Abstract
Atherosclerosis (AT) is a chronic immuno-inflammatory disease characterized by inflammatory mediators and immune activation in arterial wall. Although NF-\(\upkappa \)B and micro RNAs are involved in the atherosclerotic lesions, the pathogenesis of atherosclerosis is still unknown. The aim of this study was to investigate the association of atherosclerosis with NFKB1-rs28362491, NFKBIA-rs696, pre-miRNA-146a-rs2910164 and pre-miRNA-499-rs3746444 polymorphisms as well as the analysis of their single and combined effects on its susceptibility in a Turkish population. We analysed the distribution of NFKB1-94 ins/del ATTG (rs28362491), NFKBIA (rs696), pre-miR-146a (rs2910164) and pre-miR-499 (rs3746444) genetic polymorphisms using PCR-RFLP assay in 150 atherosclerotic patients and 145 healthy controls in a Turkish population. The data revealed no significant differences in the distribution of the genotype and alleles of rs28362491 ,whereas AA genotype of rs696 lead to a higher risk for atherosclerotic patients. TT genotype and T allele of pre-miR-499 rs3746444 were found to be associated with atherosclerosis risk. In addition, significant differences were found between atherosclerotic patients and control subjects, concerning pre-miR-146a rs2910164 polymorphism. The subjects carrying the GG genotype and G allele of rs2910164 were found to have an increased risk against AT. The results of combined genotype analysis, showed no notable differences between the multiple comparisons of rs28362491–rs696 whereas rs28362491–rs2910164 ins/ins/GG is associated with increased AT risk. The combined genotypes of rs28362491/rs3746444 ins/ins/TT, revealed a significant protective effect on AT. These findings indicate that genetic polymorphisms of NFKB1A rs696, pre-miR-146a rs2910164 and pre-miR-499 rs3746444 may represent novel markers of AT susceptibility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atherosclerosis is a progressive disorder of the arterial wall and the underlying cause of probable cardiovascular diseases such as stroke and heart attack. Today, atherosclerosis is defined as a complex disease with a strong inflammatory component (Gareus et al. 2008). Epidemiological studies have suggested that the susceptibility of such diseases is mediated by environmental factors and the association of a sort of genes, predominantly NF-\(\upkappa \)B (Fontaine-Bisson et al. 2009), although the pathophysiology of atherosclerosis is not thouroughly understood.
NF-\(\upkappa \)B, one of the major transcription factors in inflammatory processes, regulates the expression of many genes involved in the initiation and progression of atherosclerotic lesions, including adhesion molecules (VCAM-1, ICAM-1, P-selectin and E-selectin), chemokines (e.g., MCP-1) and cytokines (e.g., TNF, IL-1, IL-6) (De Winther et al. 2005). According to Gareus et al. (2008), inhibition of NF-\(\upkappa \)B resulted in reduced development of atherosclerosis in vivo in the well-established \(ApoE^{-/-}\) mouse model. Therefore, NF-\(\upkappa \)B seems to play a critical role in development of atherosclerosis. One of the most famous polymorphisms identified within the promoter of NFKB1 gene is −94 ins/del ATTG rs28362491 which is able to regulate the expression and the activity of NF-\(\upkappa \)B. Therefore, rs28362491 del allele carriers may be described as insufficient antiinflammatory, resulting in susceptibility of atherosclerosis.
Further, the \(\hbox {I}\upkappa \hbox {B}\upalpha \) (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), an inhibitory version of the NF-\(\upkappa \)B protein is encoded by NFKB1A. This gene also probably plays an important role in immunoinflammatory diseases. It was determined that the alterations in the expression of \(\hbox {I}\upkappa \hbox {B}\upalpha \) protein were related to the (rs696) polymorhism in the 3\(^{\prime }\)UTR region of NFKBIA gene changes the expression of NF-\(\upkappa \)B inhibitor and led to a change in NF-\(\upkappa \)B activity (Goto et al. 2001).
