Abstract
Asthma has significant impacts on living quality particularly in children. Long noncoding RNA (lncRNA) MALAT1 plays a crucial role in neonatal respiratory diseases. Meanwhile, MALAT1 knockdown could induce viability and attenuate apoptosis of airway-related cells. However, the role of MALAT1 in neonatal asthma, asthma-related cell, and its possible mechanism is unclear. This study aims to investigate MALAT1 level in asthma and to identify the effects of MALAT1 on bronchial/tracheal smooth muscle cells (B/TSMCs). Newborn asthma modeling rat was constructed by introducing ovalbumin (OVA). MALAT1 levels in tissues or B/TSMCs were determined by RT-qPCR. Exogenous changes of MALAT1, RyR2 or miR-133a in B/TSMCs were fulfilled by cell transfection; cell apoptosis was measured by using Cell Death Detection ELISA kit and Hochest33342; IL-6, TNF-α and IL-1β level was detected by using corresponding ELISA kit; ryanodine receptor 2 (RyR2) mRNA and miR-133a level was determined by RT-qPCR; cleaved caspase-3 (c-caspase-3) and RyR2 expression was detected by Western blot; luciferase reporter assay was performed to confirm the target regulation of miR-133a on RyR2. We found that MALAT1 was significantly upregulated in tracheal tissues of newborn asthma modeling rats. In MALAT1-silenced or -overexpressed B/TSMCs, we found a synchronous change of cell apoptosis, inflammatory factor secretion (IL-6, TNF-α, and IL-1β) or RyR2 level, but a reverse change of miR-133a level with MALAT1. Besides, MALAT1 induced B/TSMCs apoptosis and inflammation increase could be partially reversed when RyR2 was silenced or when miR-133a was overexpressed. The luciferase reporter assay confirmed that RyR2 is a direct target gene of miR-133a in B/TSMCs. Finally, we conclude that MALAT1 knockdown could protect from B/TSMCs injury via regulating miR-133a/RyR2 axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Asthma is one of the most common chronic inflammatory diseases, which is mainly consisting of recurrent airway obstruction, bronchial hyper-responsiveness, and airway inflammatory cell infiltration. The symptoms of asthma generally includes recurrent wheezing, chest tightness, cough, as well as sensation and shortness of breath (Han et al. 2016; Hsu et al. 2016). Nowadays the incidence of childhood asthma is increasing year by year. The United States reported that the incidence of asthma and wheezing in children is as high as 9. 4% (about 7 million), and the number of asthma-related deaths is 7, 000 per year (Dinakar and Chipps 2017). The pathogenesis of asthma refers to numerous genetic factors and environmental factors like allergens (Dutmer et al. 2016; Gagro 2011). For a long time, asthma has significant impacts on living quality particularly in children, in some way due to the higher incidence of allergies in them (Chang 2012). For children, asthma is generally characterized by its heterogeneity in terms of etiology, degree of inflammatory response, airway obstruction as well as pulmonary function (Gagro 2011). Therefore, timely diagnosis and treatment are essential to prevent inflammatory response, airway remodeling and pulmonary function decline. Although there are some effective strategies for asthma prevention and treatment in children, the risk factors and specific mechanisms of pediatric asthma are still unclear.
Long noncoding RNA (LncRNA) belongs to noncoding RNAs with a length about 200 nucleotides or more (Li et al. 2016), which plays important roles in a wide range of biological processes thus has relevance with multiple human diseases for example cancer and cardiovascular disease (Zhang et al. 2018). In recent years, accumulated studies have demonstrated that lncRNA is also associated with the pathogenesis of asthma (Omran et al. 2013; Zhu et al. 2018). For instance, Austin PJ et al. identified that lnRNA plasmacytoma variant translocation (PVT1) is a novel regulator of the asthmatic phenotype in human airway smooth muscle (Austin et al. 2017). The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been reported to implicate in tumor progression (Cheng et al. 2018a, b; Orlandi et al. 2019), liver damage (Sookoian et al. 2018), and osteoarthritis (Zhang et al. 2019), while rarely in cross-talk of respiratory disease. The discovery that MALAT1 knockdown could induce the viability and attenuate the apoptosis of airway related cells, made MALAT1 to be a hopeful therapeutic target of neonatal respiratory distress syndrome (NRDS) (Juan et al. 2018; Li et al. 2018a, b). Additionally, in asthma, airway remodeling may coexist with airway inflammation, causing the occurrence and persistence of chronic inflammation due to airway smooth muscle cell inflammation and proliferative hypertrophy (B Andrew 2008). Studies have revealed that some lncRNAs could regulate proliferation or inflammatory response of airway smooth muscle cells during the development of asthma, such as PVT1 (Austin et al. 2017) and BCYRN1 (Zhang et al. 2016). It has been also reported that MALAT1 can regulate the occurrence and development of diseases by participating in a variety of non-cancer cell proliferation, apoptosis, and inflammatory reactions (Li et al. 2018a, b). For example, the release of pro-inflammatory cytokine and apoptosis of human coronary artery endothelial cells were markedly elevated when MALAT1 was overexpressed (Li et al. 2018a, b). Besides, MALAT1 could also promote proliferation, angiogenesis, and immunosuppressive properties of mesenchymal stem cells by inducing vascular endothelial growth factor (VEGF) (Li et al. 2017). Currently, the role of MALAT1 in pediatric asthma or airway smooth muscle cells is still unknown.
