GhSNAC3 is a member of the ATAF subfamily of NAC proteins and may be involved in response to stresses.
GhSNAC3 is a nuclear protein
We used the Plant-mPLoc software (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/; Chou and Shen 2010) to predict the subcellular localization of GhSNAC3 and found that GhSNAC3 might be a nuclear protein. Then a GhSNAC3–GFP fusion construct was created using apBI221-GFP vector driven by a CaMV 35S promoter. Both the GhSNAC3–GFP construct and a control construct containing only GFP were transformed into onion epidermal cells by particle bombardment. Confocal imaging indicated the control protein was observed in the cytoplasm and nucleus (figure 3, i–iii), while the GhSNAC3–GFP was detected only in the nucleus (figure 3, iv–vi). These results indicated that GhSNAC3 is a nuclear protein.
GhSNAC3 has transcriptional activation function
We used a yeast system to examine the transcriptional activation of GhSNAC3. The full-length GhSNAC3 fused to the GAL4-binding domain in pGBKT7, was expressed in yeast cells and examined for their ability to activate transcription fused to the GAL4 activation domain. Cells containing either pGBKT7 or pGBKT7-GhSNAC3 could grow well on the SD medium without tryptophan (figure 4i). GhSNAC3 boosted yeast growth on the triple dropout medium (SD/–Trp–His–Ade) (figure 4ii) and showed \(\beta \)-galactosidase activity in the presence of 5-bromo-4-chloro-3-indolyl \(\beta \)-galactopyranoside (figure 4iii), while the control pGBKT7 did not (figure 4, ii&iii). These results indicated that GhSNAC3 is a transcription activator.
GhSNAC3 is induced by drought and salt stresses
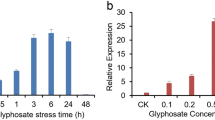
To investigate the responses of GhSNAC3 to various stresses and treatments, we isolated total RNA from leaves of the cotton cv. Lumian 418 and examined the expression patterns of GhSNAC3 under diverse stress conditions including drought, high salinity, cold (\(4{^{\circ }}\hbox {C}\)) and ABA treatment. As shown in figure 3, GhSNAC3 was significantly induced by drought (37.27-fold, \(P {<} 0.01\)), high salinity (30.91-fold, \(P {<} 0.01\)) and ABA (35.62-fold, \(P {<} 0.01\)) treatments as compared with the control. However, the expression of GhSNAC3 was not significantly affected by cold (0.95-fold, \(P {>} 0.05\)) stress (figure 5). These results indicated GhSNAC3, like other NACs from the ATAF subfamily, is a stress-responsive gene.
GhSNAC3 transgenic lines enhanced drought and salt tolerance. (i) Growth status of GhSNAC3 transgenic lines on medium with 100 mM NaCl; (ii) Growth status of GhSNAC3 transgenic lines on medium with 200 mM mannitol; (iii) Root length statistics of different transgenic; (iv) Weight per plant statistics of different transgenic lines. Values are mean and SE of three independent assays. For a given line and stress, different letters indicate significant differences between lines of tobacco; upper case for salt stress and lower case for drought stress (Tukey’s HSD, \(P< 0.01\)).
GhSNAC3 transgenic plants exhibit enhanced drought and salt tolerance
To characterize the functions of the GhSNAC3 gene under salt and drought stresses, GhSNAC3 was ectopically expressed in tobacco under the control of the CaMV 35S promoter. As a result, 11 independent transgenic lines were generated and confirmed by PCR analysis (figure 4 in electronic supplementary material). Transgenic lines G3-1, G3-4 and G3-7 exhibited higher GhSNAC3 transcript accumulation and were then selected for salt and drought assays (figure 5 in electronic supplementary material). Under normal condition, no obvious differences were observed between the WT and transgenic tobacco lines according to various phenotypic parameters (figure 6 and table 1 in electronic supplementary material). However, when one-week-old tobacco seedlings were transferred to 1/2 MS media supplemented with 100 mM NaCl and grown for 30 days, the growth of WT plants was severely affected and much slower than transgenic lines (figure 6i). Compared with WT plants, root length significantly increased by 2.82-fold (\(P<\) 0.01), 3.32-fold (\(P<\) 0.01), and 2.92-fold (\(P<\) 0.01) for transgenic lines G3-1, G3-4 and G3-7, respectively (figure 6iii). In addition, the fresh weight of transgenic lines G3-1, G3-4 and G3-7 were 2.01-fold (\(P<\) 0.01), 2.24-fold (\(P<\) 0.01) and 2.12-fold (\(P<\) 0.01), respectively, more than WT plants (figure 6iv). Under drought stress (200 mM mannitol), the WT plants showed more chlorosis and stunted phenotypes than transgenic lines (figure 6ii). Root length and fresh weight analysis indicated that significant differences were observed between WT and transgenic lines. The root length of transgenic lines G3-1, G3-4 and G3-7 were 2.16-fold (\(P<\) 0.01), 2.65-fold (\(P<\) 0.01) and 2.83-fold (\(P<\) 0.01), respectively, longer than WT plants (figure 6iii). On the basis of fresh weight, the transgenic lines G3-1, G3-4 and G3-7 were 2.32-fold (\(P<\) 0.01), 2.54-fold (\(P<\) 0.01), and 2.98-fold (\(P<\) 0.01), respectively, more than WT plants (figure 6iv). These results provided evidence that overexpression of GhSNAC3 improves salt and drought tolerances of transgenic tobacco plants.
