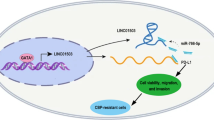
Abstract
Cervical cancer (CC) is one of the most common female malignancies in the world. Although paclitaxel (PTX) is a critical chemotherapy agent for the treatment of CC, its treatment outcome is limited by the development of drug resistance. The present study aims to define the role of long non-coding RNA (lncRNA) LINC00511 in the progression of CC with the involvement of cell proliferation, apoptosis and resistance to PTX in Hela/PTX cells. CC and adjacent normal tissue samples were collected from 84 patients with CC, and used to determine LINC0051 expression. PTX-resistant Hela/PTX cell line was constructed, in which LINC0051 was overexpressed or silenced to further investigate the effect of LINC00511 on PTX-resistant Hela/PTX cell viability, proliferation, migration, invasion, cell cycle, apoptosis and resistance of CC cells to PTX. The expression of Bcl-2, Bax, cleaved-caspase-3, matrix metalloproteinase (MMP)-2, MMP-9, multidrug resistance protein 1 (MRP1) and P-glycoprotein (P-GP) was also assessed. High-expression of LINC00511 was found in CC with its close association with the tumor stage, tumor size and lymph node metastasis. After silencing LINC00511 expression, the expression of MRP1, P-GP, Bcl-2, MMP-2 and MMP-9 was decreased, while Bax and cleaved-caspase-3 increased with more CC cells arrested at G1 phase. Furthermore, silencing of LINC00511 could suppress cell resistance to PTX, cell viability, cell proliferation, migration and invasion yet promoted cell apoptosis in PTX-resistant Hela/PTX cells. Collectively, our findings demonstrate that silencing of LINC00511 could inhibit CC cell resistance to PTX, cell proliferation, migration and invasion, and promote cell apoptosis in CC. Silencing of LINC00511 provides a novel therapeutic target for CC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cervical cancer (CC) ranks the fourth in aggressive cancers affecting females, showing a high rate of prevalence and mortality around the world (Dong et al. 2018). The crucial risk factor is human papillomavirus (HPVs) in the process of CC pathogenesis (Rodriguez et al. 2012). Patients with early-stage CC are possible to be cured, while about 67% of patients are diagnosed at advanced stage (Wei et al. 2017). In recent years, a more satisfying outcome has been revealed in patients with CC, still there are many patients who died of metastasis due to the limited and ineffective prognosis (Liu et al. 2017). A large number of promising therapeutic methods have been applied for CC treatment such as combined therapy of surgery, chemotherapy and radiotherapy, but metastatic recurrence resulted from the migrated tumor cells is a great obstacle for prognosis of CC (Zhang et al. 2016). Nowadays, anti-tumor agent paclitaxel (PTX) has been used for resistant or recurrent metastatic CC, while the resistance to PTX greatly influences its clinical effect (He et al. 2018). Therefore, an increased understanding of the molecular mechanisms of CC progression and metastasis is essential for improving clinical treatment strategies and suppressing resistance to PTX.
The association between long non-coding RNAs (lncRNAs) and chemosensitivity to PTX has been revealed in endometrial cancer, lung adenocarcinoma and advanced ovarian cancer (Li et al. 2017; Ren et al. 2017; Wang et al. 2017). LncRNAs were previously indicated to play significant regulatory roles in cancer biology, which were defined as RNAs that were longer than 200 nucleotides and were not translated into protein (Spizzo et al. 2012; Shi et al. 2015). Some lncRNAs are involved in a variety of biological processes and human diseases, and the aberrant expression of certain lncRNAs was reported to be linked with carcinogenesis (Zhang et al. 2015; Chang et al. 2016; Zhang et al. 2017). Specifically, a previous study revealed that silencing of lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) could suppress tumor growth, induce cell apoptosis, as well as regulation of radiosensitivity through interaction with microRNA-145 in CC (Lu et al. 2016). Another study demonstrated that silencing of lncRNA colon cancer-associated transcript 2 (CCAT2) exerted an inhibitory effect on CC cell proliferation and contributory effect on cell apoptosis (Wu et al. 2016). Long intergenic non-protein coding RNA 511 (LINC00511) was found to be up-regulated in pancreatic ductal adenocarcinoma (PDAC), and LINC00511 depletion is capable of preventing PDAC cells from proliferation, migration and invasion (Zhao et al. 2018). Moreover, LINC00511 was also upregulated in tongue squamous cell carcinoma (TSCC), and the knockdown of LINC00511 can inhibit TSCC cell proliferation, invasion and cell cycle via sponging microRNA-765 (Ding et al. 2018). Therefore, we made a hypothesis that LINC0051 may affect the cell proliferation, migration, invasion and apoptosis, and resistance to PTX in CC.
2 Materials and methods
2.1 Ethics statement
All patients in the present study signed informed consents. The study was approved by the ethics committee of Linyi Cancer Hospital.
2.2 Study subjects
A total of 84 CC patients aged from 30 to 70 years with the mean age of 54.57 ± 8.36 years, who received surgical resection from September 2010 to September 2012 in the Department of Obstetrics and Gynecology, Linyi Cancer Hospital (Shandong, China), were enrolled in this study. Those pregnant and breastfeeding patients as well as the patients that were combined with other malignant tumors were excluded from the study. There were 73 cases with the tumor diameter ≤4 cm and 11 cases >4 cm. The tumor staging was classified according to the 2009 International Federation of Gynecology and Obstetrics (FIGO) staging system. We observed 29 cases at Ia stage, 31 cases at Ib, 17 cases at IIa and 7 cases at IIb. Besides, 21 cases showed poor differentiation and 63 cases moderate and high differentiation. Lymph node metastasis (LNM) was found in 5 cases, and 79 cases did not show LNM. The CC tissues and adjacent normal tissues (>2 cm from the margin of the tumor) were collected during surgery. The surgical specimens were immediately stored in liquid nitrogen. All specimens were diagnosed by pathological examination. All the patients had not been treated with any radiotherapy or chemotherapy prior to surgical resection.