MicroRNAs (miRNAs) are short noncoding endogenous RNAs with a length of 18–25 nucleotides, which regulate gene-expression by binding and inhibiting mRNA post-transcriptionally (Lu and Rothenberg 2018; Simonson and Das 2015). Evidences have suggested that lncRNAs function as endogenous miRNA sponges by binding to miRNAs and then causing malfunction of various pathological and physiological processes (Ahmed et al. 2016). Previously, studies have revealed that miRNAs like MiR-3162–3p (Fang et al. 2016)and microRNA-19b (Ye et al. 2017)could regulate the functions of airway in asthma animal models. Besides, microRNA-1 downregulation could regulate smooth muscle hypertrophy during asthmatic disease (Chen et al. 2006). MicroRNA-155 knockout mice exhibited rapid airway remodeling and collagen deposition in smooth muscle cells (Nicolas et al. 2009). A discovery that microRNA-133a downregulation-mediated ras homolog gene family member A (RhoA) increase could regulate bronchial smooth muscle (BSM) contraction (Chiba and Misawa 2010), hints that miRNA-133a may implicate in smooth muscle function in asthmatics, which also aroused our interest.
In the present study, newborn asthma modeling rat was successfully constructed by introducing ovalbumin (OVA). Then, we discovered that lncRNA MALAT1 was significantly upregulated in tracheal tissues of newborn asthma modeling rats. In MALAT1-silenced or -overexpressed bronchial/tracheal smooth muscle cells (B/TSMCs), we found a synchronous change of cell apoptosis rate and inflammatory response with MALAT1 levels. With the help of bioinformatics predictions, we confirmed by experiments that MALAT1 induced B/TSMCs apoptosis and inflammatory response may be partially fulfilled via its regulation on miR-133a/RyR2 axis. Our findings identified the adverse effects of MALAT1 on B/TSMCs, an important cell line for asthma research.
2 Materials and methods
2.1 Animals and neonatal rat asthma modeling
Specific pathogen free (SPF) male Wistar rats (weighing 50±5 g at the beginning of sensitization) were purchased from Shanghai Laboratory Animal Research Center (Shanghai, China). After adaptive feeding with aseptic operation in SPF animal room (room temperature: 25 ± 3°C; relative humidity: 40–60%; daily artificial lighting: 12 h), rats were randomly divided into control group and ovalbumin (OVA) group with 8 in each. For OVA group, rats were sensitized with 1 mg OVA (Grade II; Sigma-Aldrich, St. Louis, MO, USA) and 100 mg aluminum hydroxide (Al(OH)3; Aladdin Industrial Corporation, Shanghai, China) in 1 mL saline by intraperitoneal injection on the 1st and 8th days (Ablimit et al. 2013; Cheng et al. 2018a, b); starting on the 15th day, rats were challenged with inhaled OVA aerosol (1% w/v solution diluted in saline; Grade V; Sigma-Aldrich) generated by a nebulizer for 30 min each time and repeated every other day for 6 days. For the control group, rats were received treatments in the same way; however, they were intraperitoneally injected with 1 mL saline followed by inhalation of saline aerosol. After the last challenge for 24 h, rats were anesthetized with 1.5% pentobarbital sodium (15 mg/kg; Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) by intraperitoneal injection; the lung tissues were removed and immersed in 10% formalin for fixation and then embedded in paraffin; and the tracheal tissues were exposed and isolated for use. All animal protocols were approved by the Ethics Committee on Animal Experiments of Xian Yang Central Hospital and performed according to official recommendations of the Chinese Community guidelines.
2.2 Hematoxylin-eosin (H&E) staining
The paraffin-wax sections of rat lung tissues with a thickness of 4 μm were prepared, dewaxed and stained with hematoxylin and eosin (H&E; Sigma-Aldrich) for pathological observation. H&E stained sections were observed at 200× magnification under a light microscopy (Lu et al. 2010).
2.3 Cell line and cell culture
Human primary bronchial/tracheal smooth muscle cells (B/TSMCs; cat No. PCS-130–011) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were incubated in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (Gibco), 100 μg/mL streptomycin (Sigma Aldrich) and 100 U/mL penicillin (Sigma Aldrich) at 37°C with 5% CO2.
2.4 Cell transfection
The siRNAs targeting MALAT1 (siMALAT1–1, siMALAT1–2, siMALAT1–3) and its negative control siRNA (siMALAT1-NC) were synthesized by GenePharma (Shanghai, China). Sequences of these siRNAs were as follows: siMALAT1–1, sense: 5′-GAT CCA TAA TCG GTT TCA AGG-3′, antisense: 5′-TTG AAA CCG ATT ATG GAT CAT-3′; siMALAT1–2, sense: 5′-GGC AAU GUU UUA CAC UAU UTT-3′, antisense: 5′-AAU A GU GUA AAA CAU UGC CTA-3′; siMALAT1–3, sense: 5′-GAG GUG UAA AGG GAU UUA UTT-3′, antisense: 5′-AUA AAU CCC UUU ACA CCU CTT-3′; si-MALAT1-NC, sense: 5′-UUC UCC GAA CGU GUC ACG UTT-3′, antisense: 5′-ACG UGA CAC GUU CGG AGA ATT-3′. B/TSMCs seeded in 6 well plates reaching 80% confluency were transfected with 50 nM siMALAT1–1, siMALAT1–2, siMALAT1–3, or si-MALAT1-NC using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For MALAT1 overexpression, the full length cDNA of human MALAT1 was amplified by RT-PCR with total RNA extracted from B/TSMCs, and then cloned into pcDNA3.1(+) vectors (Invitrogen, Carlsbad, CA, USA) to generate pcDNA3.1(+)-MALAT1; and the empty pcDNA3.1(+) vectors were used as its negative control (pcDNA3.1(+)-NC). B/TSMCs in 6 well plates (80% confluency) were transfected with 10 μg pcDNA3.1(+)-MALAT1 or pcDNA3.1(+)-NC, respectively. After transfection for 48 h, cells were harvested to analysis MALAT1 levels by qRT-PCR to verify transfection efficiency.
