Abstract
Auxin response factors (ARF) are transcription factors that regulate auxin responses in plants. Although the genomewide analysis of this family has been performed in some species, little is known regarding ARF genes in apple (Malus domestica). In this study, 31 putative apple ARF genes have been identified and located within the apple genome. The phylogenetic analysis revealed that MdARFs could be divided into three subfamilies (groups I, II and III). The predicted MdARFs were distributed across 15 of 17 chromosomes with different densities. In addition, the analysis of exon–intron junctions and of the intron phase inside the predicted coding region of each candidate gene has revealed high levels of conservation within and between phylogenetic groups. Expression profile analyses of MdARF genes were performed in different tissues (root, stem, leaf, flower and fruit), and all the selected genes were expressed in at least one of the tissues that were tested, which indicated that MdARFs are involved in various aspects of physiological and developmental processes of apple. To our knowledge, this report is the first to provide a genomewide analysis of the apple ARF gene family. This study provides valuable information for understanding the classification and putative functions of the ARF signal in apple.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural auxins are highly important growth regulator molecules that are known to play pivotal roles in various development processes, which range from root and shoot development to flower and fruit development in plants, including growth and development of the root and stem, formation and differentiation of vascular tissue, organ senescence, apical dominance, gravitropism, phototropism etc. (Guilfoyle and Hagen 2007; Kumar et al. 2011). At present, many highly active synthetic auxins have been developed in the agricultural and horticultural industries. Related research indicates that there are two types of transcription factor families that are required for controlling the expression of auxin response genes: type one is the auxin response factor (ARF) family, and the other is the Aux/IAA repressor family (Guilfoyle and Hagen 2007). ARF acts as a transcription factor to regulate the expression of auxin response genes by binding with specificity to TGTCTC auxin response elements (AuxRE) in promoters of primary or early auxin-responsive genes, such as Aux/IAA, Gretchen Hagen3 (GH3), and small auxin-up RNA (SAUR) family members (Guilfoyle et al. 1998).
A typical ARF protein in higher plants is composed of three modular and portable domains that can function independent of one another. Many of these proteins always contain a DNA-binding domain (DBD) (generally the B3-like DNA-binding domain) in the amino-terminal region, a middle region (MR), which consists of an activation domain (AD) or a repression domain (RD), and a protein–protein interaction domain in the carboxy-terminal region (conservative domains III/IV, CTD) (Ulmasov et al. 1999a; Tiwari et al. 2003). However, ARFs 3, 13 and 17 in Arabidopsis have amino-terminal DNA-binding domains but lack the carboxyl terminus. In composite AuxREs, the consensus sequence for a functional element is TGTCNC, in which the first four nucleotides, TGTC, are important for the combination of ARF, whereas the nucleotides at positions 5 and 6 could be substituted by others, particularly position 5 (Guilfoyle et al. 1998). In addition, to further investigate the structure of the ARF family members, a region that is enriched with glutamine, serine and leucine residues might be found in ADs of MR, whereas the conserved amino sequence of serine, proline, leucine and glycine residues might be found in RDs (Ulmasov et al. 1999b; Guilfoyle and Hagen 2007). Thus, the difference in this region might have a critical role in determining whether an ARF protein can act as an activator or repressor of the auxin response process. Further, the CTD domain of ARF, which contains domains III and IV, is similar to those domains that were found in the C-terminus of Aux/IAAs (Okushima et al. 2005b), which can form dimers with each other using this region. Recently, it was found that the auxin signal transduction process is related to the degradation of the Aux/IAA protein and to the activation of the ARF protein (Guilfoyle et al. 1998). Detailed information from a yeast two-hybrid system indicated that ARFs and Aux/IAA proteins can use their conserved carboxyl-terminal domains (primarily domains III and IV) to interact among themselves and among each other (Kim et al. 1997; Hardtke et al. 2004). ARF binds to the Aux/IAA protein and forms an inactivated heterodimer that represses the transcription of primary genes when there is no auxin or when the auxin level is low; this pathway results in no auxin response. However, the Aux/IAA repressor can be recognized by the SCFTI complex, and ubiquitin ligase is activated when auxin levels are elevated. Thus, ARF and Aux/IAA proteins form homodimers by the ubiquitin–proteasome pathway, and then bind with specificity to TGTCTC-containing auxin response elements (AuxREs) that are found in promoters of primary/early auxin response genes. The transcription of these genes is then initiated and can regulate the expression of downstream genes (Guilfoyle et al. 1998; Guilfoyle and Hagen 2007).
