Abstract
Best practices from open data science are spreading across research fields, providing new opportunities for research and education. Open data science emphasizes the view that digitalization is enabling new forms of resource sharing, collaboration and outreach. This has the potential to improve the overall transparency and efficiency of research. Microbiome bioinformatics is a rapidly developing area that can greatly benefit from this progress. The concept of microbiome data science refers to the application of best practices from open data science to microbiome bioinformatics. The increasing availability of open data and new opportunities to collaborate online are greatly facilitating the development of this field. A microbiome data science ecosystem combines experimental research data with open data processing and analysis and reproducible tutorials that can also serve as an educational resource. Here, we provide an overview of the current status of microbiome data science from a community developer perspective and propose directions for future development of the field.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Analysis of molecular profiling data obtained from high-throughput “-omics” studies is essential for unravelling large-scale patterns in microbial community composition, function and interactions between microbial organisms. The development of bioinformatics tools has been pivotal for understanding the importance of microbiome in human health (Erickson et al. 2012; Heintz-Buschart et al. 2017; Schirmer et al. 2018). Numerous tools from command line interfaces such as Mothur (Schloss et al. 2009), DADA2 (Callahan et al. 2016b), Anvi’o (Eren et al. 2015) and the Python-based QIIME and QIIME2 (Caporaso et al. 2010; Bolyen et al. 2018) to web-based tools such as Calypso (Zakrzewski et al. 2016) and MicrobiomeAnalyst (Dhariwal et al. 2017) have been designed to serve microbial research community. The methods in this field are developing rapidly, however, and the quality of research software can vary widely (Mangul et al. 2018). Open source code does not as such guarantee quality or accuracy, and the research community can be slow to correct or improve implementations. Hence, suboptimal or even erroneous algorithms may be potentially used over long periods of time.
Open collaboration and joint development of data analytical methods can accelerate the dissemination and access to latest research algorithms. The emergence of open data science (McKiernan et al. 2016; Lahti 2018) has revolutionized such collaborative research and is greatly facilitating the development and adoption of open methods and practices in data-intensive research. The availability of distributed version control systems (Wilson et al. 2017) has created new opportunities to transparently benchmark and criticize alternative approaches. In microbiome bioinformatics, much of such development is currently focused on two computational programming environments, R and Python, where researchers are now routinely sharing research software and reproducible notebooks that summarize complete data analytical procedures from raw data to the final reporting. Graphical interfaces can further support researchers by providing interactive tools for data exploration and analysis (Venables and Smith 2006).
We provide a brief overview of the current status of microbiome data science in R from a community developer perspective. While the R ecosystem is one of the main platforms for current community-driven development efforts, the key concepts apply more widely to other data science environments.
2 Microbiome data science
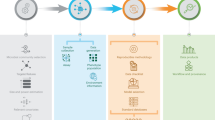
The route from the processing of raw data to final analysis and reporting relies on a vast number of methods and concepts in microbial ecology (figure 1), and an individual researcher is seldom able to master all relevant research areas. Hence, multi-disciplinary research can be greatly supported by well-designed workflows that implement best practices in the field and provide examples and guidance for choosing the methods, while maintaining the flexibility and opportunities to customize any part of the analysis workflow (Eren et al. 2015; McKiernan et al. 2016; Knight et al. 2018; Pollock et al. 2018; Schloss 2018). Research software can be efficiently communicated in the context of experimental benchmarking data and reproducible online tutorials that can be interactively tested and further modified by users. These so-called electronic notebooks have emerged to provide new educational resources as well as open collaboration platforms to facilitate methods criticism and development (see, e.g., Ragan-Kelley et al. 2013). Hence, key elements enabling microbiome data science include open data, open methods, and open collaboration (Lahti 2018).
Overview of the contemporary microbiome data science ecosystem in R. The shaded boxes highlights research areas where the demand for new analysis methods and tools is particularly topical. A microbiome data science ecosystem binds together research data and methods, and enables new forms of collaboration.