For RyR2 silencing, siRNAs targeting RyR2 (siRyR2–1, siRyR2–2, siRyR2–3) and its negative control siRNA (siRyR2-NC) were synthesized by Invitrogen. Sequences of them were as follows: siRyR2–1, 5′-CAA GCG CAT CGA GAG GGT CTA CTT T-3′; siRyR2–2, 5′-AAG TAC GAG TTG GAG ATG ACC-3′; siRyR2–3, 5′-AAG TGG TTC TGC AGT GCA CCG-3′; siRyR2-NC (proprietary by Invitrogen). B/TSMCs seeded in 6 well plates reaching 80% confluency were transfected with 20 nM siRyR2–1, siRyR2–2, siRyR2–3, or siRyR2-NC using Lipofectamine 2000, respectively. Cells were harvested to verify RyR2 levels by western blot post-transfection for 48 h. For further mechanism research, pcDNA3.1(+)-MALAT1 (10 μg) and siRyR2–3 (20 nM), or pcDNA3.1(+)-MALAT1 (10 μg) and siRyR2-NC (20 nM) co-transfected B/TSMCs were prepared by methods described above.
For miR-133a overexpression, 50 nM miR-133a mimics (miR-133a-mimic) or its negative controls (miR-mimic-NC) purchased from GenePharma were arranged to transfect B/TSMCs seeded in 6 well plates by using Lipofectamine 2000, respectively. Similarly, cells were harvested to verify miR-133a levels by qRT-PCR post-transfection for 48 h. For further mechanism research, pcDNA3.1(+)-MALAT1 (10 μg) and miR-133a-mimic (50 nM), or pcDNA3.1(+)-MALAT1 (10 μg) and miR-mimic-NC (50 nM) co-transfected B/TSMCs were prepared by methods described above.
2.5 Apoptosis assays
The apoptosis rate of B/TSMCs was evaluated by using Cell Death Detection ELISA kit (Roche Diagnostics, Penzberg, Germany). Briefly, cell suspension was prepared and centrifuged at 12000 rpm for 10 min. After removing the supernatant, 200 μL lysis buffer was added to cell pallets effecting for 30 min. Then, the acquired cytoplasmic lysates were transferred onto a streptavidin-coated plate adding with a mixture of anti-DNA-POD and anti-histone-biotin effecting for 2 h. Absorbance was measured at the wavelength of 405 nm.
2.6 Hochest33342 staining assay
B/TSMCs were seeded in 6‑well plates (3×105 cells/well) and stained with Hoechst 33342 (5 µg/ml; Thermo Fisher Scientific, Inc.) for 1 hour at 37°C. After washing twice with PBS, images of the cells were acquired by using an immunofluorescent inverted fluorescence microscope (Nikon TE2000; Nikon Corporation, Tokyo, Japan).
2.7 Reverse transcription quantitative real-time PCR (RT-qPCR)
For rat trachea tissue and prepared B/TSMCs, total RNA was extracted by using TRIzol reagent (Invitrogen). After quantitation, 5 μg total RNA from each sample was reverse-transcribed into cDNA using Reverse Transcription Reagent Kit (Promega, Madison, WI, USA) as per the manufacturer’s instructions. RT-PCR reactions were performed in triplicate on an Applied Biosystems Prism 7500 Fast Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with SYBR Green PCR Master Mix (Applied Biosystems). Reaction parameters were set as follows: 95°C for 5 minutes, 95°C for 45 s, then 57°C for 30 s, 72°C for 1 min. Gene-special primers were synthesized by Shanghai Sangon Biological Engineering and Technology Service (Shanghai, China); and its sequences were as follows: for rat, MALAT1: 5′-TGC TGC CTT TTC TGT TCC TT-3′(Forward) and 5′-AAG GTG CTG GGT AGG GAA GT-3′(Reverse), β-actin: 5′-AGA GGG AAA TCG TGC GTG AC-3′(Forward) and 5′-CAA TAG TGA TGA CCT GGC CGT-3′(Reverse); for human, MALAT1: 5′-GAC CCT TCA CCC CTC ACC-3′ (Forward) and 5′-TTA TGG ATC ATG CCC ACA AG-3′ (Reverse), RyR2: 5′-GAA TTC ATC ATG GAT ACT CTA CC-3′ (Forward) and 5′-GTC ATG CAC ATT ATC TTC TGC AT-3′ (Reverse), β-actin: 5′-CCA TCA TGA AGT GTG ACG-3′ (Forward) and 5′-GCC GAT CCA CAC GGA GTA-3′ (Reverse), miR-133a: 5′-CTG CAT TGG TCC CCT TCA AC-3′ (Forward) and 5′-CAG TGC AGG GTC CGA GGT AT-3′ (Reverse), U6 small nuclear RNA (U6 snRNA): 5′-CTC GCT TCG GCA GCA CA-3′ (Forward) and 5′-AAC GCT TCA CGA ATT TGC GT-3′ (Reverse). The level of mRNA or miRNA was calculated using the 2−ΔΔCT method. Data were normalized to β-actin (MALAT1, RyR2) or U6 (miR-133a).
2.8 Western blotting
For prepared B/TSMCs, cells were lysed with RIPA lysis buffer (Beyotime, Shanghai, China); and protein concentration for each sample was detected by using BCA Protein Assay Kit (Beyotime). A total of 50 μg protein was separated by 10% SDS-PAGE, and then electro-transferred onto polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA, USA). After blocking with 0.5% skim milk powder at room temperature for 1 hour, the obtained membranes were incubated with specific primary antibodies at 4 ˚C overnight, and then incubated with the secondary antibodies for 2 hours at room temperature. Antibodies from Abcam (Cambridge, MA, USA) were as follows: rabbit anti-human monoclonal cleaved-caspase 3 (Cat. No: ab32042; dilution: 1:500), rabbit anti-human monoclonal caspase-3 (Cat. No: ab32351; dilution: 1:4000), rabbit polyclonal RyR2 (Cat. No: ab59225; dilution: 1:1000), rabbit polyclonal β-actin (Cat. No: ab8227; dilution: 1:1500) primary antibodies; and horse radish peroxidase-conjugated goat anti-rabbit IgG secondary antibodies (Cat. No: ab7090; dilution: 1:3500). The protein bands were visualized by using Enhanced chemiluminescence reagents (Thermo-Pierce, Rockford, IL, USA). Detected bands of target protein were quantified using Gel-Pro Analyzer 4.0 software (Media Cybernetics, LP, USA), and data were normalized to β-actin or total caspase-3.