The ARF family contains numerous members, and these genes are transcribed in many tissues and organs. Bioinformatics analyses and biological experiments of common dicot and monocot plants make it possible to determine the number of ARF genes and their potential functions. In Arabidopsis, there are 22 full-length ARF genes and one partial-length gene (ARF23), which was created by a stop codon in its DBD (Remington et al. 2004; Okushima et al. 2005b). These 23 genes are distributed on all five chromosomes; however, there are 13 genes in chromosome 1 that appear to be restricted to embryogenesis/seed development (Remington et al. 2004). Using a one-hybrid screen with a core motif, the first of these transcription factors to be identified was ARF1 in Arabidopsis. ARF1 contains 665 amino acids and a sequence that stretches over ˜120 amino acids in the DBD (Ulmasov et al. 1997; Guilfoyle et al. 1998). ARF1 has been found to regulate the process of flower development, and its transcripts may decrease slightly in response to dark-induced senescence in leaves (Ellis et al. 2005). ARF2 was isolated in a yeast two-hybrid screen using the carboxyl-terminal region of ARF1 and functions in developing floral organs, as well as in light-grown and dark-grown seedlings. In addition, flowering, leaf senescence, and floral abscission were delayed in ARF2 single mutant plants (Li et al. 2004; Ellis et al. 2005; Okushima et al. 2005a; Schruff et al. 2006). ARF3 and ARF4 play important roles in developing reproductive and vegetative tissues (Sessions et al. 1997; Pekker et al. 2005), and ARF5 is involved in embryonic and vascular tissue development (Hardtke and Berleth 1998; Hardtke et al. 2004; Weijers et al. 2006; Wenzel et al. 2007), ARF8 was reported to regulate fertilization and fruit development (Tian et al. 2004; Nagpal et al. 2005; Goetz et al. 2006; Wu et al. 2006). Related research of T-DNA insertion mutations for AtARF genes indicated that AtARF1–AtARF2, AtARF3–AtARF4, AtARF5–ATARF7, AtARF6–AtARF8, and AtARF10–AtARF16 have overlapping functions during all types of growth and developmental processes in Arabidopsis (Nagpal et al. 2005; Okushima et al. 2005b; Wilmoth et al. 2005; Guilfoyle and Hagen 2007). In total, 25 OsARFs were found in rice by using all 23 predicted AtARF protein sequences as queries. A related analysis indicated that there are 24 OsARFs that have a typical DBD domain that is similar to those ARFs that were previously identified, and all of the OsARFs contain a highly conserved region that may include 320 amino acids in the N-terminal region (Wang et al. 2007). Further, 16 OsARF proteins are considered repressors, and nine members are predicted to function as activators. The transcription of OsARF1 and OsARF23 is enhanced by auxin treatment under light conditions; however, the expression of OsARF5, 14 and 21 decreases under these conditions. OsARFs 1, 2, 16, 21 and 23 can be positively regulated by dark conditions. OsARF8 functions in hypocotyl elongation and is involved in auxin homeostasis (Wang et al. 2007). In total, 17 ARFs were identified in tomato (Solanum lycopersicum) (Kumar et al. 2011). SlARF7 may act as a negative regulator of fruit set (Vriezen et al. 2008; de Jong et al. 2009), whereas SIARF10 has a higher expression level in fruit development. SlARF12 positively functions in flowering and in early stages of fruit development but negatively functions in reproductive tissues. SlARF1, SlARF9, S1ARF11, SlARF15 and SlARF16 have been detected in flower development, and SlARF16 has the highest expression level in flower buds. In addition, there are 35, 26, 20 and 39 ARF members in maize, sorghum, grapes and poplar, respectively (Kumar et al. 2011); these genes each have specific functions (Hagen and Guilfoyle 2002; Kalluri et al. 2007; Wang et al. 2007).
Apple (M. domestica) is one of the most widely cultivated fruit trees and is the most economically important woody plant in temperate regions (Lee et al. 2007). The draft genome sequence of apple has been decoded, which has provided an excellent opportunity for genomewide analyses of all of the genes that belong to specific gene families (Velasco et al. 2010). The genomewide analysis of the RING finger (Li et al. 2011), DREB (Zhao et al. 2012), LBD (Wang et al. 2013) and Hsf gene families have been reported in apple (Giorno et al. 2012). However, no genomewide information regarding the apple ARF gene family is currently available.