It is also known that the expression of NF-\(\upkappa \)B inhibitor \(\hbox {I}\upkappa \hbox {B}\upalpha \) superrepressor (\(\hbox {DNI}\upkappa \hbox {B}\upalpha \)) blocks the development of atheromatous plaques in apolipoprotein E (ApoE)-deficient mice, a well-established mouse model of atherosclerosis (Gareus et al. 2008). Recent studies also revealed that, new markers such as microRNAs (miRNAs) play critical roles in regulation of gene expression which are important in the pathogenesis of many inflammatory diseases (Huang et al. 2011). Previous studies provide hopeful evidence in support of the role of miRNAs in cardiovascular diseases (Catalucci et al. 2009). However, the mechanism data on these small molecules in AS are still missing.
Several studies have revealed that mir-146a, an NF-\(\upkappa \)B target gene, plays an essential role by enhancing the inflammatory response in inflammatory diseases (Taganov et al. 2006). Naturally, this has influenced many scientists to look into the convergence of miRNAs and their target genes with NF-\(\upkappa \)B signalling cascade that are sensitive to inflammation process. Single-nucleotide polymorphisms (SNPs) of pre-miR-146a have functional importance and can modify the expression level of mature miRNA-146a. The SNP rs2910164 (G/C) is located in stem region and leads to a decrease in the total amount of mature miRNA, which in turn affects the transcription of target genes and the pathogenesis of inflammatory diseases such as cardiovascular diseases (Jazdzewski et al. 2008; Akkız et al. 2011; Hashemi et al. 2013). As miR-146a is a transcriptional target of NF-\(\upkappa \)B, it may serve as a feedback inhibitor of NF-\(\upkappa \)B activation.
Another important miRNA, Mir-499, is a cardiac abundant miRNA that plays a critical role in the myosin gene regulation and so is strongly associated with heart diseases. The pre-miR-499 (rs3746444) polymorphism involves an A>G nucleotide substitution which leads to a mismatch in the stem structure miR-499 precursor. These variants were reported in the pathogenesis of cardiovascular diseases (Xiong et al. 2014).
In the light of this data, we hypothesized that polymorphisms in NFKB1, NFKBIA and miRNAs genes (pre-miR-146 and pre-miR-499) would be attractive candidate factors for searching AT risk. Our aim was to evaluate whether these SNPs (NFKB1 rs28362491, NFKBIA rs696, miR-146a rs2910164, miR-499rs3746444) alone or combined are associated with AT in a Turkish population.
Materials and methods
Subjects
In this study, 150 patients (mean age: 68.01) who have atherosclerotic disease requiring surgery for coronary, carotid and peripheral arteries and 145 healthy subjects (mean age: 57.66) who do not have history of cardiovascular disease were enrolled. Patients 72% and healthy subjects 70% were male. All subjects underwent cardiovascular physical examinations. Patients scheduled for coronary bypass surgery had coronary angiographies but the other group of patients including carotid and peripheral arterial diseases had either magnetic resonance angiography, computed tomographic angiography or digital subtraction angiography or sometimes only ultrasonographic examination prior to operation depending on clinical condition of the patients or they already had these investigations done before attending our clinic. All patients were operated in Istanbul University, Cerrahpasa Medical Faculty, Department of Cardiovascular Surgery. All cases and controls were inhabitants of European region of Istanbul. All study subjects have Turkish origin and provided signed informed consent prior to the sample and data collection, and study protocol was approved by the Institutional Ethical Committee of Cerrahpasa Medical Faculty, Istanbul University. The exclusion criteria included cancer, autoimmune diseases, severe kidney / hepatic disease and pregnancy.
DNA isolation
For DNA isolation the blood was collected into EDTA-containing tubes and the DNA was extracted from peripheral blood leukocytes using a commercial kit (Roche Diagnostics GmbH, Mannheim, Germany; 11796828001) according to the manufacturer’s instructions. Isolated DNA samples were stored frozen at \(-20^{\circ }\hbox {C}\). The concentration and purity of DNA samples were established using a \(\hbox {NanoDrop}^{\mathrm{TM}}\) spectrophotometer. DNA samples with OD ratio 1.8 ± 0.2 \(\mu \hbox {g/mL}\) were included in the study.