2.3 Follow-up
We performed a 60-month follow-up for these patients and the end of follow-up was September 2017. During the follow-up period, the recurrence of tumor or death of the patient was regarded as the end of follow-up. If the above events did not occur, the final follow-up time was regarded as the end of follow-up. The interval between the surgery date and the date of death of patients was defined as the overall survival (OS). The interval between the operative date and the earliest evidence of recurrence was defined as relapse free survival (RFS).
2.4 Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted by Trizol (15596026, Invitrogen, Carlsbad, CA, USA) and the integrity of RNA was identified by 1% agarose gel electrophoresis. The concentration and purity of RNA were determined by Nano-Drop ND-1000 spectrophotometer. According to the instructions of the PrimeScript RT reagent Kit (RR047A, Takara, Tokyo, Japan), the RNA was reversely transcribed into cDNA under the following conditions with a reaction system of 20 μL: 37°C for 15 min and 85°C for 5 s. Primers of LINC00511, multidrug resistance protein 1 (MRP1), matrix metalloproteinase (MMP)-2 and MMP-9, P-glycoprotein (P-GP), B-cell lymphoma/leukemia-2 (Bcl-2), Bcl-2-associated X protein (Bax) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed and synthesized by Sangon Biotech (Shanghai, China) (table 1). Reverse transcription with 20 μL system was conducted according to the EasyScript First-Strand cDNA Synthesis SuperMix instructions (AE301-02, TransGen Biotech Co., Ltd., Beijing, China). Subsequently, the reaction solution was subjected to RT-qPCR according to the instructions of SYBR® Premix Ex TaqTM II Kit (TaKaRa Biotechnology, Dalian, Liaoning, China). A total of 20 μL reaction system: consisted of 10 μL SYBR Premix, 2 μL cDNA template, 0.6 μL forward primers, 0.6 μL reverse primers and 6.8 μL diethyl pyrocarbonate (DEPC). RT-qPCR was performed using Applied Biosystems (ABI Company, Oyster Bay, NY, USA) 7500 RT-qPCR. The reaction conditions consisted of pre-denaturation at 95°C for 30 s, denaturation at 95°C for 30 s, annealing at 50°C for 20 s, extension at 72°C for 30 s, with a total of 40 cycles. 2−ΔΔCt represented the ratio of the target gene expression between the experimental group and the control group. The formula was ΔCt = Cttarget genes – CtGADPH, ΔΔCT = ΔCtexperimental group – ΔCtcontrol group. Ct referred to the amplification cycles when the real-time fluorescence intensity reached the set threshold value and the amplification entered a logarithmic growth (Arocho et al. 2006). The experiment was repeated independently three times. The expression of LINC00511, MRP1, MMP-2, MMP-9, P-GP, Bcl-2, Bax and GADPH in cells was then calculated.
2.5 Western blot analysis
CC cells or tissues were collated and treated with radioimmunoprecipitation assay (RIPA) lysate (P0013B, Shanghai Beyotime Biotechnology Co. Ltd., Shanghai, China) with moderate phenylmethylsulfonyl fluoride (PMSF) and phosphatase inhibitors, then re-suspended, further incubated on ice for 30 min and centrifuged at 4°C at 28,984 g for 10 min. The collected supernatant was the total protein of tissues or cells, and the concentration was subsequently determined using the bicinchoninic acid (BCA) kit (Shanghai Beyotime Biotechnology Co. Ltd., Shanghai, China). In the next step, the concentration was adjusted to 4 μg/μL using phosphate buffer saline (PBS). Then 30 μg of cell total protein was taken out and run using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose (NC) membrane by the wet method. The membrane was blocked with 5% skimmed milk powder for 1.5 h, which was prepared with Tris-buffered saline with Tween 20 (TBST). Then the NC membrane was incubated in a plastic dish overnight at 4°C with the addition of following primary antibodies, including MRP1 (ab180960, 1:1000), MMP-2 (ab37150, 1:2000), MMP-9 (ab73734, 1:1000), P-GP (ab103477, 1:500), Bcl-2 (ab59348, 1:1000, Bax (ab32503, 1:1000), cleaved-caspase-3 (ab2302, 1:1000) and GAPDH (ab9485, 1:2500). All the above antibodies were purchased from Abcam (Cambridge, MA, USA). The next day, the membrane was washed three times with TBST (15 min each) and then incubated with horseradish peroxidase (HRP)-labeled immunoglobulin G (IgG) goat anti-rabbit (ab205718, 1 : 2000 – 1:50,000) for 2 h. After three times TBST washing (15 min each), the membrane was colored with enhanced chemiluminescence (ECL) solution (34580, Thermo Fisher Scientific Inc., Waltham, MA, USA) and images were captured by SmartView Pro 2000 (UVCI-2100, Major Science, Saratoga, CA, USA). The Quantity One software was applied to analyze the gray value of protein bands (Alexopoulou et al. 2010).
