Abstract
A challenge in bioinformatics is to analyse volumes of gene expression data generated through microarray experiments and obtain useful information. Consequently, most microarray studies demand complex data analysis to infer biologically meaningful information from such high-throughput data. Selection of informative genes is an important data analysis step to identify a set of genes which can further help in finding the biological information embedded in microarray data, and thus assists in diagnosis, prognosis and treatment of the disease. In this article we present an unsupervised feature selection technique which attempts to address the goal of explorative data analysis, unfolding the multi-faceted nature of data. It focuses on extracting multiple clustering views considering the diversity of each view from high-dimensional data. We evaluated our technique on benchmark data sets and the experimental results indicates the potential and effectiveness of the proposed model in comparison to the traditional single view clustering models, as well as other existing methods used in the literature for the studied datasets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
A challenge in bioinformatics is to analyse volumes of gene expression data and obtain useful information. Many clustering algorithms are being developed to handle the high-dimensional data by projecting the data into a lower-dimensional subspace, e.g. principal component analysis, which depends on stringent separation requirements (Chaudhuri et al. 2009). In a typical microarray dataset, the number of genes as measured is of magnitude of several thousands, far exceeding the number of samples, with many of the genes being either correlated or irrelevant. A great deal of recent research has focused on the challenging task of selecting informative genes from microarray data. In this task, unsupervised dimensionality reduction can be used as a preprocessing step where the goal is to find the smallest gene subset that best uncovers interesting natural clusters of data (Mitra et al. 2002; Ding 2003; Jaeger et al. 2003; Varshavsky et al. 2006; Hong et al. 2008; Li et al. 2008; Sharma et al. 2012a, b).
Clustering has been used in many areas of biological data analysis (Pirim et al. 2012), the goal being to find structures in high-dimension data. Such structures are often multi-faceted owing to the nature of the problem. Traditional clustering methods seek to find a unified clustering solution and are inherently limited in achieving multi-faceted structures (Cui et al. 2007). In most biological applications, data can be interpreted in many different ways. There may exist multiple groupings of the data that are all reasonable in some perspective. This problem is often more prominent for high-dimensional data, where each object is described by a large number of features. In such cases, different feature subspaces can often warrant different ways to partition the data. Each feature subspace presents the user a different view of the data (Cui et al. 2007; Fang et al. 2010; Gupta et al. 2011).
2 Multiview clustering
Biological data is often multi-faceted by nature and may be interpreted in alternate ways (Xu et al. 2013). Multiview clustering provides multiple sets of clusters, all reasonable in some perspective, thus providing greater insights than a single solution in such analysis (Muller et al. 2012; Xu et al. 2013). Recently, many multiview-based learning methods showing the diversity of different views have been proposed in the literature. Multiviews obtained from multiple sources or different feature subsets not only obtain the views of different attributes but also ensure that each views best represent the data and ensure the efficient learning (Xu et al. 2013). The work done in the paper by Bickel and Scheffer (2004) develops and studies partitioning and agglomerative, hierarchical multiview clustering algorithms for text data. The article by Cui et al. (2007) presents a multiview framework and suggests two approaches within this framework: (1) orthogonal clustering and (2) clustering in orthogonal subspaces. Chaudhuri et al. (2009) paper consider constructing high-dimensional data projections to lower dimension using multiple views of the data, via canonical correlation analysis (CCA). A probabilistic multiview clustering model outperforming an early-fusion approach based on multiview feature correlation analysis is derived in the paper by Bruno and Marchand-Maillet (2009). A new multiview clustering method which uses clustering results obtained on each view as a voting pattern in order to construct a new set of multiview clusters is proposed in in the paper by Kim et al. (2010). The article by Chen et al. (2013) proposes TW-k-means, an automated two-level variable weighting clustering algorithm for multiview data, which can simultaneously compute weights for views and individual variables. The survey work done in articles by Sun (2013) and Xu et al. (2013) aims to provide an insightful organization of current developments in the field of multiview learning, identify their limitations, and gives suggestions for further research. Considering the three major issues, viz. diversity, compatibility and accuracy in generating multiview feature sets different strategies based on clustering, random selection and uniform band slicing have been proposed in the literature (Di and Crawford 2012; Xu et al. 2013). Xu et al. (2013) states that increase in the number of views to increase diversity, or increase in randomness to avoid noisy view, further improves the performance of the model. A multiview approach using controlled vocabularies selected from nine well-known bio-ontologies is presented by Yu et al. (2010) to retrieve biomedical knowledge. Ensemble learning can reduce the potential for over-fitting the training data (Yang et al. 2010).