Genotyping
NFKB1-94 ins/del ATTG, NFKBIA 3\(^\prime \)UTR A\(\rightarrow \)G, pre-miR-146a rs2910164 and pre-miR-499 rs3746444 genotypes were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. The primers were as follows, forward primer: TGG GCA CAA GTC GTT TAT GA; reverse primer: CTG GAG CCG GTA GGG AAG for NFKB1-94 ins/del ATTG polymorphism; forward primer: GGC TGA AAG AAC ATG GACTTG; reverse primer: GTA CAC CAT TTA CAG GGA GGG for NFKBIA 3\(^{\prime }\)UTR A\(\rightarrow \)G polymorphism; forward primer: TGC CTT CTG TCT CCA GTC TTC CAA; reverse primer: ATG GGT TGT GTC AGT GTC AGA GCT for rs2910164; forward primer: CAA AGT CTT CAC TTC CCT GCC A; reverse primer: GAT GTT TAA CTC CTC TCC ACG TGA TC for rs3746444 were chosen. PCR conditions were set at \(95^{\circ }\hbox {C}\) for 1 min initial denaturation, followed by 35 cycles of \(95^{\circ }\hbox {C}\) for 30 denaturation, \(60^{\circ }\hbox {C}\) (NFKB1) and 61\(^{\circ }\hbox {C}\) (NFKBIA) for 30 s annealing primers, \(72^{\circ }\hbox {C}\) for 1 min extension and finally \(72^{\circ }\hbox {C}\) for 5 min. On the other hand, PCR conditions for pre-miR-146a and pre-miR-499 were 5 min at \(94^{\circ }\hbox {C}\) followed by 35 cycles of 1 min at \(94^{\circ }\hbox {C}\), 1 min at \(58^{\circ }\hbox {C}\) (pre-miR-146a) and 1 min at \(67^{\circ }\hbox {C}\) (pre-miR-499), and 2 min at \(72^{\circ }\hbox {C}\), with a final step at \(72^{\circ }\hbox {C}\) for 20 min to allow a whole extension of all PCR fragments.
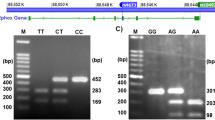
For detectig the genotypes of −94 ins/del ATTG polymorphism in NFKB1 gene, the 281 bp (deletion allele) or 285 bp (insertion allele) PCR product were digested with 3 units of restriction enzyme \(PfI\hbox {MI}\,(10 \hbox { U}/\mu \hbox {L}, \hbox { Fermentas})\). The mix was incubated for overnight at \(37^{\circ }\hbox {C}\), finally subjected to electrophoresis for 30 min at 120 V on \(3\%\) agarose gel. The wild-type (deletion) genotype did not contain PflMI (Van91I) restriction site, hence the PCR product of 281 bp remained undigested. The insertion variants were cleaved by PflMI (Van91I) restriction enzyme into two fragments of 240 bp and 45 bp. Heterozygotes showed all three bands (see figure 1). SacI (20,000 U/mL, NEB) was used for rs2910164 gene polymorphism and all samples incubated at \(37^{\circ }\hbox {C}\) for 1 h. Consequently, the genotypes were assessed as follows: a single 147 bp fragment for the mutated type homozygous alleles (GG genotype); two fragments of 122 and 25 bp for the wild type homozygous alleles (CC genotype); and three fragments of 147, 122, and 25 bp for the heterozygous alleles (GC genotype) (see figure 2).
For rs3746444 gene polymorphism, BclI (10,000 U/mL, NEB) was preferred and each of the sample incubated at \(50^{\circ }\hbox {C}\) for 1 h. The genotypes were assessed as follows: a single 146 bp fragment for the mutated type homozygous alleles (CC genotype), two fragments of 26 and 120 bp for the wild type (TT genotype); and three fragments of 26, 120 and 146 bp for the heterozygous alleles (CT genotype) (see figure 3).