2.6 Construction of PTX-resistant Hela/PTX cell line
CC cell line HeLa was donated by the Shandong Laboratory Animal Center (Jinan, Shandong, China). After revival, HeLa cell line was cultured using RPMI 1640 medium containing 10% fetal bovine serum (FBS, Gibco, Mul-grave, Victoria, Australia), 100 U/mL penicillin, and 100 mg/mL streptomycin in an incubator (Invitrogen, Carlsbad, CA, USA) with saturated humidity of 5% CO2 at 37°C. Subculture was performed when cells reached 90% confluence. With the medium removed, cells were washed two times with PBS, and treated with 0.25% trypsin until the cells became round and a gap appeared. An appropriate amount of FBS was added to stop the digestion, and the single cell suspension was made with a pipette. HeLa cells in logarithmic growth phase were incubated with PTX for 24 h, and then cultured and passaged in RPMI 1640 complete medium (without PTX). The aforementioned procedures were repeated, in which the concentration of PTX increased from 10 to 20 μg/L, 40 g/L and 500 μg/L. After continuous culture of HeLa cells for 10 months, PTX-resistant Hela/PTX cell line could be collected which maintained a good growth state in 500 μg/L PTX. The PTX-resistant Hela/PTX cell line was cryopreserved, and cell recovery occurred after 2 months.
2.7 Cell culture, grouping and transfection
PTX-resistant Hela/PTX cells were assigned into blank, sh-LINC00511 (transfected with LINC00511 knockdown plasmids), sh-LINC00511 NC (transfected with sh-LINC00511 negative control plasmids), LINC00511 (transfected with LINC00511 overexpression plasmids) and LINC00511 NC (transfected with LINC00511 negative control plasmids) groups. The above groups were lately treated with PTX at a concentration of 200 μg/L, and further allocated into blank + PTX group (treated with PTX), sh-LINC00511 + PTX group (transfected with LINC00511 knockdown plasmids and treated with PTX), sh-LINC00511 NC + PTX group (transfected with LINC00511 knockdown negative control plasmids and treated with PTX), LINC00511 + PTX group (transfected with LINC00511 overexpression plasmids and treated with PTX) and LINC00511 NC + PTX group (transfected with LINC00511 negative control plasmid and treated with PTX). The aforementioned plasmids were all purchased from Dharmacon Inc. (Lafayette, CO, USA). The HeLa/PTX cells transfected with LINC00511 knockdown plasmids were seeded in six-well plates at a density of 3 × 105 cells/well, and further cultured in fresh complete medium. Upon reaching 50–80% confluence, cells were transfected using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) kit. In specific terms, 4 μg of the target plasmids and 10 μL of Lipofectamine 2000 were diluted with 250 μL of serum-free Opti-MEM (Gibco, Mul-grave, Victoria, Australia) medium, respectively, and then gently mixed. The mixture was well mixed after placing it for 5 min at room temperature, and then added into six-well plates after placing it further for 20 min. The mixture was cultured in a 5% CO2 incubator at 37°C for 6 h and then the medium was changed into the complete medium. Finally, cells were cultured and collected after 48 h.
2.8 Cell counting kit (CCK-8) assay
2.8.1 Detection of 50% inhibition concentration (IC50)
PTX-resistant Hela/PTX cell line and HeLa cell line were seeded in 96-well plates at a density of 5 × 103 cells/well, and 200 μL of cell suspension was supplemented into each well. After 24-h culture, cells were treated with RPMI 1640 medium containing PTX with a final concentration of 5, 10, 50, 100, 500, 1000, 5000 and 10,000 μg/L, respectively, with three replicates in each well. After culturing for 24, 48 and 72 h, 10 μL of CCK-8 solution was added into each well (CK04, Dojindo Laboratories, Kumamoto, Japan) and mixed evenly. Next, cells were further cultured in a 5% CO2 incubator at 37°C for 2 h. The optical density (OD) of each well was measured at 450 nm by using an automatic microplate reader ((Multiskan MK3, Thermo, Waltham, MA, USA). The IC50 was calculated using SPSS 22.0 (IBM Corp. Armonk, NY, USA) for regression analysis (Chen et al. 2012).
2.8.2 Detection of cell viability
After transfection for 48 h, HeLa/PTX cells were washed two times with PBS, detached with 0.25% trypsin and centrifuged at 201 g for 5 min. The single cell suspension was made with a pipette. HeLa/PTX cells were then seeded in 96-well plates at a density of 2.5 × 103 cells/well, and cultured in a 5% CO2 incubator at 37°C. Each well had three replicates. After culturing for 24, 48 and 72 h, 10 μL of CCK-8 solution was added into each well (CK04, Dojindo Laboratories, Kumamoto, Japan) and mixed evenly. Next, cells were further cultured in 5% CO2 incubator at 37°C for 2 h. The optical density (OD) of each well was measured at 450 nm by using an automatic microplate reader (Multiskan MK3, Thermo, Waltham, MA, USA). Each experiment was repeated three times, and the cell viability curve was drawn with OD value of each group as ordinate and time as abscissa.
2.9 5-Ethynyl2′-deoxyuridine (EdU) assay
Cells of each group were collected and seeded in 96-well plates at a density of 1.6 × 105 cells/well for 48-h culture. Then each well was added with 50 mM EdU solution and incubated at 37°C for 4 h. After that, cells were fixed with 4% formaldehyde for 15 min at room temperature, treated with 0.5% Triton X-100 for 20 min at room temperature for permeabilization. After washing three times with PBS, 100 mL of Apollo® mixture was added to each well and incubated for 30 min at room temperature. Subsequently, cells were stained with 100 mL of Hoechst 33,342 for 30 min and photographed under a fluorescence microscope (Olympus, Tokyo, Japan). The number of EdU positive staining cells (cells stained into the red) was calculated using Image-Pro Plus (IPP) 6.0 software (Media Cybernetics, Bethesda, MD, USA). The rate of EdU incorporation was expressed as: the percentage of EdU positive staining cells/sh-LINC00511-HeLa/PTX-positive cells (cells stained into blue). The experiment was repeated three times with the mean value calculated (Guo et al. 2011).