2.9 Determination of cytokine production
The secretion level of cytokines including IL-6, TNF-α and IL-1β in cell culture supernatants was measured by using corresponding ELISA kit (Boster, Wuhan, China) according to the manufacturer’s recommendations.
2.10 Luciferase reporter assay
To construct the pGL3-RyR2–3′ untransfected region (UTR) wild-type (WT) or mutant (Mut) reporter plasmid, the 3′-UTR sequence of RyR2 predicted to interact with miR-133a or a mutated sequence within the predicted target site of miR-133a was amplified by RT-qPCR. Following, the acquired sequences were cloned into the XbaI/FseI site of pGL3 vector (Promega, Madison, WI, USA). Before transfection, cells were seeded in 24-well plates (2×105 cells/well) and cultured for 24 hours. The prepared WT or Mut reported plasmid together with Renilla luciferase pRL-TK vector (Promega) were co-transfected with miR-133a-mimic or miR-mimic-NC into B/TSMCs using Lipofectamine 2000. After co-transfection for 48 h, luciferase activity was determined using the Dual-Glo Luciferase assay system (Promega).
2.11 Statistical analysis
All the experiments were performed in triplicate. Data were described as mean ± standard deviation (SD) and analyzed by SPSS 19.0 software (SPSS; Chicago, IL, USA). Differences between two groups were analyzed by Student’s t-test. Comparisons among more than two groups were performed by One-way ANOVA followed by LSD-t test. A P-value <0.05 was considered as statistically significant difference.
3 Results
3.1 MALAT1 level is significantly increased in tracheal tissues of newborn asthma modeling rats
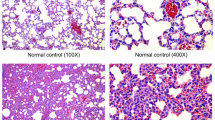
From figure 1A, an abnormal proliferation and thickening of smooth muscle layer could be obviously observed at airway position in lung tissues of rats from OVA group than that from control group. Subsequently, we discovered that MALAT1 level in rat trachea tissues was remarkably increased in OVA group when compared to control group (figure 1B, P<0.05). These data indicated that MALAT1 level was significantly increased in tracheal tissues from newborn asthma modeling rats.
MALAT1 level is significantly increased in tracheal tissues of newborn asthma modeling rats. Rat asthma modeling was constructed by using OVA. (A) H&E staining images of rat lung tissues (200× magnification). (B) Relative MALAT1 levels in rat tracheal tissues by RT-qPCR. Data in control group deemed as 1. n=8. * P<0.05 vs. control group.
3.2 MALAT1 regulates B/TSMCs apoptosis and inflammatory factor secretion
In order to investigate the effects of MALAT1 on B/TSMCs, MALAT1 was silenced and overexpressed, respectively. Figure 2A displayed that MALAT1 was successfully silenced in siMALAT1–1, siMALAT1–2, and siMALAT1–3 group (P<0.05), among which siMALAT1–3 group exhibited the best silencing efficiency thereby was applied in the following experiments. Data in figure 2B suggested that MALAT1 level was markedly upregulated in pcDNA3.1(+)-MALAT1 group (P<0.05), which meets the requirements of follow-up experiments. In addition, apoptosis-related indexs including cell apoptosis rate (figure 2C), the number of brillant blue cells stained by Hochest33342 (figure 2D) and relative cleaved-caspase 3 level (figure 2E), as well as inflammatory indexs including IL-6 level (figure 2F), TNF-α level (figure 2G) and IL-1β level (figure 2H) were significantly decreased in siMALAT1–3 group than that in control and siMALAT1-NC group (P<0.05) while dramatically increased in pcDNA3.1(+)-MALAT1 group than that in control and pcDNA3.1(+)-NC group (P<0.05). These discoveries indicated that MALAT1 could facilitate B/TSMCs apoptosis and inflammatory factor secretion.
MALAT1 regulates B/TSMCs apoptosis and inflammatory factor secretion. MALAT1 was silenced or overexpressed in B/TSMCs by cell transfection described in methods. (A) Relative MALAT1 levels in B/TSMCs by RT-qPCR (MALAT1 silencing). (B) Relative MALAT1 levels in B/TSMCs by RT-qPCR (MALAT1 overexpression). (C) Cell apoptosis rate (%). (D) Images of hochest33342 straining. (E) The levels of c-caspase3 in B/TSMCs by western blotting. (F) IL-6 levels in B/TSMCs. (G) TNF-α levels in B/TSMCs. (H) IL-1β levels in B/TSMCs. For A, B and E: data in control group deemed as 1. For (A–H): *P<0.05 vs. control or siMALAT1-NC group; #P<0.05 vs. control or pcDNA3.1(+)-NC group; n=3.
3.3 MALAT1 induced B/TSMCs apoptosis and inflammation increase could be partially reversed by RyR2 siRNA
A previous study has proved that RyR2 was a typical risk gene for allergic asthma in children (R Giampaolo et al. 2011). Whether the effects of MALAT1 on B/TSMCs apoptosis and inflammatory factor secretion are associated with RyR2 is unknown and interesting. Through RT-qPCR and western blot, the level of RyR2 mRNA (figure 3A) and protein (figure 3B) was dramatically decreased in siMALAT1–3 group when compared to control and siMALAT1-NC (P<0.05), while markedly increased in pcDNA3.1(+)-MALAT1 group when compared to control and pcDNA3.1(+)-NC group (P<0.05). Data in figure 3C suggested that RyR2 was successfully silenced in siRyR2–1, siRyR2–2, and siRyR2–3 group (P<0.05), among which siRyR2–3 group exhibited the best silencing efficiency thereby was applied in following experiments. In addition, retest results showed that pcDNA3.1(+)-MALAT1 induced increase of cell apoptosis rate and inflammatory factor level (IL-6, TNF-α and IL-1β) was significantly inhibited in pcDNA3.1(+)-MALAT1+siRyR2–3 group (figure 3D–G, P<0.05). These results revealed that MALAT1 induced B/TSMCs apoptosis and inflammation increase could be partially reversed when RyR2 was silenced.