The reaction products of pre-miR-146a and pre-miR-499 were run on electrophoresis for 30 min at 120 V, on \(4\%\) ethidium bromide-stained metaphor gel and directly visualized under ultraviolet illumination.
Similarly, PCR product of the NFKBIA 3\(^\prime \)UTR A\(\rightarrow \)G was digest with 3 unit of restriction enzyme \(Hae\hbox {III} (10 \hbox { U}/\mu \hbox {L}, \hbox {Sigma})\), and incubated for overnight at \(37^{\circ }\hbox {C}\), finally subjected to electrophoresis for 30 min at 120 V on \(3\%\) agarose gel. The PCR product (424 bp) was cleaved into two fragments of 108 bp and 316 bp. Homozygosity of the common allele (represented by AA genotype) revealed itself by 424 bp bands, while the homozygosity of the variant allele (represented by GG genotype) was represented by 316 bp and 108 bp bands (see figure 4).
Statistical analysis
GraphPad Prism 5 programs were used for the analyses of the patients and control values. Hardy–Weinberg equilibrium (HWE) was tested by chi-square analysis. Genotype and allele frequencies were compared between cases and controls by chi-square analysis. Odds ratio (OR) and respective 95% confidence intervals (CI) were reported to evaluate the effects of any difference between allelic and genotype distribution. Mann–Whitney U test (not normally distributed variables). Significance values are reported without Bonferroni correction. A two-sided P value \(\le \)0.05 was considered statistically significant.
Results
Distribution of polymorphisms in NFKB1 (rs28362491), NFKBIA (rs696), pre-miR-146a (rs2910164) and pre-miR-499 (rs3746444) genes in atherosclerosis
The distribution of genotype and allele frequencies of all polymorphisms studied here are shown in table 1. All allele and genotype frequencies were within the range of HWE. The rs28362491, rs696, rs2910164, rs3746444 were successfully genotyped in 150 with AT and 145 control subjects.
In the single genotyping study of NFKB1-94 ins/del ATTG polymorphism, no statistically significant differences were observed in the case of allele frequencies as well as genotype distribution of the rs28362491 (\(P>0.05\)).
The frequency of AA genotype of NFKBIA rs696 was considerably higher in atherosclerotic patients (\(P=0.024\), \(\hbox {OR}=2.082\), 95% CI: 1.093–3.963). According to our results, having the AA genotype of rs696 has a 2.082 times risk factor in patients with atherosclerosis. No statistical differences was detected by comprising the allele frequencies of NFKBIA rs696 (P > 0.05). In a single genotyping study of mir-146a rs2910164 polymorphism, the overall frequencies of GG, CG, CC allele combinations were 64, 28, \(8\%\) in patients group and 39, 55, \(6\%\) in the control group, respectively. The frequency of GG genotype of rs2910164 was significantly higher in patients (P < 0.0001, \(\hbox {OR}=3.168\), 95% CI: 1.926–5.211). When the allele frequencies were compared, it was found that the frequency of G allele was higher in the atherosclerotic group (\(P=0.0019\), OR \(=\) 1.782, 95% CI: 1.713–4.398). The above results suggest that the GG genotype and G allele of mir-146a rs2910164 have an increase risk for susceptibility to atherosclerosis. The recessive model (GG/CC + CG) (P < 0.0001, \(\hbox {OR}=2.745\), 95% CI: 1.713–4.398) is also found to be a risk factor for atherosclerotic patients with respect to the dominant model.
In the study of mir-499 rs3746444 polymorphism, the overall frequencies of TT, CT and CC allele combinations were \(16\%, 76\%, 8\%\) in patients group and \(15\%, 55\%, 30\%\) in the control group. The frequency of TT genotype was significantly higher in atherosclerotic patients (\(P=0.0025\), OR = 4.000, 95% CI: 1.690–9.468). When we compared the allele frequencies, we found that having rs3746444 T allele was identified as a risk factor for atherosclerosis by 1.594 times (\(P=0.0049\), OR \(=\) 1.594, 95% CI: 1.151–2.207). In addition, the dominant model (TT \(+\) CT vs. CC) is correlated with an increased risk compared to the recessive model (P < 0.0001, OR \(=\) 5.010, 95% CI: 2.518–9.970).