2.1 Data
Open availability of research data can improve the overall quality and trustworthiness of research. Moreover, convenient access to benchmarking data from published case studies can be valuable for verification, meta-analysis, and methods development. Importantly, the use of standard data formats such as phyloseq (McMurdie and Holmes 2013) can greatly facilitate the development and integration of new methods and the reproducibility of experiments, and lower the barrier for using analysis tools without expert knowledge on data processing and integration details. In microbiome research, typical data sets include counts of taxonomic units, genes, or metabolites, and complementary information on taxonomic classifications, phylogenies and nucleotide sequences. The algorithmic R packages can be complemented by so-called data packages, which can be distributed through Bioconductor, for instance. Data packages can have a larger size than the standard algorithm packages, and they provide well-documented example data sets that facilitate the development of methods, unit tests, and educational tutorials. Whereas R data packages have already a long history in bioinformatics, recently such data packages have started to emerge in the microbiome field also, providing data from recent microbiome studies at taxonomic and functional levels (see, e.g., Pasolli et al. 2017; Schiffer et al. 2019).
2.2 Analysis
R is well-suited for a variety of interactive analysis tasks and data handling operations and the contemporary R ecosystem covers dozens of packages for various aspects of microbiome data science (table 1). Most methods currently focus on 16S rRNA amplicon sequencing or assume that OTU tables are readily available from metagenomic sequencing studies. Data summarization is now facilitated by dedicated preprocessing algorithms such as DADA2 (Callahan et al. 2016b), and class structures such as phyloseq, which integrates OTU counts, taxonomic trees, and sample metadata into a single object that serves as a standardized starting point for downstream methods (McMurdie and Holmes 2013). Estimation of alpha diversity and related ecological indices including richness, evenness, dominance, and rarity, is provided by various packages (Oksanen et al. 2011; Lahti and Shetty 2017) and complemented by phylogenetic trees (Kembel et al. 2010) or co-occurrence networks (Willis and Martin 2018); the Shiny-phyloseq package provides further tools for interactive network analysis (McMurdie and Holmes 2015). Community dissimilarity, or beta diversities, can be analysed using both phylogenetic (Chen 2012) and non-phylogenetic metrics (Beals 1984). Many methods are available for differential abundance analysis (Robinson et al. 2010; Paulson et al. 2013; Fernandes et al. 2014; Love et al. 2014), and systematic benchmarking tests have indicated wide variation in the performance of alternative methods (Weiss et al. 2017). Advanced approaches consider nested hierarchies in multiple testing (Sankaran and Holmes 2014). Community-level differences between sample groups can be tested with PERMANOVA and other methods (Oksanen et al. 2011; Anderson and Walsh 2013) and further complemented by unsupervised analyses (Sankaran and Holmes 2018a; Singh et al. 2019) such as Dirichlet Multinomial Mixtures (DMMs) (Ding and Schloss 2014; Harris et al. 2014). Further tools are available for the analysis of phylogenetic trees (Paradis et al. 2004; Wright 2016; Stevens et al. 2017; Washburne et al. 2017), co-occurrence networks (Schwager et al. 2014; Kurtz et al. 2015), metabolic interactions (Cao et al. 2016), and microbiome function (Aßhauer et al. 2015). Visualization tools span from amplicon sequencing data (Andersen et al. 2018) to unsupervised ordination by incorporating phylogenetic structure (Fukuyama 2017) to network analysis (Csardi and Nepusz 2006), phylogenetic trees (Paradis et al. 2004), taxonomic diversity (Foster et al. 2017), and geospatial analysis (Charlop-Powers and Brady 2015). Many generic utilities for microbiome profiling data are also available (Chen et al. 2016; Korpela 2016; Lagkouvardos et al. 2017; Lahti and Shetty 2017). However, general-purpose package can be more challenging to maintain and develop in the long term compared to packages with a more specific scope. R packages have also been created to access taxonomic information (Chamberlain et al. 2014) and to support interoperability with other systems such as the Python-based QIIME and the Biological Observation Matrix (BIOM) format (Bittinger 2014; McMurdie and Paulson 2016). The MultiAssayExperiment provides utilities for parallel multi-omics profiling (Ramos et al. 2017), and further class structures are available for generic time series but these opportunities have not yet been fully exploited in the microbiome data science.
R packages in microbiome data science are mainly distributed through four channels, which have varying levels of software review. Github is a generic open source development platform that does not pose any formal review requirements for new R packages; CRAN has strict technical checks for package consistency, and rOpenSci (Boettiger et al. 2015) and Bioconductor (Gentleman et al. 2004) have implemented comprehensive human-curated software review procedures that can improve the overall quality of the research software, including source code, compliance to standards, and documentation.