2.10 Annexin V- fluoresceinisothiocyanate (FITC)/propidium (PI) double staining
The transfected cells in each group were collected, detached with 0.25% trypsin and made into the single cell suspension. Subsequently, cells were washed two times with PBS and centrifuged with the supernatant discarded. Then cells were fixed with pre-cooled 70% ethanol overnight at 4°C, resuspended and washed two times with pre-cooled PBS after centrifugation. Lately, cells were resuspended in 100 μL of PBS, and a final concentration of cells was adjusted to 1 mg/mL with the addition of RNase. The cells were treated with a water bath at 37°C for 30 min. The PI staining solution was added to adjust cell suspension to a final concentration of 50 μg/mL, after which cells were stained in the dark for 40 min at 4°C followed by PBS washing. Finally, the DNA content at different cell cycles was measured at >575 nm to calculate the percentage of the cell cycle.
2.11 TdT-mediated dUTP-biotin nick end-labeling (TUNEL) staining
Cells in each group were used to perform TUNEL staining according to the Click-iT TUNEL Alexa Fluor Imaging Assay Protocol (C10245, Thermo Fisher Scientific Inc., Waltham, MA, USA). Cell apoptosis was observed under an optical microscope and the images were captured. Apoptotic cells referred to those positive staining cells whose nucleus was stained into green. Ten high-magnification fields in each section were randomly collected and observed. The number of apoptotic cells in each field was counted. Apoptosis rate was expressed as: percentage of apoptotic cells/total number of cells and the mean value was regarded as an apoptotic index (AI).
2.12 Transwell assay
Matrigel gel (356234, BD Biosciences, Franklin Lakes, NJ, USA) stored at -80°C was melted into a liquid at 4°C overnight. At 4°C, 200 μL Matrigel gel was added to 200 μL serum-free medium and fully mixed and diluted. Then, 50 μL of diluted Matrigel was added into the chamber of each Transwell plate and incubated in an incubator for 2–3 h until the gel became solid. The cells were detached and counted, and made into cell suspension with medium containing 20% FBS. The cell suspension of 200 μL was added to the apical chamber of each well, and the 800 μL conditioned medium containing 20% FBS was added to the basolateral chamber. The chambers were incubated at 37°C for 20–24 h. The Transwell plate was taken out for PBS rinsing twice, and soaked in formaldehyde for 10 min, and rinsed with clean water 3 times. The gel was stained with 0.1% crystal violet, placing at room temperature for 30 min followed by rinsing with PBS twice and wiping the cells on the upper surface with cotton balls. The cells were observed under an inverted microscope, photographed and counted. The Transwell migration experiment did not need Matrigel, and the incubation period was 16 h.
2.13 Statistical analysis
SPSS 22.0 software (IBM Corp. Armonk, NY, USA) was used for statistical analysis. The measurement data were displayed as mean ± standard deviation, and those with skewed distribution or uneven variance were represented by the interquartile range. First, the normality and homogeneity of variance were tested. Later the paired t-test was used for comparison between the experimental data of CC tissues and adjacent normal tissues with normal distribution and even variance. Comparisons among multiple groups were performed with one-way analysis of variance (ANOVA), and comparisons between two groups were assessed by the post-hoc test. When the data did not show normal distribution or equal variance, the rank-sum test was conducted. The expression of significant difference was set as p<0.05.
3 Results
3.1 LINC00511 is highly expressed in CC
Initially, we performed RT-qPCR to determine the expression of LINC00511 in CC tissues and adjacent normal tissues of 84 cases. As shown in figure 1A, the expression of LINC00511 was significantly elevated in CC tissues compared with that in adjacent normal tissues (p<0.05). In order to further investigate the relationship between the expression of LINC00511 and prognosis of CC patients, the expression of LINC00511 in CC and adjacent normal tissues and the detailed information of CC patients were collected. The results of Kaplan-Meier analysis revealed that a much lower OS rate and RFS was revealed in patients with high LINC00511 expression (p<0.05) (figure 1B and C). These results showed that LINC00511 was expressed at a high level in CC tissues.
CC patients with high expression of LINC00511 exhibited poor survival. (A) Expression of LINC00511 was higher in 84 cases of CC tissues than adjacent normal tissues according to RT-qPCR; *p<0.05 vs the CC tissues. (B) High expression of LINC00511 in CC patients was associated with a poor OS rate. (C) High expression of LINC00511 in CC patients was associated with a poor RFS. The results of RT-qPCR cell experiment were measurement data, expressed as mean ± standard error. Comparisons among multiple groups were performed with the paired t-test for statistical analysis. The experiment was repeated three times; CC, cervical cancer; LINC00511, long intergenic non-protein coding RNA 511; RT-qPCR, reverse transcription quantitative polymerase chain reaction; OS, overall survival; RFS, relapse-free survival.
3.2 High expression of LINC00511 is linked to tumor stage, tumor size, LNM and HPV16 infection in CC
Subsequently, to explore the relationship between LINC00511 and clinicopathological features, we collected corresponding data. As shown in table 2, the median expression of LINC00511 in CC was 0.996, which was regarded as a demarcation to classify the 84 cases into highly expressed and poorly expressed groups. Based on clinicopathological data, we found that high expression of LINC00511 was closely related to tumor stage, tumor size, LNM and HPV16 infection (all p<0.05), and had no significant relationship with age, histological type, differentiation degree and metastasis (p > 0.05).
3.3 HeLa/PTX cells exhibit resistance to PTX
Then CCK-8 assay was applied to measure the inhibitory effect of different concentrations of PTX on proliferation of HeLa cells and HeLa/PTX cells. As shown in figure 2, it revealed that the inhibitory rate of HeLa cells and HeLa/PTX cells was enhanced with the increase of PTX concentration. According to the results of SPSS 22.0 statistical analysis, the IC50 value of HeLa and HeLa/PTX cells were 8.383 μg/L and 383.800 μg/L, respectively. The results showed that HeLa/PTX might have significant resistance to PTX.