Combined genotypes of NFKB1 (rs28362491), NFKBIA (rs696), pre-miR-146a (rs2910164) and pre-miR-499 (rs3746444) genes polymorphisms in atherosclerosis
Tables 2, 3 and 4 summarizes combined genotype analysis of polymorphisms in NFKB1 rs28362491, NFKBIA rs696, and miRNAs genes pre-miR-146a rs2910164 and pre-miR-499 rs3746444 in atherosclerotic patients and the controls. Table 2 summarizes the association study among the combined genotypes of rs28362491/rs696 and the overall risk for AT. There were no significant differences between the multiple comparisons of NFKB1 rs28362491/NFKBIA rs696 in patients with atherosclerosis and the control subjects.
According to combined genotype analysis of NFKB1 rs28362491/pre-miR-146a rs2910164 (ins/ins/GG) combined genotype frequencies are to be a risk factor for AT (\(P=0.0146\), OR \(=\) 3.184, 95% CI: 1.338–7.580) (see table 3).
Table 4 represents the analysis of combined genotype analysis of NFKB1 rs28362491 and pre-miR-499 rs3746444. The combined genotype analysis of NFKB1 rs28362491/pre-miR-499 rs3746444 (ins/ins/TT) had protective effect in the case of AT (\(P=0.0005\), \(\hbox {OR}= 0.114\), 95% CI: 0.003–0.4).
Discussion
Atherosclerosis is a chronic inflammatory disease which is a result of the compound effects mediated by risk factors, chronic inflammation and genetic compounds (Prins et al. 2012). The nuclear factor kapa-light-chain enhancer of activated B cells signalling pathway plays a key role in the regulatory network of inflammation (Dabek et al. 2010). We chose to study the NFKB1 −94ins/del ATTG (rs28362491) polymorphism because of its proven functionality and known biological effects (Karban et al. 2004; Park et al. 2007). Deletion of one ATTG repeat in the promoter region of NFKB1 removes the binding of a nuclear factor as shown in colon tissue and in cell lines and leads to less p50 biosynthesis and less promoter transcriptional activity in vitro (Karban et al. 2004). This paper also seeks to address the impact of NFKB1-94 ATTG rs28362491 polymorphism on the risk of AT in a sample of Turkish population. We indicated that rs28362491 polymorphism did not correlate with AT in this population.
\(\hbox {I}\upkappa \hbox {B}\upalpha \) is the inhibitory version of NF-\(\upkappa \)B, which binds the NF-\(\upkappa \)B and influences the transcriptional activity of NF-\(\upkappa \)B in the cytoplasm (Hung et al. 2010). In the last decade, many researchers have also pointed that variations within the NFKBIA which encodes \(\hbox {I}\upkappa \hbox {B}\upalpha \) protein could potentially influence the function of NF-\(\upkappa \)B protein and in turn the process of inflammation. Some studies reported evidence for an anti-inflammatory effect of \(\hbox {I}\upkappa \hbox {B}\upalpha /\hbox {NF-}\upkappa \hbox {B}\) dependent mechanism in the alveolar epithelial cells. Özbilüm et al. (2013) show that NF-KB1A -826 C/T promoter polymorphism is a risk factor for the development of coronary artery disease (CAD) whereas no significant differences was detected in NF-KB1A-297C/T and -881A/G in allele and genotype frequencies of CAD. One of the most famous polymorphism known within the gene NFKB1A is in 3\(^\prime \)UTR region. NFKBIA 3\(^\prime \)UTR A/G(rs696) polymorphism may also have a genetic effect on the development of a variety of inflammatory diseases associated with altered immune response (Yenmis et al. 2015). According to our results, the AA genotype of rs696 in the NFKBIA gene was significantly higher in atherosclerotic patients than in controls. AA genotype had twice risk factor for the development of AT. By contrast, the combined genotypes of rs28362491 and rs696 seems to play no role together on the pathogenesis of atherosclerosis.