Thus, multiview learning is considered to be more effective, more promising and shows better generalization ability, as each view forms alternative solutions to the given problem, representing different perspectives on the data and thus gives greater insight than only one solution or single view (Muller et al. 2012; Sun 2013; Xu et al. 2013). Generating multiple views needs decomposition of the original feature set into multiple disjoint feature subsets each corresponding to different views. Traditional machine learning solution for the multiview problem is to consider all multiple views into one single view to effectively define the learning model (Xu et al. 2013). This approach of multiview selection may result in over-fitting when training sample size is small and it ignores the distinct statistical property of each view (Sun 2013; Xu et al. 2013). Although exhaustive work has been done in this field, a wide variety of applications, viz. high-dimensional microarray data, still require further research to be done in this topic (Xu et al. 2013). In genomics, one gene may have multiple functions, and each cluster may form alternative solutions to the given problem, representing different perspectives on the data (Muller et al. 2012).
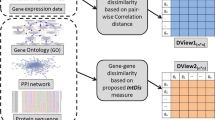
In this paper, we suggest a graph-based unsupervised feature/gene selection (GUFS) technique and apply it to obtain multiview clustering from microarray datasets. The graph-based technique creates multiple views, each involving varying number of genes that are automatically obtained. This is natural in genomic data where gene groups are important in deciding alternate interpretations of the microarray data considering the diversity, thereby facilitating gene subset selection which are informative genes in regard to different views. We tested our gene selection model on benchmark datasets, viz. B-cell chronic lymphocytic leukemia (B-CLL) and interstitial lung disease (ILD). The experimental results indicate the potential and effectiveness of the proposed model in comparison to the traditional single view clustering models, as well as other existing methods used in literature for the studied datasets.
This work extends our earlier work on unsupervised feature selection (Mitra and Swarnkar 2012) and concept of this multiview clustering has been used in integration with protein–protein interaction network in a conference paper (Swarnkar et al. 2014). The remaining sections of the paper are organized as follows: section 3 presents related materials and methods used for our proposed graph-based multiview model for feature selection and section 4 discusses results and comparisons. Finally, section 5 presents our conclusion and discussion.
3 Materials and methods
Block diagram in figure 1 represents the schematic work flow of the proposed multiview feature selection model GUFS. To assess presence of outliers that can skew the expression result in the study sample (B-CLL and ILD), we have filtered the genes, based on their variance across the samples, and thus considered the genes with variance less than tenth percentile for further processing (Kohane et al. 2002). Further, the 10, 000 permutation’s t-test (Dudoit et al. 2002) is used and the genes with p-value cut-off of 0.05 are considered to have statistical significance (Huang et al. 2009; Xiao et al. 2014) and are used for subsequent analysis. This set of data are normalized using the mean column intensity, and the raw intensities are transformed to the range of [0, 1] values for each sample.
3.1 Graph-based unsupervised feature selection (GUFS)
We describe below the steps of graph-based feature/gene selection algorithm on microarray data in obtaining gene subsets and resultant multiview clusters.
-
(i)
Construct gene profile network for each gene/feature based on the expression level of that specific gene over the samples. The network has samples as its vertices and similarity of expression of that specific gene determines the existence of an edge between two samples. For each gene in the dataset there exists a gene profile network.
-
(ii)
A gene correlation network, i.e. pair-wise distance measure matrix of genes, using symmetric difference between the edge set is constructed in this step. Here, we have used the XOR, (i.e., the number of edges present in one network but absent in the other), as symmetric difference measure for its computational simplicity and effectiveness. The resultant graph is a gene correlation network with genes as vertices and above distance as edge weight, where edge weight represents the degree of co-expression between two genes. Lower the edge weight, higher the degree of co-expression between two genes.