Discussion
Environmental stresses, such as drought and high salinity, severely constrain plant growth and development. However, plants have evolutionally developed complex mechanisms, such as changes in gene expression or growth, to survive drought and salinity stresses (Nuruzzaman et al. 2013). Interestingly, some NAC genes were demonstrated to play important roles in responses to drought and salinity stresses (Tran et al. 2004; Fang et al. 2015). Rice NAC transcription factor SNAC1 was determined to be involved in drought, salt and cold tolerance. Overexpression SNAC1 significantly increased drought tolerance in transgenic rice plants and yielded 2–34% higher seed setting than the control under severe drought conditions during flowering (Hu et al. 2006). In this study, we isolated a drought-inducible and salt-inducible NAC gene, GhSNAC3, from young leaves of upland cotton and overexpressed GhSNAC3 in transgenic tobacco to further reveal its functional roles in response to drought and salt stresses. Our results show that transgenic tobacco plants have increased tolerance to drought and salt stress (figure 6).
Gene structure analysis shows GhSNAC3 contains three exons and two introns (figure 1). Meng et al. (2009) pioneered to identify six NAC genes (GhNAC1–GhNAC6) from upland cotton and found all of them shared the same gene structure, namely three exons and two introns. Shang et al. (2013) predicted 145 NAC genes from the genome of diploid cotton species G. raimondii and revealed the majority of them were composed of two introns and three exons. The same genomic organization is also shared by most NAC genes in Arabidopsis and rice (Ooka et al. 2003; Nuruzzaman et al. 2010), indicating the three-exons / two-introns structure is common to NAC genes.
Extensive efforts revealed that NAC genes from the ATAF subfamily play important roles in responses to abiotic and biotic stresses (Tran et al. 2004; Nuruzzaman et al. 2013). In Arabidopsis, the ANAC002/ATAF1, ANAC019, ANAC032, ANAC055, ANAC072/RD26, ANAC081/ATAF2 and ANAC102 genes constitute the ATAF subfamily. The ANAC019, ANAC055 and ANAC072 genes were induced by drought, high salinity and ABA. Overexpressing either ANAC019, ANAC055 or ANAC072 conferred transgenic Arabidopsis plants enhanced drought tolerance and altered the expression of several genes responsive to drought and high salinity stresses (Tran et al. 2004). ATAF1 was reported to be involved in both abiotic and biotic stress responses. Transgenic Arabidopsis plants overexpressing ATAF1 exhibited enhanced drought tolerance and attenuated resistance to the necrotrophic pathogen Botrytis cinerea and hemibiotrophic pathogen Pseudomonas syringae (Wu et al. 2009). ATAF2 was involved in wound, salicylic acid, methyl jasmonate and salinity responses. Overexpression of ATAF2 in transgenic Arabidopsis plants resulted in increased susceptibility to the fungal pathogen Fusarium oxysporum and decreased the expression of some pathogenesis-related genes (Delessert et al. 2005). ANAC102 was induced by low-oxygen stress; though overexpression of ANAC102 in transgenic Arabidopsis plants did not affect seed germination under low-oxygen conditions. However, downregulation of ANAC102 altered the expression patterns of a number of genes involved in low-oxygen response and significantly reduced seed germination under low-oxygen stress (Christianson et al. 2009). In this study, GhSNAC3 has high sequence similarity with ANAC019, ANAC055 and ANAC072 and falls into the ATAF subgroup of NAC protein family (figure 2). Expression analysis indicated that GhSNAC3, like ATAF1, ANAC019, ANAC055 and ANAC072, was also induced by drought, high salinity and ABA treatments (figure 5). Overexpression of GhSNAC3 in transgenic tobacco plants resulted in enhanced drought and salt tolerance (figure 6). These results suggest NAC genes from the ATAF subfamily share conserved roles in stress responses. Recently, the ANAC002/ATAF1, ANAC019, ANAC032, ANAC055, ANAC072/RD26, ANAC081/ATAF2 and ANAC102 genes were reported to be involved in ABA-inducible leaf senescence (Takasaki et al. 2015). In addition, ATAF1 and ATAF2 are involved in biotic stress responses. Therefore, further experiments will be required to determine the other biological functions of GhSNAC3.