Inhibitory effect of PTX on HeLa and HeLa/PTX cell proliferation was facilitated with the increased concentration of PTX according to CCK-8 assay. The OD values were measurement data, expressed as mean ± standard error. The experiment was repeated three times; PTX, paclitaxel; HeLa/PTX, PTX-resistant human cervical carcinoma HeLa cell line; CCK-8, cell counting kit 8; OD, optical density.
3.4 Silenced LINC00511 declines the viability of PTX-resistant Hela/PTX cells and inhibits cell proliferation
CCK-8 assay and EdU staining were followed to assess the effect of LINC00511 on the viability and proliferation of PTX-resistant Hela/PTX cell line. The result in figure 3 indicated that compared with the blank group, the sh-LINC00511 group exhibited notably reduced cell viability and proliferation. The proportion of PTX-resistant Hela/PTX cell significantly increased at G1 phase while significantly decreased at S phase. Additionally, the LINC00511 group showed an opposite trend (all p<0.05), and no significant difference was found in the sh-LINC00511 NC and LINC00511 NC groups (p > 0.05). The results revealed that silencing of LINC00511 could suppress the viability and proliferation of PTX-resistant Hela/PTX cell line and arrested the cell cycle at G1 phase.
Viability and proliferation of PTX-resistant Hela/PTX cell line were impeded by silencing of LINC00511. (A) Viability of PTX-resistant Hela/PTX cells was reduced by silencing of LINC00511 according to CCK-8 assay. (B) Red-stained cells were proliferated PTX-resistant Hela/PTX cells, and less proliferated PTX-resistant Hela/PTX cells were revealed after silencing of LINC00511 according to the results of EdU staining (200×). (C) Proliferation of PTX-resistant Hela/PTX cells was depleted by silencing of LINC00511. (D) Cell cycle of PTX-resistant Hela/PTX cells after silencing of LINC00511 was arrested at G1 phase according to the results of flow cytometry. (E) Statistical histogram for the cell cycle of PTX-resistant Hela/PTX cells in each group. The OD values of cells in each group, red-stained positive cells after transfection and the number of cells in different cell cycle were measurement data, expressed as mean ± standard error. The data of panels A and E were analyzed by repeated measures of ANOVA, and data of panel C were analyzed by one-way ANOVA. The experiment was repeated three times. *p<0.05 vs the blank group; NC, negative control; PTX-resistant Hela/PTX, PTX-resistant human cervical carcinoma HeLa cell line; LINC00511, long intergenic non-protein coding RNA 511; sh-LINC00511, short-hairpin LINC00511; CCK-8, cell counting kit 8; EdU, 5-ethynyl2′-deoxyuridine.
3.5 Silenced LINC00511 inhibits migration and invasion of PTX-resistant Hela/PTX cells
To detect the effects of LINC00511 on the migration and invasion of PTX-resistant Hela/PTX cells, we conducted Transwell assay (figure 4A and B). Cell migration ability was measured by cell migration distance and cell invasion ability was measured by cell penetration number. The mRNA and protein expression of MMP-2 and MMP-9 was investigated by RT-qPCR and western blot analysis (figure 4C and D). In contrast to the blank group, cell migration and invasion of PTX-resistant Hela/PTX cells were obviously increased, and the mRNA and protein expression of MMP-2 and MMP-9 was increased in the LINC00511 group. In addition, the sh-LINC00511 group exhibited an opposite trend (all p<0.05). The above findings showed that the migration and invasion of PTX-resistant Hela/PTX cells could be suppressed through silencing of LINC00511.
Inhibited migration and invasion were exhibited in the PTX-resistant Hela/PTX cells after silencing of LINC00511. (A) Cell migration distance was shortened after silencing of LINC00511 according to the Transwell migration results; (B) PTX-resistant Hela/PTX cell invasion was suppressed by silencing of LINC00511; (C) mRNA expression of MMP-2 and MMP-9 was decreased after silencing of LINC00511 as detected by RT-qPCR, with the blank group as normalized; (D) and (E) protein expression of MMP-2 and MMP-9 was decreased in PTX-resistant Hela/PTX cell treated with silenced LINC00511, and the gray value of the corresponding protein band/the gray value of the GAPDH protein band represented the relative protein expression. *p<0.05 vs the blank and NC group. The measurement data were expressed as mean ± standard error and analyzed by one-way ANOVA. The experiment was repeated three times. NC, negative control; PTX-resistant Hela/PTX, PTX-resistant human cervical carcinoma HeLa cell line; LINC00511, long intergenic non-protein coding RNA 511; sh-LINC00511, short-hairpin LINC00511; RT-qPCR, reverse transcription quantitative polymerase chain reaction; MMP, matrix metalloproteinase.
3.6 Silenced LINC00511 promotes apoptosis of PTX-resistant Hela/PTX cells
To detect the effects of LINC00511 on the apoptosis of PTX-resistant Hela/PTX cells, we conducted TUNEL staining. RT-qPCR and western blot analysis were also adopted to determine the mRNA and protein level of Bcl-2, Bax and cleaved-caspase-3. As shown in figure 5, in contrast to the blank group, cell apoptosis obviously increased in the sh-LINC00511 group with increased expression of Bax and cleaved-caspase-3 and decreased Bcl-2. In addition, the LINC00511 group exhibited an opposite trend (all p<0.05), and no significant difference was found between the sh-LINC00511 NC and LINC00511 NC groups as compared with the blank group (p > 0.05). The above findings showed that the apoptosis of PTX-resistant Hela/PTX cells could be accelerated through silencing of LINC00511.