MALAT1 induced B/TSMCs apoptosis and inflammation increase could be partially reversed by RyR2 siRNA. For mechanism research, pcDNA3.1(+)-MALAT1 and RyR2 siRNA co-transfected B/TSMCs were prepared. (A) Relative RyR2 mRNA levels in B/TSMCs by RT-qPCR. (B) and (C): The levels of RyR2 protein in B/TSMCs by western blotting. (D) Cell apoptosis rate (%). (E) IL-6 levels in B/TSMCs. (F) TNF-α levels in B/TSMCs. (G) IL-1β levels in B/TSMCs. *P<0.05 vs. control or siMALAT1-NC group; #P<0.05 vs. control or pcDNA3.1(+)-NC group; &P<0.05 vs. control or siRyR2-NC group; ▽P<0.05 vs. pcDNA3.1(+)-MALAT1 group; n=3. For A, B and C, data in control group deemed as 1.
3.4 MALAT1 triggered miR-133a downregulation contributed to MALAT1’s effects on B/TSMCs apoptosis and inflammatory factor secretion
Through bioinformatics analysis by TargetScanHuman 7.2, it was predicted that miR-133a contains binding sequence to MALAT1 (figure. 4A). By using RT-qPCR, we found that the relative miR-133a level in B/TSMCs was significantly increased in siMALAT1–3 group than that in control and siMALAT1-NC group (figure 4B, P<0.05), meanwhile significantly decreased in pcDNA3.1(+)-MALAT1 group than that in pcDNA3.1(+)-NC and control group (figure 4B, P<0.05), which suggested that MALAT1 could negatively regulated miR-133a level in B/TSMCs. Besides, by introducing miR-133a-mimic, a marked increase of miR-133a level was observed in miR-133a-mimic group than that in control and miR-mimic-NC group (figure 4C, P<0.05), indicating a successful transfection. After re-testing cell apoptosis rate (figure 4D), IL-6 level (figure 4E), TNF-α level (figure 4F) and IL-1β level (figure 4G), we confirmed that MALAT1 overexpression induced increases of these indexs (P<0.05) also emerged partial reversions when exogenous miR-133a-mimic was introduced (P<0.05). These results indicated that MALAT1 triggered miR-133a downregulation contributed to MALAT1’s effects on B/TSMCs apoptosis and inflammatory factor secretion.
MALAT1 triggered miR-133a downregulation contributed to MALAT1’s effects on B/TSMCs apoptosis and inflammatory factor secretion. For further mechanism research, pcDNA3.1(+)-MALAT1 and miR-133a-mimc co-transfected B/TSMCs were prepared. (A) Matched sequence between MALAT1 and miR-133a by bioinformatics analysis. (B) and (C): Relative miR-133a levels in B/TSMCs by RT-qPCR. (D) Cell apoptosis rate (%). (E) IL-6 levels in B/TSMCs. (F) TNF-α levels in B/TSMCs. (G) IL-1β levels in B/TSMCs. *P<0.05 vs. control or siMALAT1-NC group; #P<0.05 vs. control or pcDNA3.1(+)-NC group; &P<0.05 vs. control or miR-mimic-NC group; ▽P<0.05 vs. pcDNA3.1(+)-MALAT1 group; n=3. For B and C, data in control group deemed as 1.
3.5 MiR-133a/RyR2 axis is a downstream signal of MALAT1 in B/TSMCs
Similarly, through bioinformatics analysis, the 3′UTR sequence of RYR2 was predicted to contain complementary sequence with miR-133a (figure 5B). Confirmed by experiments, in miR-133a-mimic treated B/TSMCs, RyR2 protein levels exhibited a significant decrease when compared to control and miR-mimic-NC group (figure 5A, P<0.05). At this point, we have reasons to speculate that MALAT1 may regulate miR-133a/RyR2 axis in B/TSMCs. Meanwhile, a luciferase reporter assay verified that RyR2 is a direct target gene of miR-133a (figure 5B). These results demonstrated that miR-133a/RyR2 axis is a downstream signal of MALAT1 in B/TSMCs. In summary, we conclude that MALAT1 knockdown could protects from bronchial/tracheal smooth muscle cell injury via regulating miR-133a/RyR2 axis.
MiR-133a/RyR2 axis is a downstream signal of MALAT1 in B/TSMCs. To confirm whether RyR2 is a target gene of miR-133a, luciferase gene reporter assay was performed. (A) The levels of RyR2 protein in B/TSMCs by western blot (miR-133a-mimic introduction). *P<0.05 vs. control or miR-mimic-NC group; data in control group deemed as 1; n=3. (B) Matched sequence between miR-133a and RyR2 and relative luciferase activity. *P<0.05 vs. corresponding miR-mimic-NC group; data in miR-mimic-NC group deemed as 1; n=3.
4 Discussion
Pediatric asthma belongs to a long course disease characterized by high incidence and repeated attacks (Shi et al. 2011). Understanding the pathogenesis of asthma and seeking new approaches to treat asthma possess significant values. In our study, the effects of MALAT1 on B/TSMCs apoptosis and inflammatory response were clarified. Herein, B/TSMC is a common cell line during asthma-related researches, and its dysfunction is a crucial factor for bronchial asthma (Trian et al. 2007). In addition, we found out the possible molecular mechanism for MALAT1 to exert effects in B/TSMCs by experiments with the help of bioinformatics predictions.