2.3 Workflows and tutorials
Sharing of technical knowledge and best practices can be greatly facilitated by open online resources (table 1) (Callahan et al. 2016a; Schloss 2018). Open practices facilitate community-driven development work on methods and algorithms, thus facilitating free and open knowledge sharing and helping to democratize microbiome data science by limiting monopolies of power. Some good practices in microbiome data science workflows include routine application of automated unit tests and crowd-sourced quality control in the form of issue reports and case studies on reproducible notebooks (Ram 2013; Wilson et al. 2017). Various examples of such case studies have been made openly available through common software repositories (see e.g. Baxter et al. 2016; Proctor et al. 2018). Availability of source code can greatly facilitate the reproducibility, verification and further use of the algorithms and has the potential to increase the overall efficiency and impact of research.
3 Discussion
Microbiome data science facilitates collaborative development of algorithms and methods. In the collaborative development model, independent research groups contribute to the same methods base for instance through shared version control systems. This has facilitated access to various algorithmic methods in microbial ecology. Whereas we have provided a brief overview of the current microbiome data science ecosystem in R, many complementary methods are available in Python and other environments. Recent developments towards integrating R and Python, two widely used data science programming environments have been remarkable and new packages such as reticulate have emerged to allow fluent exchange of information between Python from R (Allaire et al. 2018). The subsequent ability to perform open data analysis in a single environment can greatly support the trustworthiness, reproducibility and reusability of research outcomes. Most currently available R packages are heavily focused on 16S rRNA gene analysis, and often contain overlapping functionality whose performance has not yet been comprehensively benchmarked. At the same time, the demand is now specifically increasing for methods that could facilitate the analysis and integration of deep metagenomic and multi-omics profiling data and multivariate time series, both in the context of targeted case studies as well as large population cohorts.
References
Allaire J, Ushey K, Tang Y, Eddelbuettel D, Lewis B and Geelnard M 2018 reticulate: interface to ‘Python’. R package version 1
Andersen K, Kirkegaard R, Karst S and Mads A 2018 ampvis2 an R package to analyse and visualise 16S rRNA amplicon data bioRxiv 299537
Anderson MJ and Walsh DCI 2013 PERMANOVA, ANOSIM and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 83 557–574
Aßhauer KP, Wemheuer B, Daniel R and Meinicke P 2015 Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31 2882–2884
Baxter NT, Ruffin MT, Rogers MA and Schloss PD 2016 Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 8 37
Beals EW 1984 Bray-Curtis Ordination: An Effective Strategy for Analysis of Multivariate Ecological Data. In: Advances in Ecological Research. MacFadyen A and Ford ED (eds) (Academic Press) pp. 1–55
Bittinger K 2014 qiimer: Work with QIIME output files in R R package version 0.9.2
Boettiger C, Chamberlain S, Hart E and Ram K 2015 Building software, building community: lessons from the rOpenSci project. J. Open Res. Software 3(1) e8
Bolyen E, Rideout J, Dillon M, Bokulich N, Abnet C, Al-Ghalith GA et al. 2018 QIIME 2 Reproducible, interactive, scalable and extensible microbiome data science. PeerJ Preprints 6 e27295v1
Bravo H, Chelaru F, Wagner J, Kancherla J and Paulson J 2017 metavizr: R Interface to the metaviz web app for interactive metagenomics data analysis and visualization. Version 1.8.0. R/Bioconductor package. https://doi.org/10.18129/B9.bioc.metavizr
Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ and Holmes SP 2016a Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Res 5 1492
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA and Holmes SP 2016b DADA2 high-resolution sample inference from Illumina amplicon data. Nat. Methods 13 581
Cao Y, Wang Y, Zheng X, Li F and Bo X 2016 RevEcoR: an R package for the reverse ecology analysis of microbiomes. BMC Bioinformatics 17 294
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. 2010 QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335
Chamberlain S, Szocs E, Boettiger C, Ram K, Bartomeus I, Baumgartner J et al. 2014 taxize: Taxonomic information from around the web. R Package Version 30
Charlop-Powers Z and Brady SF 2015 phylogeo: an R package for geographic analysis and visualization of microbiome data. Bioinformatics 31 2909–2911