PTX-resistant Hela/PTX cells exhibited promoted apoptosis after silencing of LINC00511. (A) Green-stained cells were apoptotic cells, and more apoptotic cells revealed after silencing of LINC00511 according to the results of TUNEL staining (400×). (B) PTX-resistant Hela/PTX cell apoptosis was facilitated by silencing of LINC00511. (C) mRNA expression of apoptotic-related gene Bax, cleaved-caspase-3 and Bcl-2 by RT-qPCR, with the blank group normalized; (D) gray value of apoptotic-related proteins Bax, cleaved-caspase-3 and Bcl-2; (E) the expression of apoptotic-related proteins Bax, cleaved-caspase-3 and Bcl-2 by western blot analysis, and the gray value of the corresponding protein band/the gray value of the GAPDH protein band represented the relative protein expression. The green-stained positive cells after different transfections and results of RT-qPCR and western blot analysis were measurement data, expressed as mean ± standard error and analyzed by one-way analysis of variance. The experiment was repeated three times. *p<0.05 vs the blank group; NC, negative control; PTX-resistant Hela/PTX, PTX-resistant human cervical carcinoma HeLa cell line; LINC00511, long intergenic non-protein coding RNA 511; sh-LINC00511, short-hairpin LINC00511; Bcl-2, B-cell lymphoma 2; TUNEL, TdT-mediated dUTP-biotin nick end-labeling; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
3.7 Silenced LINC00511 attenuates resistance of PTX-resistant Hela/PTX cells to PTX
Finally, we performed CCK-8 assay, RT-qPCR and western blot analysis to evaluate the effects of LINC00511 on IC50 value, mRNA expression and protein level of resistance-related proteins MRP1 and P-GP. As shown in figure 6, the IC50 value, expression of LINC00511 and mRNA expression and protein level of MRP1 and P-GP apparently declined in the sh-LINC00511 + PTX group, and PTX-resistant Hela/PTX cells conferred significantly enhanced sensitivity to PTX in comparison with the blank + PTX group (all p<0.05). Additionally, the LINC00511 + PTX group exhibited a contrary tendency (p<0.05), the results between the sh-LINC00511 NC + PTX and LINC00511 NC + PTX groups did not differ significantly (p > 0.05). Together, the findings confirmed that the resistance of PTX-resistant Hela/PTX cells to PTX could be inhibited by silencing of LINC00511.
Resistance of PTX-resistant Hela/PTX cells to PTX was inhibited after silencing of LINC00511. (A) IC50 value decreased in PTX-resistant Hela/PTX cells after silencing of LINC00511 according to the results of CCK-8 assay. (B) mRNA expression of LINC00511, MRP1 and P-GP in PTX-resistant Hela/PTX cells was reduced by silencing of LINC00511 according to the results of RT-qPCR with the blank group normalized. (C) Gray value of MRP1, P-GP and GADPH protein bands. (D) Protein levels of LINC00511, MRP1 and P-GP in PTX-resistant Hela/PTX cells were reduced by silencing of LINC00511 according to the results of western blot analysis, and the gray value of the corresponding protein band/the gray value of the GAPDH protein band represented the relative protein expression. The results of RT-qPCR and western blot analysis were measurement data, expressed as mean ± standard error and analyzed by one-way ANOVA. The experiment was repeated three times. *p<0.05 vs the blank group; NC, negative control; PTX-resistant Hela/PTX, PTX-resistant human cervical carcinoma HeLa cell line; LINC00511, long intergenic non-protein coding RNA 511; sh-LINC00511, short-hairpin LINC00511; MRP1, multidrug resistance protein 1; P-GP, P-glycoprotein; GADPH, glyceraldehyde 3-phosphate dehydrogenase; CCK-8, cell counting kit 8; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
4 Discussion
CC is one of the most commonly seen malignant tumors in women with a high mortality and it is necessary to identify more effective prognostic and diagnostic biomarkers to further improve therapeutic effect (Boromand et al. 2018). Moreover, aberrant expression of lncRNAs was indicated to influence CC progression, for example, up-regulation of Hox transcript antisense intergenic RNA (HOTAIR) was revealed to be linked with LNM and poor OS rate of CC (Kim et al. 2015; Peng et al. 2016). In our study, the results showed that silencing of LINC00511 can suppress CC growth, suggesting a therapeutic potential in the treatment of CC patients.
Initially, we determined the expression of LINC0051 and observed a higher LINC0051 expression in CC. Up-regulation of LINC0051 was revealed in several cancers and played as an oncogenic lncRNA by driving tumorigenesis in cancers, such as non-small-cell lung cancer (NSCLC), TSCC, pancreatic ductal adenocarcinoma (PDAC) and lung adenocarcinoma, among which knockdown of LINC0051 could exert inhibitory effect on cell proliferation, migration and invasion yet promote cell apoptosis (Sun et al. 2016; Wei and Zhang 2016; Ding et al. 2018; Zhao et al. 2018). LINC0051 was reported to be associated with cell adhesion, cell migration and extracellular signal-regulated kinases 1/2 (ERK1/2) cascade regulation in triple-negative breast cancer according to the results of gene ontology analysis (Xu et al. 2017). More importantly, increased expression of LINC00511 was exhibited in metastatic basal breast cancer cell lines according to a validation (Oh et al. 2016). In the present study, we found that LINC0051 was up-regulated in CC. Subsequently, we established PTX-resistant HeLa/PTX cell line to evaluate the effect of LINC0051 on cell proliferation, cell cycle, apoptosis and resistance to PTX.