Given the importance of ARF during various developmental processes that range from root and shoot development to flower and fruit development in plants, in the present study, we performed a systematic analysis of the apple ARF family for the first time. We then carefully analysed the chromosome location and gene structure of the putative ARF genes that were predicted by genomewide surveys of the apple genomic sequences. Additionally, the putative MdARFs were subjected to phylogenetic analyses with their Arabidopsis counterparts. These comparisons have enabled the identification of gene orthologues and clusters of orthologous groups that can be studied for further functional characterization. Further, we analysed the expression patterns of these ARFs using qRT-PCR and performed expression profile analyses in different tissues (root, stem, leaf, flower and fruit). To our knowledge, this report is the first genomewide analysis of the apple ARF gene family, which would provide valuable information for understanding the classification and putative functions of MdARFs. Ultimately, these findings will lead to potential applications for the improvement of stress resistance in apple via genetic engineering.
Materials and methods
Identification of MdARFs in apple
To identify members of the ARF gene family, multiple database searches were performed. The Arabidopsis ARF gene sequences were used as queries to perform repetitive BLAST searches against the GDR database (Genome Database for Rosaceae: http://www.rosaceae.org/). Additionally, all protein sequences were then used as queries to perform multiple database searches against proteome and genome files that were downloaded from the GDR database. Stand-alone versions of BLASTP and TBLASTN (basic local alignment search tool: http://blast.ncbi.nlm.nih.gov), which are available from NCBI (National Center for Biotechnology Information), were used, with the e-value cutoff set to 1e-003. Moreover, the predicted ARF gene family sequences were downloaded from the Apple GFDB database (Apple Gene Function and Gene Family Database: http://www.applegene.org/). All protein sequences that were derived from the candidate ARF genes that were collected were examined using the domain analysis programs Pfam (Protein family: http://pfam.sanger.ac.uk/) and SMART (Simple Modular Architecture Research Tool: http://smart.embl-heidelberg.de/), with the default cutoff parameters. The isoelectric point (pI) and molecular weight of MdARFs were obtained with the assistance of proteomics and sequence analysis tools on the ExPASy Proteomics Server (http://expasy.org/).
The chromosomal location and gene structure of MdARF genes
The chromosomal locations and gene structures were retrieved from the apple genome data that were downloaded from the GDR database. The remaining genes were selected using a Perl-based program and were mapped to the chromosomes using the software MapDraw. Additionally, the gene structures of the MdARFs were generated with the server GSDS (Gene Structure Display Server: http://gsds.cbi.pku.edu.cn/).
Sequence alignment and phylogenetic analysis
MdARF sequences were aligned using the program ClustalX, with the BLOSUM30 matrix as the protein weight matrix. The multiple sequence comparison by log-expectation (MUSCLE) program (ver 3.52) was also used to perform multiple sequence alignments to confirm the ClustalX data output (http://www.clustal.org/) (Edgar 2004). Phylogenetic trees that were based on the protein sequences of the MdARFs were constructed using the neighbor-joining (NJ) method of the program MEGA5 (molecular evolutionary genetics analysis) with p-distance and the complete deletion option parameters engaged (Saitou and Nei 1987; Tamura et al. 2011). The reliability of the trees that were obtained was tested using bootstrapping with 1000 replicates. Images of the phylogenetic trees were also drawn using the program MEGA5 (http://www.megasoftware.net/).
Expression analysis of MdARFs in microarray
The microarray data of gene expression in apple fruit during the fruit ripening process were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) using the GSE series accession number GSE24523. The sequences of identified MdARF-containing genes were used as queries to blast against a probe sequence (GPL11164) to find corresponding unigene IDs that were used in the microarray data. The microarray data during rootstock–scion interactions (GSE4762) was also downloaded from the Gene Expression Omnibus database. Moreover, MdARF and MdARF-containing genes were used as queries to blast against the probe platform (GPL3715) to discover corresponding unigene IDs that were used in the microarray data. The phylogenetic analysis was performed to determine the corresponding unigene IDs when sequences of high similarity were acquired. The microarray data were organized into a database using a Perl-based program and then clustered using the program Cluster3.0 with Euclidean distances and the hierarchical cluster method of complete linkage clustering. The clustering tree was constructed and viewed in the Java application TreeView.
Plant materials and gene expression analysis
Root, stem, leaf, flower and fruit samples from apple (M. hupehensisRehd.) were collected from a five-year-old apple tree that was grown under natural conditions in the Shandong province of China and stored at −80°C. RNA was extracted from triplicate biological replicates of the above samples using the CTAB method, which was followed by treatment with Turbo DNase I (Ambion, Austin, USA) and reverse transcription of 3 μg RNA from each sample. First-strand cDNA was synthesized using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The transcript levels of the MdARF genes were determined using semiquantitative RT-PCR. Primer details are listed in table 1 in electronic supplementary material at http://www.ias.ac.in/jgenet/.