On the other hand, NF-\(\upkappa \)B is also regulated by miRNA146. Combined with functionality and NF-\(\kappa \)B inhibition assays, miRNA146a was the first found NF-\(\upkappa \)B regulated, proinflammatory miRNA identified in the human central nervous system (Li et al. 2011a). An increasing number of studies have revealed that rs2910164 polymorphisms in pre-miR-146a affects the pathogenesis of several human diseases, such as rheumatoid arthritis (Hashemi et al. 2013), large-artery atherosclerotic stroke (Zhu et al. 2014), CAD (Xiong et al. 2014), multiple sclerosis (Fenoglio et al. 2012), tuberculosis (Li et al. 2011b), SLE (Zhang et al. 2011) and Behcet’s disease (Yenmis et al. 2015). Currently many studies have shown that the polymorphism rs2910164 involves a G>C nucleotide substitution that leads to change from a G:U pair to C:U in the stem structure of the pre-miR-146a precursor which affects the specificity of mature mi-RNA in binding to its target genes such as NF-\(\upkappa \)B (Hashemi et al. 2013). Pre-miR-146a rs2910164 has an important role in inflammation and immune response. According to Xiong et al. (2014) increased expression of mir-146a may promote the occurance and progression of CAD and the G to C change in the precursor of mir-146a tends to cause increased expression of mature mir-146a in CAD patients. Elevetad levels of mature mir-146a expression enhances the function of TH-1 cells which is critical in the progression of the acute coronary syndrome (Guo et al. 2010). Xiong et al. (2014) revealed that C allele of the SNP rs2910164 is associated with genetic susceptibility to CAD. According to our results, carrying GG genotype and G allele of rs2910164 were found to be a risk factor against atherosclerosis and in the recessive model of pre-miR-146a rs2910164, a significant association—an increased risk for atherosclerosis—was found. When we evaluated two polymorphisms (rs28362491/rs2910164) together, combined genotype (ins/ins/GG + ins/ins/GG) seemed to be associated with the risk of AT (table 3). The range of 95% CI is so wide on table 3, as it confirms that atherosclerosis is a multifactorial disease mainly depending on the factors such as smoking, hyperlipidemia, gender, diabetes mellitus, hypertension, inflammation, obesity, hyperhomocysteinemia. These factors play role in different levels in each patients with atherosclerosis and thus are responsible for this inhomogeneity. Because of this multifactorial distribution-driven inhomogeneity, statistical corrections/modifications are not plausible to make 95% CI range better.
MiR-499 activates the production of C-reactive protein (CRP) and regulates the expression of many cytokines, including IL-23a, IL-2R, IL-6, IL-2 and IL-18R (Yang et al. 2012; Hashemi et al. 2013). Therefore it is the key factor in the development of chronic inflammation in atherosclerotic disease. Recently, an important polymorphism in the pre-miR-499 with an A to G change (rs3746444) was identified. This polymorphism was located in the stem region of the miR-499 and may damage the secondary structure stability and thus affect the miRNA maturation process and binding affinities to its target genes (Hu et al. 2011), thus has a correlation with the risk of variety of inflammatory diseases such as CAD (Zhi et al. 2012), myocardial infarction (Chen et al. 2014) and ischemic stroke (Liu et al. 2014). Our results implied that TT genotype of pre-miR-499 rs3746444 may play important roles in the development of AT. These findings are similar to Lu et al. (2014) result of overall association of autoimmune diseases with homozygote TT genotype (Lu et al. 2014). According to combined genotype analysis of rs28362491 and rs3746444 polymorphisms, the ins/ins/TT combined genotype has a protective role on the development of AT so that microRNA -499 may regulate NF-\(\upkappa \)B activity.