Accumulated evidences have suggested that MALAT1 plays important roles in alternative splicing, nuclear organization, and epigenetic modulating of gene expression (Tripathi et al. 2010). A study by Cardenas et al. found that MALAT1 participated in and mediated the function of vascular smooth muscle cells (VSMCs) in thoracic aortic aneurysm (Cardenas et al. 2018). Furthermore, MALAT1 knockdown could modulate proliferation, autophagy and migration of smooth muscle cell, thereby leading to cell cycle arrest in G2 phase (Song et al. 2017). In a constructed newborn asthma model rat, our study discovered that MALAT1 level was significantly increased in trachea tissues of asthma modeling rats. Furthermore, the introduction of exogenous MALAT1 could lead to the increase of apoptosis rate and inflammatory factor levels (IL-6, TNF-α, and IL-1β) in B/TSMCs, while opposite results could be observed when MALAT1 was silenced in B/TSMCs. These results hint that MALAT1 may involve in the injury and inflammatory responses in asthma relying on its effects on B/TSMCs.
MALAT1 has been proved to be a key lncRNA in regulating cellular inflammation. For instance, Zhao et al. found that MALAT1 could inhibit lipopolysaccharide (LPS)-induced inflammatory response in macrophages through its interaction with nuclear factor-κB (NF-κB) (Zhao et al. 2016). Contrarily, Puthanveetil et al. (2015) found that MALAT1 level was upregulated in high glucose-stimulated endothelial cells, which enhanced the expression of IL-6 and TNF-α by stimulating serum amyloid protein A3 (SAA3) to exert proinflammatory effect. It has also been reported that MALAT1 level was upregulated in LPS-stimulated cardiomyocyte leading to the increase of IL-6 and further promoting TNF-α secretion via regulating SAA3 (Zhuang et al. 2017). Hence, it’s worth noting that the proinflammatory or inhibitory inflammatory role of MALAT1 in complex inflammatory factor networks remains complicated and inconsistent, which may be associated with different tissue or cell type, or physiological processes. In our study, MALAT1 was identified to play a proinflammatory role in B/TSMCs.
The molecular mechanism of MALAT1 in regulating B/TSMCs apoptosis and inflammatory response was further clarified. A newly study by Shao et al. reported that miR-133a could alleviate airway remodeling in asthma through PI3K/AKT/mTOR signaling pathway by targeting IGF1R (Y Shao et al. 2019), indicating miR-133a may be benefit for the control of asthma. Previous studies have demonstrated that miR-133a implicates in cell apoptosis and inflammatory response in other cell types. For instance, Li et al. showed that the overexpression of miR-133a could inhibit ischemia-reperfusion-induced cardiomyocyte apoptosis by targeting Death Associated Protein Kinase 2 (Li et al. 2015a, b). Similarly, Li et al. found that miR-133a significantly suppressed the expression of apoptotic proteins caspase-8, caspase-9, and caspase-3, while improved the expression of Bcl-2 in cardiomyocytes (Li et al. 2015a, b). Additionally, it was reported that miR-133a increases caspase-1 p10 and IL-1β p17 cleavage activation through suppressing mitochondrial uncoupling protein 2 (Bandyopadhyay et al. 2013). Besides, Law et al. showed that expression dysregulation of miR-133α could regulate the proinflammatory signaling in human colonic epithelial cells and in experimental colitis (Ka et al. 2015). In our study, for further mechanism research, with the help of bioinformatics predictions, miR-133a was found to contain binding sequence to MALAT1. Through experiments, we confirmed that MALAT1 could negatively regulate miR-133a level in B/TSMCs. Meanwhile, the introduction of miR-133a mimic could partially reverse MALAT1-induced cell apoptosis and inflammatory factor secretion (IL-6, TNF-α, IL-1β) of B/TSMCs, also hinting the possible inhibitory role of miR-133a in asthma. These results suggested that miR-133a is a functional target of MALAT1 in B/TSMCs.
Ryanodine receptors (RyRs), intracellular calcium release channels essential for skeletal and cardiac muscle contraction, are also expressed in various types of smooth muscle cells (Du et al. 2005). It is well known that ryanodine receptor 2 (RyR2) functions as the major component of a calcium channel, and regulates mitochondrial metabolism, gene expression, cell survival, and so on (Bround et al. 2013). RyR2 has been identified to be associated with allergic asthma onset risk in an independent pediatric population (Giampaolo et al. 2011). Previously, it was confirmed that RyR2 can be a target gene of other miRNAs. For instance, miR-106b-25 cluster inhibited RyR2 by mediating post-transcriptional regulation of RyR2, which was a potential molecular mechanism involving in paroxysmal atrial fibrillation pathogenesis (Chiang et al. 2014). Besides, Belevych et al. found that miR-133a could modulate the function of RyR2 via directly target protein phosphatase 2A (PP2A) gene in heart failure (Belevych et al. 2011). The discoveries that inhibition of RyR2 by dantrolene (RyR antagonist) could inhibit compound K-induced endoplasmic reticulum (ER) stress-related apoptosis in human lung cancer cells (Shin et al. 2018) and alleviate skeletal muscle ischemia reperfusion injury by inhibiting TNF-α and inducing IL-10 (HP Lin et al. 2018), suggested a promoter role of RyR2 in apoptosis and inflammatory response. Consistent with the above findings, RyR2 was also identified to play a promoter in B/TSMCs at the aspects of apoptosis and inflammatory response. Furthermore, we found that MALAT1 induced B/TSMCs apoptosis and inflammation increase could be partially reversed when RyR2 was silenced or when miR-133a was overexpressed. Meanwhile, RyR2 was proved to be a direct target gene of miR-133a in B/TSMCs through luciferase reporter assay. Hence, we have basis to speculate that miR-133a/RyR2 axis could mediate the apoptosis and inflammatory response of B/TSMCs, thereby may take effects during the development of asthma.