Results
Identification of ARF genes in apple
In total, 31 typical ARF genes that contained full ORFs were further manually analysed, and we named the genes by their location on the chromosomes (table 1). The open reading frame (ORF) length ranged from 816 bp (MdARF6) to 3450 bp (MdARF1 and MdARF26), with an average ORF length of 2468 bp. The identified MdARF genes encode proteins ranging from 271 (MdARF6) to 1149 (MdARF1 and MdARF26) amino acids (aa) in length, with an average of 822 aa, a predicted molecular mass range of 30263 to 128510 Da and with protein pIs ranging from 5.31 (MdARF22) to 8.75 (MdARF4) (table 1).
ARFgenes in land plants ranging from mosses to eudicots also make these genes interesting candidates for the evolution of plant development. To date, a conserved number of ARF genes have been identified and functionally characterized thus far in both model and crop plants, such as Arabidopsis, rice, maize and tomato. In this study, we first identified 31 ARFcontaining full ORFs in apple using a genomewide analysis. The ARF gene family is conserved in plants, with 23 ARFs in Arabidopsis, 25 ARFs in rice, 17 ARFs in tomato and 39 ARFs in maize. The ARFgene family in apple is larger compared with the estimates for Arabidopsis, rice and tomato, which indicates that the ARF gene family in apple has expanded. It can be speculated that the presence of more ARF genes in the apple genome may reflect a greater requirement for these genes in the complicated transcriptional regulation in this woody perennial species.
Phylogenetic relations of the ARF gene family in apple
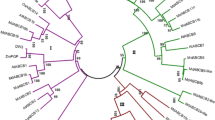
To evaluate the evolutionary relations among the MdARF proteins, full-length amino acid sequences of 31 MdARFs from Arabidopsis were subjected to a multiple sequence alignment using the MEGA4 program. The multiple sequence alignment file was then used for the construction of an unrooted phylogenetic tree. As shown in figure 1, the phylogenetic tree divided the ARFs into four groups (classes I, II, III and IV) as monophyletic clades with at least 50% bootstrap support, and classes I and II were divided into two subgroups. In total, 13 sister pairs of paralogous MdARFgenes were found, which were marked by a red shadow, and had strong bootstrap support (> 90%) (figure 2). Class Ib and IIb both contain four apple genes, and class IIa constitutes the largest clade, with 11 MdARFs, followed by class Ia (seven genes) and III (five genes).
Phylogenetic relation between Arabidopsis and apple ARF genes. The phylogenetic tree was constructed based on a complete protein sequence alignment of ARFs in Arabidopsis and in apple by the neighbor-joining method with bootstrapping analysis (1000 replicates). The subgroups are marked by a colorful background. Scale bar represents 0.05 amino acid substitution per site.
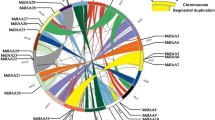
Positions of ARF gene family members on the apple chromosomes. The chromosome number is indicated at the 15 of each chromosome representation. To simplify the presentation, we named the putative ARF family genes from MdARF1 to MdARF31 from the gene order on Chr01 to Chr17 (however, MdARF24 is not found), respectively. Sister paralogous pairs are indicated by a blue line. The scale bar represents a 10 Mb chromosomal distance.
In addition, an unrooted tree, which is shown in figure S1 in electronic supplementary material was constructed from alignments of the full MdARF amino acid sequences to intuitively examine the evolutionary relations among the 31 MdARFs. The results of this work indicated 13 duplicated gene pairs (MdARF9 and MdARF10, MdARF30 and MdARF31, MdARF1 and MdARF26, MdARF23 and MdARF29, MdARF13 and MdARF22, MdARF20 and MdARF21, MdARF11 and MdARF24, MdARF15 and MdARF16, MdARF2 and MdARF25, MdARF8 and MdARF18, MdARF19 and MdARF27, MdARF14 and MdARF28, MdARF3 and MdARF4), which included 26 members, and these paralogous gene pairs represented ∼84% of the apple ARF gene family members. These pairs have strong support (100%).