Chen J 2012 GUniFrac: generalized UniFrac distances. R package version 1 2012
Chen W, Simpson J and Levesque C 2016 RAM: R for amplicon-sequencing-based microbial-ecology. R Package Version 1
Csardi G and Nepusz T 2006 The igraph software package for complex network research. Int. J. Complex Syst. 1695 1–9
Dhariwal A, Chong J, Habib S, King IL, Agellon LB and Xia J 2017 MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45(W1), W180–W188
Ding T and Schloss PD 2014 Dynamics and associations of microbial community types across the human body. Nature 509 357
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML and Delmont TO 2015 Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3 e1319
Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C et al. 2012 Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PloS One 7 e49138
Faust K, Bauchinger F, Laroche B, de Buyl S, Lahti L, Washburne AD et al. 2018 Signatures of ecological processes in microbial community time series. Microbiome 6 120
Fernandes AD, Reid JN, Macklaim JM, McMurrough TA, Edgell DR and Gloor GB 2014 Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2 15
Foster ZS, Sharpton TJ and Grünwald NJ 2017 Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS computational biology 13 e1005404
Fukuyama J 2017 Adaptive gPCA: A method for structured dimensionality reduction arXiv [statME]
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. 2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 R80
Hahsler M and Nagar A 2014 rRDP: Interface to the RDP Classifier. R/Bioconductor package. https://doi.org/10.18129/B9.bioc.rRDP
Harris K, Parsons TL, Ijaz UZ, Lahti L, Holmes I and Quince C 2017 Linking statistical and ecological theory: Hubbell’s unified neutral theory of biodiversity as a hierarchical Dirichlet process. Proc. IEEE 105(3) 516–529
Heintz-Buschart A, May P, Laczny CC, Lebrun LA, Bellora C, Krishna A et al. 2017 Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nature Microbiol. 2 16180
Jeganathan P and Holmes S 2018 bootLong: The block bootstrap method for longitudinal count data arXiv:1809.01832
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. 2010 Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26 1463–1464
Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. 2018 Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16 410–422
Korpela K 2016 mare: Microbiota Analysis in R Easily. R Package Version 1.0
Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ and Bonneau RA 2015 Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11 e1004226
Lagkouvardos I, Fischer S, Kumar N and Clavel T 2017 Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 5 e2836
Lahti L 2018 Open data science; in Advances in Intelligent Data Analysis XVII Lecture Notes in Computer Science 11191 Leo Lahti (ed) (Springer Nature India)
Lahti L and Shetty SA 2017 Tools for microbiome analysis in R. Microbiome package version 1.7.21. R/Bioconductor http://microbiome.github.com/microbiome
Le Cao K-A, Costello M-E, Lakis VA, Bartolo F, Chua X-Y, Brazeilles R and Rondeau P 2016 MixMC: a multivariate statistical framework to gain insight into microbial communities. PloS One 11 e0160169
Liland KH, Vinje H and Snipen L 2017 microclass: an R-package for 16S taxonomy classification. BMC Bioinformatics 18 172
Love M, Sanders S and Huber W 2014 Differential analysis of count data–the DESeq2 package. Genome Biol. 15 10–1186
Mangul S, Mosqueiro T, Duong D, Mitchell K, Sarwal V, Hill B, et al. 2018 A comprehensive analysis of the usability and archival stability of omics computational tools and resources. BioRxiv 452532
Manimaran S, Bendall M, Diaz S, Castro E, Faits T, Zhao Y and Johnson W 2018 PathoStat: PathoStat statistical microbiome analysis package. R Package Version 1.6.1
McKiernan EC, Bourne PE, Brown CT, Buck S, Kenall A, Lin J, et al. 2016 Point of view: How open science helps researchers succeed. Elife 5 e16800
McMurdie P and Holmes S 2013 phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 e61217
McMurdie P and Holmes S 2015 Shiny-phyloseq: Web application for interactive microbiome analysis with provenance tracking. Bioinformatics 31 282–283
McMurdie P and Paulson J 2016 biom format: An interface package for the BIOM file format. Package Version 10
Morgan M 2017 DirichletMultinomial R package. Published on-line (Bioconductor)
Murali A, Bhargava A and Wright ES 2018 IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6 140
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB et al. 2011 vegan: Community ecology package. R Package Version 117–118
Paradis E, Claude J and Strimmer K 2004 APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20 289–290
Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT et al. 2017 Accessible, curated metagenomic data through ExperimentHub. Nat. Methods 14 1023
Paulson JN, Pop M and Bravo HC 2013 metagenomeSeq: Statistical analysis for sparse high-throughput sequencing. Published on-line (Bioconductor)
Piccolo BD, Wankhade UD, Chintapalli SV, Bhattacharyya S, Chunqiao L and Shankar K 2018 Dynamic assessment of microbial ecology (DAME): a web app for interactive analysis and visualization of microbial sequencing data. Bioinformatics 34 1050–1052
Pollock J, Glendinning L, Wisedchanwet T and Watson M 2018 The madness of microbiome: attempting to find consensus “Best Practice” for 16S microbiome studies. Appl. Environ. Microbiol. 84 e02627–17
Price J, Woloszynek S, Rosen G and CM, S 2018 theseus - An R package for the analysis and visualization of microbial community data BioRxiv 295675
Proctor DM, Fukuyama JA, Loomer PM, Armitage GC, Lee SA, Davis NM, et al. 2018 A spatial gradient of bacterial diversity in the human oral cavity shaped by salivary flow. Nat. Commun. 9 681
Ragan-Kelley B, Walters WA, McDonald D, Riley J, Granger BE, Gonzalez A, Knight R, Perez F and Caporaso JG 2013. Collaborative cloud-enabled tools allow rapid, reproducible biological insights. ISME J. 7 461
Ram K 2013 Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med. 8 7
Ramos M, Schiffer L, Re A, Azhar R, Basunia A, Rodriguez C, et al. 2017 Software for the Integration of Multiomics Experiments in Bioconductor. Cancer Res. 77 e39–e42
Roberts DW 2007 labdsv: Ordination and multivariate analysis for ecology. R Package Version 1
Robinson MD, McCarthy DJ and Smyth GK 2010 edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140
Rohart F, Gautier B, Singh A and Lê Cao K-A 2017 mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13 e1005752
Sankaran K and Holmes S 2014 structSSI: simultaneous and selective inference for grouped or hierarchically structured data. J. Stat. Software 59 1
Sankaran K and Holmes S 2018a Latent variable modeling for the microbiome. Biostatistics kxy018
Sankaran K and Holmes S 2018b Interactive visualization of hierarchically structured data. J. Comput. Graph Stat. 27 553–563
Schiffer L, Azhar R, Shepherd L, Ramos M, Geistlinger L, Huttenhower C, Dowd JB, Segata N and Waldron L 2019 HMP16SData: efficient access to the human microbiome project through Bioconductor. Am. J. Epidemiol. 188(6) 1023–1026
Schirmer M, Franzosa EA, Lloyd-Price J, McIver LJ, Schwager R, Poon TW, et al. 2018 Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 3 337–346
Schloss PD 2018 The Riffomonas reproducible research tutorial series. Int. J. Occup. Saf. Ergon. 1 13
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. 2009 Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541
Schwager E, Weingart G, Bielski C and Huttenhower C 2014 CCREPE: compositionality corrected by permutation and renormalization R/Bioconductor
Singh A, Shannon CP, Gautier B, Rohart F, Vacher M, Tebbutt SJ and Lê Cao KA 2019 DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. bty1054, https://doi.org/10.1093/bioinformatics/bty1054
Stevens JR, Jones TR, Lefevre M, Ganesan B and Weimer BC 2017 SigTree: a microbial community analysis tool to identify and visualize significantly responsive branches in a phylogenetic tree. Comput. Struct. Biotechnol. J. 15 372–378
Venables WN and Smith DM 2006 An Introduction to R (R Foundation for Statistical Computing, Vienna, Austria)
Washburne AD, Silverman JD, Leff JW, Bennett DJ, Darcy JL, Mukherjee S et al. 2017 Phylogenetic factorization of compositional data yields lineage-level associations in microbiome datasets. PeerJ 5 e2969
Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A et al. 2017 Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5 27
Willis A and Bunge J 2016 Breakaway R package (CRAN)
Willis AD and Martin BD 2018 DivNet: Estimating diversity in networked communities. BioRxiv 305045
Wilson G, Bryan J, Cranston K, Kitzes J, Nederbragt L and Teal TK 2017 Good enough practices in scientific computing. PLoS Comput Biol 13 e1005510
Wright ES 2016 Using DECIPHER v2. 0 to analyze big biological sequence data in R. R Journal 8(1) 352–359
Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion M-J, Berger B and Krause L 2016 Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics 33 782–783
Acknowledgements
LL was funded by the Academy of Finland (Grants 295741; 307127). Part of this research was by the SIAM Gravitation Grant 024.002.002 and UNLOCK project of the Netherlands Organization for Scientific Research (NWO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shetty, S.A., Lahti, L. Microbiome data science. J Biosci 44, 115 (2019). https://doi.org/10.1007/s12038-019-9930-2
Published:
DOI: https://doi.org/10.1007/s12038-019-9930-2