Taken together, our study mainly identified the adverse effects of MALAT1 on B/TSMCs, a common cell line for asthma research. To be special, MALAT1 was found to facilitate B/TSMCs apoptosis and inflammatory response, which was later proved to be fulfilled partially by MALAT1’s regulation on miR-133a/RyR2 axis. Finally, we conclude that MALAT1 knockdown could protect from bronchial/tracheal smooth muscle cell injury via regulating miR-133a/RyR2 axis.
References
Ablimit A, Hasan B, Lu W, Qin W, Wushouer Q, et al. 2013 Changes in water channel aquaporin 1 and aquaporin 5 in the small airways and the alveoli in a rat asthma model. Micron 45 68–73
Ahmed B, Amer S, Zuzana B, Jian-Peng T, James W, et al. 2016 Crosstalk between long noncoding RNAs and MicroRNAs in health and disease. Int. J. Mol. Sci. 17 356
Andrew B 2008 How early do airway inflammation and remodeling occur? Allergol. Int. 57 11–19
Austin PJ, Tsitsiou E, Boardman C, Jones SW, Lindsay MA, et al. 2017 Transcriptional profiling identifies the long noncoding RNA plasmacytoma variant translocation (PVT1) as a novel regulator of the asthmatic phenotype in human airway smooth muscle. J. Allergy Clin. Immunol. Pract. 139 780–789
Bandyopadhyay S, Lane T, Venugopal R, Parthasarathy PT, Cho Y, et al. 2013 MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2. Biochem. Biophys. Res. Commun. 439 407–412
Belevych AE, Sansom SE, Radmila T, Hsiang-Ting H, Yoshinori N, et al. 2011 MicroRNA-1 and -133 Increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLoS One 6 e28324
Bround MJ, Wambolt R, Luciani DS, Kulpa JE, Rodrigues B, et al. 2013 Cardiomyocyte ATP production, metabolic flexibility, and survival require calcium flux through cardiac ryanodine receptors in vivo. J. Biol. Chem. 288 18975–18986
Cardenas CLL, Kessinger CW, Cheng Y, Macdonald C, MacGillivray T, et al. 2018 An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun. 9 1009
Chang C 2012 Asthma in children and adolescents: a comprehensive approach to diagnosis and management. Clin. Rev. Allergy Immunol. 43 98–137
Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, et al. 2006 The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38 228–233
Cheng Y, Imanirad P, Jutooru I, Hedrick E, Jin UH, et al. 2018a Role of metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) in pancreatic cancer. PLoS One 13 e0192264
Cheng Z, Wang X, Dai LL, Jia LQ, Jing XG, et al. 2018b Thymic stromal lymphopoietin signaling pathway inhibition attenuates airway inflammation and remodeling in rats with asthma. Cell Physiol. Biochem. 47 1482–1496
Chiang DY, Kongchan N, Beavers DL, Alsina KM, Voigt N, et al. 2014 Loss of MicroRNA-106b-25 Cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 expression and calcium release. Circ. Arrhythm. Electrophysiol. 7 1214–1222
Chiba Y and Misawa M 2010 MicroRNAs and their therapeutic potential for human diseases: MiR-133a and bronchial smooth muscle hyperresponsiveness in asthma. J. Pharmacol. Sci. 114 264–268
Dinakar C and Chipps BE 2017 Clinical tools to assess asthma control in children. Pediatrics 139 e20163438
Du WL, Stiber JA, Rosenberg PB, Meissner G and Eu JP 2005 Ryanodine receptors in muscarinic receptor-mediated bronchoconstriction. J. Biol. Chem. 280 26287–26294
Dutmer CM, Mcgraw MD and Liu AH 2016 Inner-city asthma: special considerations for management. Curr. Opin. Allergy Clin. Immunol. 16 148–156
Fang C, Lu W, Li C, Peng X, Wang Y, et al. 2016 MiR-3162–3p Is a novel MicroRNA that exacerbates asthma by regulating β-catenin. PLoS One 11 e0149257
Gagro A 2011 Asthma in children. Acta Med. Croatica. 65 169–179
Giampaolo R, Annalisa A, Daniel R, Francesca C, Serena F, et al. 2011 Pooled genome-wide analysis to identify novel risk loci for pediatric allergic asthma. PLoS One 6 e16912
Han YY, Forno E, Gogna M and Celedón JC 2016 Obesity and rhinitis in a nationwide study of children and adults in the United States. J. Allergy Clin. Immunol. 137 1460–1465
Hsu J, Qin X, Beavers SF and Mirabelli MC 2016 Asthma-related school absenteeism, morbidity, and modifiable factors. Am. J. Prev. Med. 51 23–32
Juan C, Wang Q, Mao Y, Cao Q, Li S, et al. 2018 Knockdown of LncRNA MALAT1 contributes to cell apoptosis via regulating NF-κB/CD80 axis in neonatal respiratory distress syndrome. Int. J. Biochem. Cell B 104 138–148
Ka MLI, Kyriaki B, Christos P and Angelos O 2015 Neurotensin-regulated miR-133a is involved in proinflammatory signalling in human colonic epithelial cells and in experimental colitis. Gut 64 1095–1104
Li AY, Yang Q and Yang K 2015a miR-133a mediates the hypoxia-induced apoptosis by inhibiting TAGLN2 expression in cardiac myocytes. Mol. Cell. Biochem. 400 173–181
Li H, Zhu H and Ge J 2016 Long Noncoding RNA: Recent updates in atherosclerosis. Int. J. Biol. Sci. 12 898–910