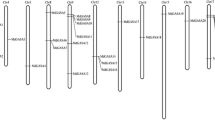
Exon and intron organization of ARF genes
Structural analyses are thought to provide valuable information concerning duplication events when interpreting phylogenetic relations within gene families. In the MdARFgene family, the number of introns varied from one (MdARF28) to 15 (MdARF25 and MdARF30) (figure 3). Additionally, most members within the same subfamily shared a similar exon/intron structure and gene length. In class Ia, MdARF16 consisted of four exons, and other genes have more than 10 exons. Moreover, MdARFs belonging to class Ib possess eight to 15 exons. Class IIa and IIb MdARFs are composed of more than 10 exons, except MdARF28. The class III MdARFs consisted of no more than seven exons. Overall, most closely related members in same subfamilies share similar exon/intron structures in terms of the intron number and exon length.
Analysis of conserved domains of MdARFs
The amino acid sequences of related proteins were used to process the structure of MdARFs members. Notably, typical features of ARF proteins are also exhibited in MdARFs. Most members always contain three conservative amino acid residues: a DNA-binding domain, Auxin-res and AUX-IAA (figure 4). All of the ARF proteins contain a DNA-binding domain, which is generally a B3-type DBD in amino-terminal regions, this region can specifically combine with AuxREs with the consensus sequence TGTCNC. The middle region is Auxin-resp, which also exists in all apple ARF proteins. However, for the AUX-IAA region, the phenomenon is different. Eight members lack this region: two members (MdARF20/21) in class aII, one member (MdARF17) in class bI, three members (MdARF8/18/28) in class bII and one member (MdARF14) in class III.
Chromosomal location of ARFs on apple genomes
Chromosomal location analyses showed that 31 MdARF genes are present on 15 of 17 chromosomes, which were dispersed throughout their respective genomes (figure 2). Only one MdARF gene was found on each of chromosomes 4, 6, 7, 9, 10, 12, 13 and 16, whereas two genes were found on chromosomes 2 and 17, and 3 or more were found on chromosomes 3, 5, 8, 11 and 15. Interestingly, five pairs of MdARFs(MdARF3/4, MdARF9/10, MdARF15/16, MdARF20/21 and MdARF30/31) were tightly colocalized in the apple genome. Therefore, it is suggested that segmental duplication and transposition events all have played roles in the evolution of the ARF gene family in apple. Gene duplications have an important role not only in genomic rearrangement and expansion but also in the diversification of gene function. A recent GWD event was reported to have occurred in apple 60–65 million years ago, which resulted in the expansion of several gene classes. According to this previous report, gene duplication caused the expansion of some gene families (e.g. Ring, LBD, HSP and DREB).
Expression pattern of ARFs from microarray
The expression pattern of genes is significant to some degree for determining their functions. Thus, from this perspective, a microarray analysis of ARFs was completed and is presented in figures 5 and 6 with published microarray data of GEO (GSE24523). Using a BLAST search against the apple UniGene database from the GDR database, 24 samples, GSM618107-GSM618130, were selected from four weeks before ripening to the ripening of apple. Twenty of the 31 apple ARF genes were identified, whereas probes for the other 11 ARF genes were not found. Based on the consequent hierarchical clustering, the expression patterns for ARF genes are divided into four groups: groups I, II, III and IV. Some members of each group show similar expression patterns. Group I contains three members (MdARF5, MdARF13 and MdARF22), and the expression levels of these genes exhibited higher expression levels at week-0 (ripening) and week-2 (before ripening than week-4 (before ripening). There are seven members (MdARF3, MdARF4, MdARF23, MdARF29, MdARF9, MdARF10 and MdARF31) in group II. Based on the similarity of their expression patterns, three pairs of these members are produced in this group (MdARF3–MdARF4, MdARF23–MdARF29 and MdARF9–MdARF10). However, all members of this group are highly expressed before ripening, particularly two weeks before ripening. Group III contains three members (MdARF15, MdARF12 and MdARF8); these three genes have their own expression pattern and exhibit active expression levels in week-0 (ripening). Group IV contains seven members; (MdARF7, MdARF18, MdARF30, MdARF27, MdARF19, MdARF11 and MdARF24), MdARF11 and MdARF24 show similar expression patterns. All of the seven members can be detected in the fruit development stage which contains ripening and proripening, suggesting that the members of this group may positively control fruit development.
The expression patterns of the MdARFs from a microarray during rootstock–scion interactions. The colour scale, which represents the relative signal values, is shown above (green refers to low expression, black refers to medium expression, and red refers to high expression). Of the 27 samples (GSM107608-GSM107659) that are shown, 10 functioned in rootstock–scion interactions (Ambrosia/B9, Gala/B9, Melrose/B9, Gala/B9, Gala/G30, Gala/M111, Gala/M27, Gala/M7, Gala/M9 and Gala/Supporter).