GhSNAC3 is a nuclear protein and has transcriptional activation activity. However, we could not find the nuclear location signal (NLS) in the protein sequence of GhSNAC3 using Signal P 4.1 Sever (http://www.cbs.dtu.dk/services/SignalP/, Petersen et al. 2011) and Prediction Protein software (https://www.predictprotein.org/home, Yachdav et al. 2014). Recently, MfNACsa from Medicago falcata was reported to be involved in drought response. MfNACsa belongs to the ATAF subfamily and harbours the NLS, IKKALVFYAGKAPKGVKTN, in the D subdomain (Duan et al. 2017). In soybean, a stress-induced NAC transcription factor GmNAC30 also contains the same NLS in the D subdomain (Mendes et al. 2013). According to Arabidopsis and rice NAC proteins, subdomain D was tightly conserved (Ooka et al. 2003). We then made an alignment between GhSNAC3 and MfNACsa and found the NLS of GhSNAC3, IKKALVFYVGKAPKGVKTN, located in the IV subdomain (figure 3 in electronic supplementary material).
Previous studies showed that ANAC019, ANAC055 and ANAC072 could recognize conserved consensus cis-acting elements, CATGT and CACG. Overexpression of either ANAC019, ANAC055 or ANAC072 in Arabidopsis resulted in enhanced tolerance to drought stress. Various stress-inducible genes were upregulated in transgenic plants by microarray analysis. Moreover, most upregulated genes contain the CACG core motif in their promoter region (Tran et al. 2004). Therefore, to understand the mechanism via which GhSNAC3 enhances drought and salt tolerance in plants, it is necessary to identify the target genes regulated by GhSNAC3 in future studies.
References
Bjellqvist B., Hughes G. J., Pasquali C., Paquet N., Ravier F., Sanchez J. C. et al. 1993 The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031.
Blom N., Gammeltoft S. and Brunak S. 1999 Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362.
Christianson J. A., Wilson I. W., Llewellyn D. J. and Dennis E. S. 2009 Thelow-oxygen-inducible NAC domain transcription factor ANANC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149, 1724–1738.
Chou K. C. and Shen H. B. 2010 Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5, e11335.
Delessert C., Kazan K., Wilson I. W., van der Straeten D., Manners J., Dennis E. S. and Dolferus R. 2005 The transcription factor ATAF2 represses the expression of pathogenesis–related genes in Arabidopsis. Plant J. 43, 745–757.
Duan M., Zhang R., Zhu F., Zhang Z., Gou L., Wen J. et al. 2017 A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 29, 1748–1772.
Fang Y., Liao K., Du H., Xu Y., Song H., Li X. and Xiong L. 2015 A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 66, 6803–6817.
Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L. et al. 2015. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285.
He X. J., Mu R. L., Cao W. H., Zhang Z. G., Zhang J. S. and Chen S. Y. 2005 AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916.
Horsch R. B., Fry J. E., Hoffmann N. L., Eichholtz D., Rogers S. G. and Fraley R. T. 1985 A simple and general method for transferring genes into plants. Science 227, 1229–1231.
Hu B., Jin J., Guo A. Y., Zhang H., Luo J. and Gao G. 2015 GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297.
Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q. and Xiong L. 2006 Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Nat. Acad. Sci. USA 103, 12987–12992.
Huang G. Q., Li W., Zhou W., Zhang J. M., Li D. D., Gong S. Y. and Li X. B. 2013 Seven cotton genes encoding putative NAC domain proteins are preferentially expressed in roots and in responses to abiotic stress during root development. Plant Growth Regul. 71, 101–112.
Ji S. J., Lu Y. C., Feng J. X., Wei G., Li J., Shi Y. H. et al. 2003 Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31, 2534–2543.
Kong X., Luo Z., Dong H., Enji A. E., Li W. and Lu H. 2013 Gene expression profiles deciphering leaf senescence variation between early- and late-senescence cotton lines. PLoS One 8, e69847.
Kumar S., Stecher G. and Tamura K. 2016 MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H. et al. 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R. J. et al. 2015 Genome sequence of cultivated upland cotton (Gossypium hirsutum TM–1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530.