Li S, Sun Y, Zhong L, Xiao Z, Yang M, et al. 2018a The suppression of ox-LDL-induced inflammatory cytokine release and apoptosis of HCAECs by long noncoding RNA-MALAT1 via regulating microRNA-155/SOCS1 pathway. Nutr. Metab. Cardiovasc. Dis. 28 1175–1187
Li S, Xiao F, Shan P, Su L, Chen D, et al. 2015b Overexpression of microRNA-133a inhibits ischemia-reperfusion-induced cardiomyocyte apoptosis by targeting DAPK2. J. Hum. Genet. 60 709–716
Li X, Song Y, Liu F, Liu D, Miao H, et al. 2017 Long noncoding RNA MALAT1 promotes proliferation, angiogenesis, and immunosuppressive properties of mesenchymal stem cells by inducing VEGF and IDO. J. Cell Biochem. 118 2780–2791
Li Z, Zhang Q, Wu Y, Hu F, Gu L, et al. 2018b lncRNA Malat1 modulates the maturation process, cytokine secretion and apoptosis in airway epithelial cell-conditioned dendritic cells. Exp. Ther. Med. 16 3951–3958
Lin HP, Zheng YQ, Zhou ZP and Wang GX 2018 Ryanodine receptor antagonism alleviates skeletal muscle ischemia reperfusion injury by modulating TNF-α and IL-10. Clin. Hemorheol. Micro. 70 1–8
Lu TX and Rothenberg ME 2018 MicroRNA. J. Allergy Clin. Immunol. 141 1202–1207
Lu Y, Liu M, Shi S, Jiang H, Yang L, et al. 2010 Effects of stress in early life on immune functions in rats with asthma and the effects of music therapy. J. Asthma 47 526–531
Nicolas P, Maurin T, Chevalier B, Puisségur MP, Lebrigand K, et al. 2009 Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One 4 e6718
Omran A, Elimam D and Yin F 2013 MicroRNAs: new insights into chronic childhood diseases. Biomed. Res. Int. 2013 1–13
Orlandi E, Zanot C, Poli A, Nicolis M, Rodolfo M, et al. 2019 Lack of association of metastasis-associated lung adenocarcinoma transcript 1 variants with melanoma skin cancer risk. Melanoma Res. 29 660–663
Puthanveetil P, Chen S, Feng B, Gautam A and Chakrabarti S 2015 Long noncoding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 19 1418–1425
Shao Y, Chong L, Lin P, Li H, Zhu L, et al. 2019 MicroRNA-133a alleviates airway remodeling in asthtama through PI3K/AKT/mTOR signaling pathway by targeting IGF1R. J. Cell Physiol. 234 4068–4080
Shi YH, Shi GC, Wan HY, Jiang LH and Zhang BY 2011 Coexistence of Th1/Th2 and Th17/Treg imbalances in patients with allergic asthma. Chinese med j-peking 124 1951–1956
Shin DH, Leem DG, Shin JS, Kim JI, Kim KT, et al. 2018 Compound K induced apoptosis via endoplasmic reticulum Ca2+ release through ryanodine receptor in human lung cancer cells. J. Ginseng Res. 42 165–174
Simonson B and Das S 2015 MicroRNA Therapeutics: the next magic bullet? Med. Chem. 15 467–474
Song TF, Huang LW, Yuan Y, Wang HQ, He HP, et al. 2017 LncRNA MALAT1 regulates smooth muscle cell phenotype switch via activation of autophagy. Oncotarget 9 4411–4426
Sookoian S, Flichman D, Garaycoechea ME, San Martino J, Castao GO, et al. 2018 Metastasis-associated lung adenocarcinoma transcript 1 as a common molecular driver in the pathogenesis of nonalcoholic steatohepatitis and chronic immune-mediated liver damage. Hepatol. Commun. 2 654–665
Trian T, Benard G, Begueret H, Rossignol R, Girodet PO, et al. 2007 Bronchial smooth muscle remodeling involves calcium-dependent enhanced mitochondrial biogenesis in asthma. J. Exp. Med. 204 3173–3181
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, et al. 2010 The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39 925–938
Ye L, Mou Y, Wang J and Jin ML 2017 Effects of microRNA-19b on airway remodeling, airway inflammation and degree of oxidative stress by targeting TSLP through the Stat3 signaling pathway in a mouse model of asthma. Oncotarget 8 47533–47546
Zhang J, Zhu Y and Wang R 2018 Long noncoding RNAs in respiratory diseases. Histol. Histopathol. 33 747–756
Zhang XY, Zhang LX, Tian CJ, Tang XY, Zhao L, et al. 2016 LncRNAs BCYRN1 promoted the proliferation and migration of rat airway smooth muscle cells in asthma via upregulating the expression of transient receptor potential 1. Am. J. Transl. Res. 8 3409–3418
Zhang Y, Wang F, Chen G, He R and Yang L 2019 LncRNA MALAT1 promotes osteoarthritis by modulating miR-150–5p/AKT3 axis. Cell Biosci. 9 54
Zhao G, Su Z, Song D, Mao Y and Mao X 2016 The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 590 2884–2895
Zhu YJ, Mao D, Gao W and Hu H 2018 Peripheral whole blood lncRNA expression analysis in patients with eosinophilic asthma. Medicine 97 e9817
Zhuang YT, Xu DY, Wang GY, Sun JL, Huang Y, et al. 2017 IL-6 induced lncRNA MALAT1 enhances TNF-α expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur. Rev. Med. Pharmacol. Sci. 21 302–309
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor : Tapas Kumar Kundu
Rights and permissions
About this article
Cite this article
Yang, M., Wang, L. MALAT1 knockdown protects from bronchial/tracheal smooth muscle cell injury via regulation of microRNA-133a/ryanodine receptor 2 axis. J Biosci 46, 28 (2021). https://doi.org/10.1007/s12038-021-00149-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-021-00149-3