-
(iii)
The resultant gene correlation network is now clustered into k different non-overlapping partitions, where k is user defined parameter. Choice of k takes into consideration the cluster or view quality, as well as the size of the views. This step we call it as, gene network clustering. The proposed model here uses hierarchical agglomerative clustering with edge weight of the gene correlation network as the distance measure. We tested for different values of k and for the dataset being considered here we got the best result for k = 10.
Gene subset profile network are constructed for each cluster obtained above, expecting that each gene group is having some biological functional similarity. We denote this as a view (V) and there exists, k clusters of the data (V ⊆ V) each corresponding to a set of related genes. This network has sample as its vertices and edge weights represents the Euclidean distance between two samples considering only the expression of the gene subset belonging to that particular gene network cluster.
To measure the class performance of each of the views obtained in above step, we partion each of these k networks into l sample clusters. For this we have used hierarchical clustering, as well as kNN. Thus, each of these k graphs (V ⊆ V) gives rise to a separate clustering set of l clusters each. For our dataset l is same as that of the true class label count, otherwise it can be dependent on the cluster quality or domain knowledge.
Each of these clustering may lead to multiview interpretation of the microarray expression data. The number of these informative genes is very small in comparison to the actual number of genes present in the training data set; it may vary from view to view.
It may be noted that most of the noisy features is seen to get accumulated in one of the view, and it has been observed during analysis that this view do not give any significant information in regard to statistical analysis. We discarded this largest cluster from our analysis.
In our experiment, predictive accuracy of the multiview clusters were measured in terms of specificity, sensitivity, precision, overall accuracy and the number of correctly classified instances in comparison to the known true classes. The biological significance of the views were measured by finding the percentage of known disease related genes in these views or finding the true positives in these views in regard to ground truth. Further, we studied the dominance of certain biological processes in these selected views.
3.2 Cluster validation
We aimed to measure the accuracy of the proposed model ability to select the relevant features to find structure in the data (cluster). In our evaluation we assumed the ground truth or true clusters were provided. These true clusters were referred as ‘class labels’. These labels were used only during validation of the proposed model and were not used in selecting features and discovering clusters.
To evaluate the proposed models ability to select “relevant” features, we report the overall accuracy, sensitivity, specificity, precision, f-measure and result in comparison to the known true classes (Ji et al. 2014), and are respectively defined by equations 1–6 stated as follows:
where TP is the number of true positive samples, TN is the count for true negative samples, FP is the number of false-positive samples and FN is the number of false-negative samples. These measures have been adopted for statistical analysis and comparison with existing methods in the literature. Samples were considered to be divided in two categories, namely positive samples (diseased) and negative samples (non-diseased or normal). We used the National Center for Biotechnology Information (NCBI) database ( http://www.ncbi.nlm.nih.gov/gene/ ) as our reference to collect disease related genes. A record may include nomenclature, Reference Sequences (RefSeqs), maps, pathways, variations, phenotypes, and link to genome, phenotype, and locus-specific resources worldwide. Considering this set of genes as actual data we calculated the number of hits or true positive in each clusters or gene sets. True positive or number of hits is the count of correctly classified genes as disease related.
Study of the biological relevance in the form of gene-to-annotation is a promising high-throughput strategy that helps the researchers to identify biological processes most pertinent to their study (Huang et al. 2009; Sharma et al. 2012a, b). To study the pertinent or enriched biological process of genes in each cluster, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 ( http://david.abcc.ncifcrf.gov/home.jsp ) (Huang et al. 2009) as our biological tool. The tool provides a comprehensive set of functional annotation tools for investigators to understand biological meaning behind large list of genes (Dennis et al. 2003). A set of user’s input genes is highly associated with certain terms, which is statistically measured by the Fisher Exact in DAVID system. Fisher Exact p − value = 0 represents perfect enrichment; usually p-value is equal or smaller than 5 × 10− 2 to be considered strongly enriched in the annotation categories (Huang et al. 2008, 2009). Fold change enrichment (FE) measure the magnitude of enrichment for a specific annotation category. Let x ij and y ij denote the log2 expression levels of gene i in sample j in the control and disease, respectively, then the fold-change for gene i is defined as FE i = x i − y i (Tibshirani and Witten 2007). Thus, FE score ranks the enriched terms in a more comprehensive manner, FE 1.5 and above are suggested to be considered as interesting (Huang et al. 2008). FuncAssociate ( http://llama.med.harvard.edu/funcassociate ), a Web-based application which discovers properties enriched in lists of genes or proteins that emerge from large-scale experimentation (Berriz et al. 2009) is also used for biological significance measurement. Further, gene-card ( http://www.genecards.org/ ) (Safran et al. 2010) is used to study the biological functions of individual gene.