Taking into account these considerations, our study demonstrates that NFKBIA rs696, pre-miR-499 rs374644 and pre-miR-146a rs2910164 may affect susceptibility to atherosclerosis and increase the risk for developing the disease. Also, NFKBIA rs28362491-pre-miR-146a rs2910164 combined genotype is associated with the risk of atherosclerosis whereas NFKB1 rs28362491-pre-miR-499 rs3746444 has a protective effect on the disease according to the multiple comparisons of combined genotypes of them. NF-\(\upkappa \)B seems to modulate the immune response in atherosclerosis although how exactly pathway is abnormal in disease pathogenesis is to be researched. Moreover, different population studies are necessary to understand whether these variants play considerable roles in the pathogenesis of atherosclerosis.
References
Akkız H., Bayram S., Bekar A., Akgöllü E., Usküdar O. and Sandıkçı M. 2011 No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene 486, 104–109.
Catalucci D., Gallo P. and Condorelli G. 2009 MicroRNAs in cardiovascular biology and heart disease. Circ. Cardiovasc. Gene 2, 402–408.
Chen C., Hong H., Chen L., Shi X., Chen Y. and Weng Q. 2014 Association of microRNA polymorphisms with the risk of myocardial ınfarction in a Chinese population. Tohoku J. Exp. Med. 233, 89–94.
Dabek J., Kulach A. and Gasior Z. 2010 Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB): a new potential therapeutic target in atherosclerosis? Pharmacol. Rep. 62, 778–783.
De Winther M. P., Kanters E., Kraal G. and Hofker M. H. 2005 Nuclear factor kappaB signaling in atherogenesis. Arterioscler. Thromb. Vasc. Biol. 25, 904–914.
Fenoglio C., Ridolfi E., Galimberti D., and Scarpini E. 2012 MicroRNAs as active players in the pathogenesis of multiple sclerosis. Int. J. Mol. Sci. 13, 13227–13239.
Fontaine-Bisson B., Wolever T. M., Connelly P. W., Corey P. N., and El-Sohemy A. 2009 NF-kappaB -94Ins/Del ATTG polymorphism modifies the association between dietary polyunsaturated fatty acids and HDL-cholesterol in two distinct populations. Atherosclerosis 204, 465–470.
Gareus R., Kotsaki E., Xanthoulea S., van der Made I., Gijbels M J., Kardakaris R. et al. 2008 Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab. 8, 372–383.
Goto Y., Yue L., Yokoi A., Nishimura R., Uehara T., Koizumi S. et al. 2001 A novel singlenucleotide polymorphism in the \(3^{\prime }\)-untranslated region of the human dihydrofolate reductase gene with enhanced expression. Clin. Cancer Res. 7, 1952–1956.
Guo M., Mao X., Ji Q., Lang M., Li S., Peng Y. et al. 2010 miR-146a in PBMCs modulates Th1 function in patients with acute coronary syndrome. Immunol. Cell Biol. 88, 555–564.
Hashemi M., Eskandari-Nasab E., Zakeri Z., Atabaki M., Bahari G., Jahantigh M. et al. 2013 Association of pre-miRNA-146a rs2910164 and pre miRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol. Med. Rep. 7, 287–291.
Hu Z., Liang J., Wang Z., Tian T., Zhou X., Chen J. et al. 2011 Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 30, 79–84.
Huang Y., Shen X J., Zou Q., Wang S P., Tang S. M. and Zhang G. Z. 2011 Biological functions of microRNAs: a review. J. Physiol. Biochem. 67, 129–139.
Hung Y. H., Wu C. C., Ou T. T., Lin C. H., Li R. N., Lin Y. C. et al. 2010 IkappaBalpha promoter polymorphisms in patients with Behçet’s disease. Dis. Markers 28, 55–62.
Jazdzewski K., Murray E. L., Franssila K., Jarzab B., Schoenberg D. R. and De La Chapelle A. 2008 Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 105, 7269–7274.