The results of the study demonstrated that silencing of LINC0051 significantly decreased cell viability, proliferation, migration, invasion and more cells arrested at G1 phase. Additionally, the mRNA and protein level of apoptosis-related proteins Bax, cleaved-caspase-3 elevated and Bcl-2 decreased, and that of invasion-related proteins MMP2 and MMP-9 was decreased by silencing of LINC0051. Bcl-2, Bax and caspase are important apoptotic proteins, in which Bcl-2 protein family regulated mitochondria-mediated apoptosis and activation of caspases triggered cellular death through recruitment of apoptosis-related mediators (Wang et al. 2018). In was indicated that increased Bax expression and declined Bcl-2 expression by Inotodiol were able to contribute to enhanced CC cell apoptosis (Zhao et al. 2014). A study revealed that Paris saponin I induced NSCLC cell apoptosis through the increased ratio of Bax and Bcl-2 and expression of caspase-3 (Jiang et al. 2014). In CC, alteration of Bcl-2 and Bax was reported to be associated with cellular resistance to cisplatin, which could be reversed by suppression of REV3L, and thus contribute to promoting sensitivity to cisplatin (Yang et al. 2015). MMPs, especially MMP-2 and MMP-9, were secreted by CC and were reported to play significant roles in CC cell invasion and metastasis (Roomi et al. 2010). MMP-2 and MMP-9 activities could be prompted by E6-HPV16 or E7-HPV16, and inhibited MMP-2/9 all contributed to the inhibition of CC cell migration and invasion (Zhu et al. 2015). Moreover, Kaplan-Meier analysis found that silencing of LINC00511 may promote the OS and RFS rate, which is consistent with the migration, invasion and proliferation results.
Finally, the mRNA and protein levels of MRP1 and P-GP were assessed to explore whether silencing of LINC00511 could confer suppressive effect on resistance to PTX of PTX-resistant HeLa/PTX cell line. Chemosensitivity to PTX was closely linked to lncRNAs that had been illustrated in several cancers, such as endometrial cancer, lung adenocarcinoma and advanced ovarian cancer (Li et al. 2017; Ren et al. 2017; Wang et al. 2017). MRPs belong to subfamily C of the human ABC (ATP-binding cassette) superfamily (Slot et al. 2011), among which MRP1 has the ability to confer multidrug resistance in lung cancer cells by excluding antineoplastic agents, thus achieving reduction of drug accumulation and promotion of resistance in tumor cells (Cole 2014). Additionally, high expression of MRP1 in breast cancer gave rise to clinical drug resistance, and it may be of great importance in resistance to adjuvant cyclophosphamide, methotrexate and fluorouracil (CMF) chemotherapy (Filipits et al. 2005). In HeLa cells, cancerous inhibitor of protein phosphatase 2A (CIP2A) is reported to be involved in regulating multidrug resistance of cervical adenocarcinoma upon chemotherapy by enhancing P-GP expression through E2F transcription factor 1 (E2F1) (Liu et al. 2016). In the present study, it is concluded that silencing of LINC00511 could inhibit resistance to PTX of PTX-resistant HeLa/PTX cell line, and the CC cells in the sh-LINC00511 + PTX group presented enhanced sensitivity to PTX with decreased expression of MRP1 and P-GP.
In summary, LINC00511 plays a carcinogenic role in CC while silencing of LINC00511 could inhibit CC cell viability, proliferation and resistance to PTX with induced cell apoptosis rate and arrested cell cycle at G1 phase. Therefore, our present study provided evidence that silencing of LINC00511 might be an alternative target in inhibiting resistance to PTX in CC. Due to the unclear molecular mechanism of LINC00511 in vitro, further detailed studies are required to explore the potential therapeutic function of LINC00511 in CC.
References
Alexopoulou AN, Leao M, Caballero OL, Da Silva L, Reid L, Lakhani SR, Simpson AJ, Marshall JF, Neville AM and Jat PS 2010 Dissecting the transcriptional networks underlying breast cancer: NR4A1 reduces the migration of normal and breast cancer cell lines. Breast Cancer Res. 12 R51
Arocho A, Chen B, Ladanyi M and Pan Q 2006 Validation of the 2-∆∆Ct calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 15 56–61
Boromand N, Hasanzadeh M, ShahidSales S, Farazestanian M, Gharib M, Fiuji H, Behboodi N, Ghobadi N, Hassanian SM, Ferns GA and Avan A 2018 Clinical and prognostic value of the C-Met/HGF signaling pathway in cervical cancer. J. Cell. Physiol. 233 4490–4496
Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q and Liu Z 2016 Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol. Med. Rep. 13 1541–1550
Chen Y, Zhao Y, Wang C, Xiao X, Zhou X and Xu G 2012 Inhibition of p38 MAPK diminishes doxorubicin-induced drug resistance associated with P-glycoprotein in human leukemia K562 cells. Med. Sci. Monit. 18 BR383–BR388
Cole SP 2014 Targeting multidrug resistance protein 1 (MRP1, ABCC1): past, present, and future. Annu. Rev. Pharmacol. Toxicol. 54 95–117
Ding J, Yang C and Yang S 2018 LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J. Oral Pathol. Med. 47 468–476
Dong J, Wang Q, Li L and Xiao-Jin Z 2018 Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol. Biochem. 45 2086–2094
Filipits M, Pohl G, Rudas M, Dietze O, Lax S, Grill R, Pirker R, Zielinski CC, Hausmaninger H, Kubista E, Samonigg H and Jakesz R 2005 Clinical role of multidrug resistance protein 1 expression in chemotherapy resistance in early-stage breast cancer: the Austrian Breast and Colorectal Cancer Study Group. J. Clin. Oncol. 23 1161–1168