The expression patterns of the MdARFs from a microarray during fruit development in apple. The colour scale, which represents the relative signal values, is shown above (green refers to low expression, black refers to medium expression, and red refers to high expression). HC, honeycrisp; CP, cripps pink. Week-4, 4 weeks before ripening; week-2, 2 weeks before ripening; week-0, ripening. In total, 24 samples named GSM618107-618130 (GEO accession numbers) are shown.
In addition, to investigate the expression of MdARFs in the entire plant, the expression patterns of 31 MdARF-containing probes from a microarray during rootstock–scion interactions were found using a BLAST search against the probe sequences (GPL3715). These probes can also be divided into four groups based on the hierarchical clustering. Both groups I and II contains only one member, MdARF14 and MdARF15, respectively. Although these probes have different expression patterns, MdARF14 shows higher expression in some samples (GSM107608–GSM107616); however, MdARF15 displays converse expression patterns in those samples. Group III has eleven members (3/4/8/5/21/20/13/12/18/19/27), and there are 18 members in group IV. Further, most members of this group have high expression levels in samples GSM107646 to GSM107659.
Expression pattern of ARFs in various tissues as determined by semi-quantitative RT-PCR analyses
To study the function of MdARF genes in different plant tissues (roots, stems, leaves, flowers and fruits), eight MdARFs were selected to perform qRT-PCR. As shown in figure 7, eight genes were expressed in five tissues that were tested, except the root, with various expression levels. MdARF17 has high expression levels in the stem and fruit, and MdARF1, MdARF13, MdARF24 and MdARF26showed high transcript abundances in the flower. Overall, these results indicated that MdARFs play multiple roles in apple development.
Because the gene expression patterns can provide important clues for gene function, we examined the expression of all the ARF genes in the root, stem, leaf, flower and fruit tissues using semi-quantitative RT-PCR. Eight selected genes were expressed in five tissues that were tested, except the root, with various expression levels. These results indicated that MdARF genes play a major role in apple development. In future studies, more research will be required to determine the functions of MdARF genes in apple. Overall, our results will be helpful in laying the foundation for the functional characterization of the ARF gene family and for further understanding of the structure–function correlation between these family members. Additionally, our study provides comprehensive information and novel insights into the evolution and divergence of the ARF genes in plants. Moreover, these studies may potentially aid in the understanding of the molecular basis of many agronomically important traits of apple.
Discussion
Auxin signalling plays important roles in plant growth and in development processes, such as tropic responses, lateral root formation, vascular differentiation, embryo patterning and shoot elongation (Finet et al. 2013). Auxin response factors (ARF) are transcription factors that regulate the expression of auxin response genes. To date, the members of ARF cascades from Arabidopsis, rice, tomato and maize have been identified. Analysis of the Arabidopsis genome revealed the presence of 23 ARFs, and an in silico analysis of the rice genome revealed 25 ARF genes (Wang et al. 2007). More recently, 17 and 39 ARF genes were identified in tomato and maize, respectively (Kumar et al. 2011; Xing et al. 2011). In this study, 31 ARF genes were identified in apple, and various types of information for this family were collected and collated; the number of members of this family is larger than that of Arabidopsis. Genomic rearrangements and expansions that occurred in the apple genome might explain the additional members. However, the fundamental mechanisms include extensive duplication and diversification that were generated from the long evolutionary history of the organism. Generally, whole genome and chromosomal segment duplications occur in almost all organisms and are the main contributors of expansion. As mentioned above, there are 13 duplicated gene pairs in apple, among these 13 MdARF sister pairs, five pairs (MdARF1 and MdARF26, MdARF23 and MdARF29, MdARF13 and MdARF22, MdARF2 and MdARF5, MdARF19 and MdARF29) have been identified as formed by tandem duplication. This pathway of duplication has also occurred in Arabidopsis, but has not been found in rice. Further, the highly similar microarray expression modes of those genes indicate that these genes may have similar functions.