Karban A. S., Okazaki T., Panhuysen C. I., Gallegos T., Potter J. J., Bailey-Wilson J. E. et al. 2004 Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum. Mol. Genet. 13, 35–45.
Li D., Wang T., Song X., Qucuo M., Yang B., Zhang J., et al. 2011a Genetic study of two single nucleotide polymorphisms within corresponding microRNAs and susceptibility to tuberculosis in a Chinese Tibetan and Han population. Hum. Immunol. 72, 598–602.
Li Y. Y., Cui J. G., Dua P., Pogue A. I., Bhattacharjee S. and Lukiw W. J. 2011b Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci. Lett. 499, 109–113.
Liu Y., Ma Y., Zhang B., Wang S. X., Wang X. M. and Yu J. M. 2014 Genetic polymorphisms in pre-microRNAs and risk of ischemic stroke in a Chinese population. J. Mol. Neurosci. 52, 473–480.
Lu L., Tu Y., Liu L., Qi J. and He L. 2014 MicroRNA-499 rs3746444 polymorphism and autoimmune diseases risk: a meta-analysis. Mol. Diagn. Ther. 18, 237–242.
Özbilüm N., Arslan S., Berkan Ö., Yanartaş M., and Aydemir E I. 2013 The role of NF-\({\rm \kappa }\)B1A promoter polymorphisms on coronary artery disease risk. Basic Clin. Pharmacol. Toxicol. 113, 187–192.
Park J. Y., Farrance I. K., Fenty N. M., Hagberg J. M., Roth S. M., Mosser D. M. et al. 2007 \(\text{ NF }{\rm \kappa }\text{ B }1\) promoter variation implicates shear-induced NOS3 gene expression and endothelial function in prehypertensives and stage I hypertensives. Am. J. Physiol. Heart Circ. Physiol. 293, H2320–H2327.
Prins B. P., Lagou V., Asselbergs F. W., Snieder H. and Fu J. 2012 Genetics of coronary artery disease: genome-wide association studies and beyond. Atherosclerosis 225, 1–10.
Taganov K. D., Boldin M. P., Chang K.-J., and Baltimore D. 2006 NF-\(\kappa \)B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 103, 12481–12486.
Xiong X. D., Cho M., Cai X. P., Cheng J., Jing X., Cen J. M. et al. 2014 A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat. Res. Fund. Mol. M. 761, 15–20.
Yang B., Chen J., Li Y., Zhang J., Li D., Huang Z. et al. 2012 Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Hum. Immunol. 73, 101–106.
Yenmis G., Oner T., Cam C., Koc A., Kucuk O. S., Yakicier M. C. et al. 2015 Association of NFKB1 and NFKBIA polymorphisms in relation to susceptibility of Behçet’s disease. Scand J. Immunol. 81, 81–86.
Zhang J., Yang B., Ying B., Li D., Shi Y., Song X., et al. 2011 Association of pre-microRNAs genetic variants with susceptibility in systemic lupus erythematosus. Mol. Biol. Rep. 38, 1463–1468.
Zhi H., Wang L., Ma G., Ye X., Yu X., Zhu Y., et al. 2012 Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clin. Res. Cardiol. 101, 289–296.
Zhu R., Liu X., He Z., and Li Q. 2014 miR-146a and miR-196a2 Polymorphisms in patients with ıschemic stroke in the northern Chinese Han population. Neurochem. Res. 39, 1709–1716.
Acknowledgements
This work was supported by the Research Fund of The University of Istanbul, project number 34521.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Oner T., Arslan C., Yenmis G., Arapi B., Tel C., Aydemir B. and Sultuybek G. K. 2017 Association of NFKB1A and microRNAs variations and the susceptibility to atherosclerosis. J. Genet. 95, XX–XX]
Corresponding editor: Kunal Ray
Rights and permissions
About this article
Cite this article
Oner, T., Arslan, C., Yenmis, G. et al. Association of NFKB1A and microRNAs variations and the susceptibility to atherosclerosis. J Genet 96, 251–259 (2017). https://doi.org/10.1007/s12041-017-0768-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-017-0768-9