3.3 Datasets used
The DNA microarray datasets for Homo sapiens, collected from NCBI’s Gene Expression Omnibus. are utilized in our study to show the effectiveness of the proposed model. The database Web link is http://www.ncbi.nlm.nih.gov/geo/ . The description of the datasets is given as follows:
Leukemia dataset: B-cell chronic lymphocytic leukemia (B-CLL) is the most common adult leukaemia and is characterized by accumulation of monoclonal B cells in the blood, marrow, and secondary lymphoid tissues. The clinical outcome of patients with CLL is highly variable, some of the patients usually have no clinical symptoms for many years and do not require treatment, the other half the disease is relatively aggressive and require therapy soon after diagnosis or else patient dies due to causes related to CLL. It is a heterogeneous disease with a pronounced variation in the clinical course. Although, several methods have facilitated the identification of a number of prognostically and diagnostically important genetic markers for CLL, the genetic mechanism that result in the development and progression of CLL are mainly unknown (Fält et al. 2005; Codony et al. 2009; Chuang et al. 2012). The dataset consists of lymphocytes from patients with indolent B-CLL are compared to those with progressive B-CLL, and consists of intensities of genes in 11 B-CLL patients with stable and 10 patients with clinically progressive disease.
Lung Cancer dataset: This dataset contains samples from patients with different types of interstitial lung disease (ILD) which represent a broad category of restrictive lung disorders, exhibit cellular infiltration and distortion of the interstitium and alveolar gas units. To better understand the disease, the molecular pathways involved in the ILDs needs a detail analysis, as this disease is associated with biological processes, viz. aberrant wound repair, scarring, apoptosis, or fibrosis at tissue or cell levels and with dysregulation of a complex set of cytokines, growth factors, and signalling molecules at molecular level (Cho et al. 2011; Cottin 2013). It consists of intensities of genes in 12 normal and 23 ILD.
3.4 Related dimensionality reduction algorithms compared
We compared the proposed GUFS algorithm with two other popular dimensionality reduction schemes developed in this study.
Principal component analysis (PCA) has been widely applied dimensionality reduction technique and has been widely applied on datasets in all scientific domains (Boutsidis et al. 2008).
Relief is a popular feature selection scheme which searches for nearest neighbours of instances of different classes and weights features according to how well they differentiate instances of different classes (Yu and Liu 2004).
These two methods were used as dimensionality reduction to get single views and the result obtained from these were compared with our multiview approach. We also compared our approach with existing single view supervised learning models for B-cell chronic lymphocytic leukemia (B-CLL), viz. Weighted Voting classification (WtVoting) and Linear Discriminant Analysis (LDA) (Fält et al. 2005), and for interstitial lung disease (ILD), we considered the method being used from (Cho et al. 2011).
4 Results and comparisons
We have used different evaluation methodologies, all focusing on the aspect of detecting multiple views. The results presented are as follows:
-
1.
Baseline cluster quality considering all features.
-
2.
Cluster quality of the best view for each data set obtained using GUFS
-
3.
Effectiveness of the multiview representation obtained using GUFS considering top k views.
-
4.
Biological process dominance in the views.
-
5.
Effectiveness of GUFS as a gene selection technique as compared to related dimensionality reduction schemes.
-
6.
Effectiveness of GUFS as a biologically relavent gene selection technique as compared to related gene selection schemes.
4.1 Baseline cluster quality measure
In table 1 we have summarized the results for two benchmark datasets in terms of sensitivity, specificity, results and model accuracy when the original or all the features/genes are used for clustering the data. The hierarchical clustering is applied on the normalized data set to get the required l number of clusters depending upon the true class label count or the domain knowledge.