Based on the phylogenetic tree of apple and Arabidopsis, an apparent phenomenon should be noticed, i.e., the absence of any MdARF gene in class Ib and a related study indicates that seven AtARFs of this class originally generated from one AtARF gene. This phenomenon also occurs in the phylogenetic tree of rice–Arabidopsis and tomato–Arabidopsis, which suggests that these genes might execute specific functions and play important roles in Arabidopsis. More importantly, a new group of genes (MdARF6, MdARF21, MdARF20 and MdARF5) have been found in apple and classified into class IV, which does not include any AtARF genes. These four genes may also have originally been derived from one AdARFgene, which provides a reasonable explanation for the existence of a larger ARF family in apple. The result also suggests that there are more complex auxin response processes in apple. Compared with other phylogenetic trees (rice–Arabidopsis, tomato–Arabidopsis and so on), there are two unusual types of grouping of both Arabidopsis and apple that should be noticed in this study. All of the groups contain more than two orthologous genes, of which only one belongs to the AtARF family and the others belong to the MdARF family. Nine MdARF-MdARF-AtARF groups are observed in the combined phylogenetic tree, including MdARF1-MdARF26-AtARF19, MdARF13- MdARF22-AtARF5, MdARF9-MdARF10-AtARF6, MdARF 14-MdARF28-AtARF17, MdARF8-MdARF18-AtARF3, MdA RF19-MdARF27-AtARF4, MdARF2-MdARF25-AtARF9, MdARF11-MdARF24-AtARF2 and MdARF15-MdARF16-AtARF1, revealing a 3:1 orthologous relation among these apple and Arabidopsisgenes. A 4:1 orthologous relation is found among MdARF3, MdARF4 and MdARF7 and AtARF16. The above-mentioned results indicate that these genes may originate from one common ancestor and may have conserved functions. However, this phenomenon of multiple MdARFs and a single AtARF in each group also explains the amplification of the ARF family in apple. Thus, diversity in the functions of these genes indicates that there are different levels of regulation in auxin response actions that occur in different species.
The conserved domain in the carboxyl-terminal region of both ARF and auxin-induced Aux/IAA proteins is highly similar; ARFs may act as repressors or activators depending on which partner an ARF binds to through its carboxyl-terminal domain. This interaction may play a role in auxin-regulated gene expression (Guilfoyle et al. 1998). However, MdARF6/MdARF20/MdARF21/MdARF17/MdARF8/MdA RF18/MdARF28/MdARF14 lack this region, and the same situation exists in AtARF3/AtARF13/AtARF17 and in SlARF2/SlARF3/SlARF7/SlARF13 (Okushima et al. 2005b; Kumar et al. 2011). Nevertheless, those genes that lack the carboxyl-terminal region can also be regulated in an auxin-independent manner.
Tissue-specific expression profiles and the expression patterns from a microarray for the MdARF genes are very frequently used to predict the functions of unknown genes in specific species. Thus, expression levels of MdARF genes were examined using qRT-PCR in apple roots, stems, leaves, flowers and fruits. As the above results indicated, nearly all of the genes (MdARF1/ MdARF13/MdARF9/MdARF10/MdARF24/MdARF11/MdAR F26) that were selected for analysis have higher expression levels in the flower and fruit, particularly in the flower, and only MdARF17 is expressed in both the stem and fruit. In the microarray experiment, most members of this family are highly expressed before ripening, particularly two weeks to four weeks before ripening. From the above-mentioned results, it is clear that those genes may be involved in fruit set and in early cell division stages of the fruit in apple. Further, there are many types of similar expression models for some MdARF genes, which may indicate their possible overlapping functions during various developmental processes in plants.
References
Altschul S. F., Gish W., Miller W., Myers E. W. and Lipman D. J. 1990 Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R. et al. 2002 The Pfam protein families database. Nucleic Acids Res. 30, 276–280.
de Jong M., Wolters-Arts M., Feron R., Mariani C. and Vriezen W. H. 2009 The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, 160–170.
Edgar R. C. 2004 MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Ellis C. M., Nagpal P., Young J. C., Hagen G., Guilfoyle T. J. and Reed J. W. 2005 AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574.
Finet C., Berne-Dedieu A., Scutt C. P. and Marletaz F. 2013 Evolution of the ARF gene family in land plants: old domains, new tricks. Mol. Biol. Evol. 30, 45–56.
Giorno F., Guerriero G., Baric S. and Mariani C. 2012 Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13, 639.
Goetz M., Vivian-Smith A., Johnson S. D. and Koltunow A. M. 2006 AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18, 1873–1886.
Guilfoyle T. J. and Hagen G. 2007 Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460.
Guilfoyle T. J., Ulmasov T. and Hagen G. 1998 The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell Mol. Life Sci. 54, 619–627.
Hagen G. and Guilfoyle T. 2002 Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385.
Hardtke C. S. and Berleth T. 1998 The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411.