Guo T, Wang W, Zhang H, Liu Y, Chen P, Ma K and Zhou C 2011 ISL1 promotes pancreatic islet cell proliferation. PLoS One 6 e22387
He L, Yang H, Zhou S, Zhu H, Mao H, Ma Z, Wu T, Kumar AK, Kathera C, Janardhan A, Pan F, Hu Z, Yang Y, Luo L and Guo Z 2018 Synergistic antitumor effect of combined paclitaxel with FEN1 inhibitor in cervical cancer cells. DNA Repair (Amst) 63 1–9
Jiang H, Zhao PJ, Su D, Feng J and Ma SL 2014 Paris saponin I induces apoptosis via increasing the Bax/Bcl-2 ratio and caspase-3 expression in gefitinib-resistant non-small cell lung cancer in vitro and in vivo. Mol. Med. Rep. 9 2265–2272
Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW and Kim YT 2015 Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int. J. Oncol. 46 521–530
Li W, Li H, Zhang L, Hu M, Li F, Deng J, An M, Wu S, Ma R, Lu J and Zhou Y 2017 Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J. Biol. Chem. 292 5801–5813
Liu J, Wang M, Zhang X, Wang Q, Qi M, Hu J, Zhou Z, Zhang C, Zhang W, Zhao W and Wang X 2016 CIP2A is associated with multidrug resistance in cervical adenocarcinoma by a P-glycoprotein pathway. Tumour Biol. 37 2673–2682
Liu P, Ma S, Liu H, Han H and Wang S 2017 HCFU inhibits cervical cancer cells growth and metastasis by inactivating Wnt/beta-catenin pathway. J. Cell Biochem. https://doi.org/10.1002/jcb.26570
Lu H, He Y, Lin L, Qi Z, Ma L, Li L and Su Y 2016 Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 37 1683–1691
Oh TG, Wang SM, Acharya BR, Goode JM, Graham JD, Clarke CL, Yap AS and Muscat GEO 2016 The nuclear receptor, RORgamma, regulates pathways necessary for breast cancer metastasis. EBio Med. 6 59–72
Peng L, Yuan X, Jiang B, Tang Z and Li GC 2016 LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 37 2779–2788
Ren K, Xu R, Huang J, Zhao J and Shi W 2017 Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol. 80 243–250
Rodriguez JA, Galeano L, Palacios DM, Gomez C, Serrano ML, Bravo MM and Combita AL 2012 Altered HLA class I and HLA-G expression is associated with IL-10 expression in patients with cervical cancer. Pathobiology 79 72–83
Roomi MW, Monterrey JC, Kalinovsky T, Rath M and Niedzwiecki A 2010 In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol. Rep. 23 605–614
Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS, Tuo L, Wang S and Cao XF 2015 Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 36 2501–2507
Slot AJ, Molinski SV and Cole SP 2011 Mammalian multidrug-resistance proteins (MRPs). Essays Biochem. 50 179–207
Spizzo R, Almeida MI, Colombatti A and Calin GA 2012 Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 31 4577–4587
Sun CC, Li SJ, Li G, Hua RX, Zhou XH and Li DJ 2016 Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids 5 e385
Wang L, Hu Y, Xiang X, Qu K and Teng Y 2017 Identification of long non-coding RNA signature for paclitaxel-resistant patients with advanced ovarian cancer. Oncotarget 8 64191–64202
Wang X, Yi Y, Lv Q, Zhang J, Wu K, Wu W and Zhang W 2018 Novel 1,3,5-triazine derivatives exert potent anti-cervical cancer effects by modulating Bax, Bcl2 and caspases expression. Chem. Biol. Drug Des. 91 728–734
Wei Y and Zhang X 2016 Transcriptome analysis of distinct long non-coding RNA transcriptional fingerprints in lung adenocarcinoma and squamous cell carcinoma. Tumour Biol. 37 16275–16285
Wei Y, Dong J, Li F, Wei Z and Tian Y 2017 Knockdown of SLC39A7 suppresses cell proliferation, migration and invasion in cervical cancer. EXCLI J. 16 1165–1176
Wu L, Jin L, Zhang W and Zhang L 2016 Roles of long non-coding RNA CCAT2 in cervical cancer cell growth and apoptosis. Med. Sci. Monit. 22 875–879
Xu S, Kong D, Chen Q, Ping Y and Pang D 2017 Oncogenic long noncoding RNA landscape in breast cancer. Mol. Cancer 16 129
Yang L, Shi T, Liu F, Ren C, Wang Z, Li Y, Tu X, Yang G and Cheng X 2015 REV3L, a promising target in regulating the chemosensitivity of cervical cancer cells. PLoS One 10 e0120334
Zhang H, Zhu C, Zhao Y, Li M, Wu L, Yang X, Wan X, Wang A, Zhang MQ, Sang X and Zhao H 2015 Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget 6 43770–43778
Zhang F, Wang C, Cui Y, Li S, Yao Y, Ci Y, Wang J, Hou W, Wu A and Li E 2016 Effects of propofol on several membrane characteristics of cervical cancer cell lines. Cell Physiol. Biochem. 40 172–182
Zhang HY, Yang W, Zheng FS, Wang YB and Lu JB 2017 Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in lung squamous cell carcinoma. Biomed. Pharmacother. 90 650–658
Zhao LW, Zhong XH, Yang SY, Zhang YZ and Yang NJ 2014 Inotodiol inhabits proliferation and induces apoptosis through modulating expression of cyclinE, p27, bcl-2, and bax in human cervical cancer HeLa cells. Asian Pac. J. Cancer Prev. 15 3195–3199
Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y, Lin Q, Fu Z and Rufu C 2018 Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J. Cell Mol. Med. 22 655–667
Zhu D, Ye M and Zhang W 2015 E6/E7 oncoproteins of high risk HPV-16 upregulate MT1-MMP, MMP-2 and MMP-9 and promote the migration of cervical cancer cells. Int. J. Clin. Exp. Pathol. 8 4981–4989
Acknowledgements
We would like to show sincere appreciation to the reviewers for critical comments on this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Sorab Dalal
Rights and permissions
About this article
Cite this article
Mao, BD., Xu, P., Zhong, Y. et al. LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J Biosci 44, 44 (2019). https://doi.org/10.1007/s12038-019-9851-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-019-9851-0