4.2 Cluster quality measure for different set of views
Next we study the effectiveness of the views V ⊆ V in terms of cluster quality. Figure 2 presents the cluster performance for only the best view V ⊆ V, in each dataset. Note that the number of views that may be obtained is user defined. Figure 2 shows the effect of varying numbers of views in a view set V being selected for different datasets. We studied the performance of view sets V with size k as 5, 10, 15 and found that the optimal number of views is data specific, and does effect the clustering quality. For our studied datasets, we got view set V for k=10, showing best performance in terms of overall accuracy and cluster size, as seen from figure 2.
Comparison of best accuracy of GUFS for different number of views V ⊆ V selection for Leukemia and Lung dataset; k denotes the size of view set V and V represents the gene set with different genes (g) and gene count (Leukemia (chronic lymphocytic leukemia) and Lung (interstitial lung disease (ILD)).
4.3 Effectiveness of the multiview considering top k views
Figure 3 shows the performance of the views V ⊆ V with varying number of genes, in terms of accuracy. After observing the performance of the best views V from Figure 3, in terms of number of genes present in V and accuracy measure, we studied the effectiveness of multiview representation of data. Table 2 summarizes the performance of top 3 of 10 different views V ⊆ V, shown in figure 3, explored in terms of model accuracy and other evaluation measures for Leukemia and Lung Cancer datasets. On the basis of this performance measure, the top three views we have considered for our further analysis are views 8, 1 and 4 for Leukemia, and view 8, 4 and 5 for Lung data from figure 3. In each case, a small number of genes are involved in each view V, yet each view achieves a significant overall model accuracy as compared to the baseline, as seen from tables 2 and 1, respectively. Thus, the advantage of multiview along with dimensionality reduction is clearly visible from the comparison of results from tables 1 and 2. This demonstrates that GUFS can significantly reduce the number of redundant features in high-dimensional data set and retain highly informative features/genes, which is essential for clustering and/or classification.
4.4 Biological functional association of genes in views
The weight age in terms of percentage of genes related with specific relevant biological process in each view V ⊆ V are shown in tables 3 and 4 for Leukemia and Lung data, respectively. The biological significance of the genes belonging to an enriched functional category can be measured in terms of p-value (Ghosh et al. 2014). The results are validated using p-value statistics and fold enrichment, of enriched attributes/functions (EA), the p-value cut-off of 5 × 10− 2 and FE 1.5 is being considered in our study. The enrichment of the functional association of the gene sets V ⊆ V is evaluated in three top views considered from section 4.3. Tables 3 and 4 reports the top three enriched gene sets of Leukemia and Lung, respectively, from table 2 with their respective gene count, DAVID gene ID count, the functionally EA’s (enriched attributes), and its number, percentage of EA (% of EA) in a view V for a specific biological process and the fold change enrichment (FE) of these related genes.
DAVID gene ID refers to the percentage of DAVID genes in the list associated with particular annotation term. Since DAVID gene ID is unique per gene, it is more accurate to use DAVID ID to present the gene-annotation association by removing any redundancy in user gene list, i.e. two user's IDs represent same gene (Dennis et al. 2003). The threshold of minimum gene counts belonging to an annotation term, has been considered to be equal or greater than 5 (default is 2), as we do not trust the term only having one gene involved. Interestingly, for the database leukemia, view (V) I and III shows relatively large number of EA’s satisfying our p-value and FE cut-off. The relative good percentage of genes is involved in each enriched category as seen from table 3. Similar trend can be seen from table 4 for lung database, specifically, in View-III, where the number of enriched attributes are 13, as per the threshold considered in our study.
Thus, from tables 3 and 4 we can say that our proposed graph-based multiview clustering feature selection is able to select strongly correlated genes, corresponding to those responsible for certain related biological processes. The presence of these important biological processes with higher weightage in terms of percentage as seen from tables 3 and 4 in each views gives an insights for further in depth study of biological relevance of the genes present in these views.
4.5 Comparison of GUFS with existing methods as a gene selection technique
Finally, we studied the effectiveness of GUFS as a gene selection technique. We compared the performance of GUFS with two other feature selection techniques in terms of the quality of clusters obtained using only the selected genes. For each of the dataset we ran all the three feature selection algorithms, namely PCA, Relief and GUFS, in comparison and obtained the best selected genes for each algorithm. We then applied the hierarchical agglomerative clustering on both the original dataset and each of the datasets with only selected features, and we have reported the overall accuracy of the model in tables 1 and 5 respectively. The accuracy results were also compared with the other existing methods from the literature.