Hardtke C. S., Ckurshumova W., Vidaurre D. P., Singh S. A., Stamatiou G., Tiwari S. B. et al. 2004 Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089–1100.
Kalluri U. C., Difazio S. P., Brunner A. M. and Tuskan G. A. 2007 Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant. Biol. 7, 59.
Kim J., Harter K. and Theologis A. 1997 Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Aca. Sci. USA 94, 11786–11791.
Kumar R., Tyagi A. K. and Sharma A. K. 2011 Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genomics 285, 245–260.
Lee S. Y., Chang S. S., Shin J. H. and Kang D. H. 2007 Membrane filtration method for enumeration and isolation of Alicyclobacillus spp. from apple juice. Lett. Appl. Microbiol. 45, 540– 546.
Letunic I., Doerks T. and Bork P. 2012 SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, 302–305.
Li H., Johnson P., Stepanova A., Alonso J. M. and Ecker J. R. 2004 Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 193–204.
Li Y., Wu B., Yu Y., Yang G., Wu C. and Zheng C. 2011 Genome-wide analysis of the RING finger gene family in apple. Mol. Genet. Genomics 286, 81–94.
Nagpal P., Ellis C. M., Weber H., Ploense S. E., Barkawi L. S., Guilfoyle T. J. et al. 2005 Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118.
Okushima Y., Mitina I., Quach H. L. and Theologis A. 2005a AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46.
Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C. et al. 2005b Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463.
Pekker I., Alvarez J. P. and Eshed Y. 2005 Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–2910.
Remington D. L., Vision T. J., Guilfoyle T. J. and Reed J. W. 2004 Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752.
Saitou N. and Nei M. 1987 The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schruff M. C., Spielman M., Tiwari S., Adams S., Fenby N. and Scott R. J. 2006 The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251– 261.
Sessions A., Nemhauser J. L., McColl A., Roe J. L., Feldmann K. A. and Zambryski P. C. 1997 ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491.
Tamura K., Peterson D., Peterson N., Stecher G., Nei M. and Kumar S. 2011 MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Tian C. E., Muto H., Higuchi K., Matamura T., Tatematsu K., Koshiba T. and Yamamoto K. T. 2004 Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40, 333–343.
Tiwari S. B., Hagen G. and Guilfoyle T. 2003 The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543.
Ulmasov T., Hagen G. and Guilfoyle T. J. 1997 ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868.
Ulmasov T., Hagen G. and Guilfoyle T. J. 1999a Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319.
Ulmasov T., Hagen G. and Guilfoyle T. J. 1999b Activation and repression of transcription by auxin-response factors. Proc. Natl. Aca. Sci. USA 96, 5844–5849.
Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A. et al. 2010 The genome of the domesticated apple (Malus x domestica Borkh.) Nat. Genet. 42, 833–839.
Vriezen W. H., Feron R., Maretto F., Keijman J. and Mariani C. 2008 Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 177, 60–76.
Wang D., Pei K., Fu Y., Sun Z., Li S., Liu H. et al. 2007 Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24.
Wang X., Zhang S., Su L., Liu X. and Hao Y. 2013 A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PloS one 8, e57044.
Weijers D., Schlereth A., Ehrismann J. S., Schwank G., Kientz M. and Jurgens G. 2006 Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265–270.
Wenzel C. L., Schuetz M., Yu Q. and Mattsson J. 2007 Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 49, 387–398.
Wilmoth J. C., Wang S., Tiwari S. B., Joshi A. D., Hagen G., Guilfoyle T. J., Alonso J. M. et al. 2005 NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130.
Wu M. F., Tian Q. and Reed J. W. 2006 Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218.
Xing H., Pudake R. N., Guo G., Xing G., Hu Z., Zhang Y. et al. 2011 Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12, 178.
Zhao T., Liang D., Wang P., Liu J. and Ma F. 2012 Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol. Genet. Genomics 287, 423–436.
Acknowledgement
This work was supported by the National Natural Science Foundation in China (Grant No. 31401822).
Author information
Authors and Affiliations
Corresponding author
Additional information
[Luo X.-C., Sun M.-H., Xu R.-R., Shu H.-R., Wang J.-W. and Zhang S.-Z. 2014 Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 93, xx–xx]
Xiao-Cui Luo and Mei-Hong Sun contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
LUO, XC., SUN, MH., XU, RR. et al. Genomewide identification and expression analysis of the ARF gene family in apple. J Genet 93, 785–797 (2014). https://doi.org/10.1007/s12041-014-0462-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-014-0462-0