Livak K. J. and Schmittgen T. D. 2001 Analysis of relative gene expression data using real–time quantitative PCR and the 2\(^{-\Delta \Delta {{\rm Ct}}}\) method. Methods 25, 402–408.
Mendes G. C., Reis P. A. B., Calil I. P., Carvalho H. H., Aragão F. J. L. and Fontes E. P. B. 2013 GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress– and osmotic stress–induced cell death responses through a vacuolar processing enzyme. Proc. Nat. Acad. Sci. USA 110, 19627–19632.
Meng C., Cai C., Zhang T. and Guo W. 2009 Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 176, 352–359.
Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K. and Tran L. S. 2010 LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics 26, 290–291
Nakano Y., Yamaguchi M., Endo H., Rejab N. A. and Ohtani M. 2015 NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6, 288.
Nuruzzaman M., Manimekalai R., Sharoni A. M., Satoh K., Kondoh H., Ooka H. and Kikuchi S. 2010 Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44.
Nuruzzaman M., Sharoni A. M. and Kikuchi S. 2013 Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248.
Olsen A. N., Ernst H. A., Leggio L. L. and Skriver K. 2005 NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87.
Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K. et al. 2003 Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247.
Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., Jin D. et al. 2012 Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers. Nature 492, 423–427.
Petersen T. N., Brunak S., von Heijne G. and Nielsen H. 2011 SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786.
Permingeat H. R., Romagnoli M. V. and Vallejos R. H. 1998 A simple method for isolating high yield and quality DNA from cotton (Gossypium hirsutum L.) leaves. Plant Mol. Biol. Rep. 6, 1–6.
Shah S. T., Pang C., Fan S., Song M., Arain S. and Yu S. 2013 Isolation and expression profiling of GhNAC transcription factor genes in cotton (Gossypium hirsutum L.) during leaf senescence and in response to stresses. Gene 531, 220–234.
Shah S. T., Pang C., Hussain A., Fan S., Song M., Zamir R. and Yu S. 2014 Molecular cloning and functional analysis of NAC family genes associated with leaf senescence and stresses in Gossypium hirsutum L. Plant Cell Tiss. Organ. Cult. 117, 167–186.
Shang H., Li W., Zou C. and Yuan Y. 2013 Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J. Integr. Plant Boil. 55, 663–676.
Shen H., Yin Y. B., Chen F., Xu Y. and Dixon R. A. 2009 A bioinformatics analysis of NAC genes for plant cell wall development in relation to ligocellulosic bioenergy production. Bioenerg. Res. 2, 217–232.
Sigrist C. J., de Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A. et al. 2013 New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347.
Souer E., van Houwelingen A., Kloos D., Mol J. and Koes R. 1996 The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170.
Takasaki H., Maruyama K., Takahashi F., Fujita M., Yoshida T., Nakashima K. et al. 2015 SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84, 1114–1123.
Tran L. S., Nakashima K., Sakuma Y., Simpson S. D., Fujita Y., Maruyama K. et al. 2004 Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498.
Uauy C., Distelfed A., Fahima T., Blechl A. and Dubcovsky J. 2006 A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301.
Wang C. S., Jiang Q. T., Ma J., Wang X. Y., Wang J. R., Chen G. Y. et al. 2016 Characterization and expression analyses of the \(\text{ H }^{+}\)-pyrophosphatase gene in rye. J. Genet. 95, 565–572.
Wang X., Basnayake B. M., Zhang H., Li G., Li W., Virk N. et al. 2009 The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe Interact. 22, 1227–1238.
Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B. et al. 2009 Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290.
Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T. et al. 2014 PredictionProtein-an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, W337–W343.
Yokotani N., Tsuchida-Mayama T., Ichikawa H., Mitsuda N., Ohme-Takagi M., Kaku H. et al. 2014 OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense–related genes. Mol. Plant Microbe Interact. 27, 1027–1037.
Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J. et al. 2015 Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM–1) provides a resource for fiber improvement. Nat. Biotech. 33, 531–537.
Acknowledgements
We thank Mollee Dworkin from the Delaware State University for her critical editing and comments on the manuscript. The present study was supported by National Natural Science Foundation of China (no. 31571755) and Natural Science Foundation of Shandong Province (no. BS2015NY014).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Manoj Prasad
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, ZJ., Li, F., Wang, LG. et al. Molecular characterization of a stress-induced NAC gene, GhSNAC3, from Gossypium hirsutum. J Genet 97, 539–548 (2018). https://doi.org/10.1007/s12041-018-0944-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-0944-6