Table 5 shows the comparative accuracy of our proposed graph-based multiview clustering feature selection and other studied single view feature selection methods. It can be seen that GUFS, with small number of features (we report only one view from each dataset with best predictive accuracy among all V ⊆ V), performs well in terms of accuracy for leukemia in comparison to weighted voting. Another supervised learning method linear discriminant analysis shows better accuracy in comparison to GUFS for leukemia data. Table 5 shows the accuracy measure for lung dataset is either same or comparable to GUFS when we consider number of genes to be 25 in PCA and 20 for Relief, whereas the study shows that the PCA and Relief shows zero accuracy if the number of feature are considered to be same as GUFS, i.e. 18 for lung data. The PCA is known to be the most popular single view algorithm for mixture model and it requires more stringent separation requirements (Chaudhuri et al. 2009). The accuracy obtained in work done by (Cho et al. 2011) using hierarchical clustering with 1423 genes shows the same accuracy as GUFS. Thus, for lung data all the methods show 100% accuracy. This shows that GUFS on average selects small number of features with higher prediction accuracy.
4.6 Comparison of GUFS with existing methods as a biologically relevant gene selection technique
Finally, we studied the correlation between the selected genes and the studied disease, in each view V ⊆ V, in comparison to other existing methods. To find genes associated with a phenotype or disease, NCBI Gene option was used which integrates information from a wide range of species as stated in section 3.2. Figure 4 shows that the GUFS selects more number of correlated genes in a view, or we can say that GUFS automatically selects groups of genes which have some biological functional similarities. GUFS shows very good result for leukemia and is comparable or better for lung in terms of disease correlated gene selection; this shows the effectiveness of GUFS to select genes, known to be closely associated with a disease.
Comparison in terms of biological significance of views obtained by the proposed GUFS method (considering the three most enriched clusters separately) and other methods for datasets leukemia (chronic lymphocytic leukemia) and lung (interstitial lung disease). RFGS: random forest gene selection; SVST: Support vector sampling technique; SOM: Self-organizing map; GUFS: proposed graph-based multiview clustering feature selection; View I, View II and View III: first, second and third most enriched clusters obtained by GUFS respectively.
5 Conclusion and discussion
The goal of explorative data analysis is to extract the underlying structure of a given set of data. This may be multi-faceted by nature. The proposed graph-based multiview gene selection algorithm (GUFS) attempts to address this problem by extracting multiple clustering views from high-dimensional data. GUFS facilitate gene subset selection from multiple views considering the diversity of each view.
The framework was evaluated through experiments comparing with two popular single view clustering algorithms and other existing methods of gene selection. It is observed that the method can select a small gene subset that provides satisfactory performance in terms of clustering and is able to identify the subset of genes that are biologically significant or correlated. A subsequent analysis of the views is done and found that GUFS shows a very promising result in terms of disease correlated gene selection in comparison to existing methods. These results may facilitate the biologists in unfolding many biological significance questions related with the disease. Using a single source of data limits our understanding of complete biological model. The integration of various kinds of data including gene expression profiles, gene ontology, etc., may provide further insights into the fundamental biology and pathogenesis of the disease and will uncover the collective behaviour of genes.
References
Berriz GF, Beaver JE, Cenik C, Tasan M and Roth FP 2009 Next generation software for functional trend analysis. Bioinformatics 25 3043–3044
Bickel S and Scheffer T 2004 Multi-view clustering; in Proceedings of the Fourth IEEE International Conference on Data Mining, ICDM '04, IEEE Computer Society pp 19–26
Boutsidis C, et al. 2008 Unsupervised feature selection for principal components analysis; in Proc. of the 14th ACM SIGKDD Int. Conf. on Knowledge discovery and data mining pp 61–69
Bruno E and Marchand-Maillet S 2009 Multiview clustering: a late fusion approach using latent models; in SIGIR (ACM) pp 736–737
Chaudhuri K, Kakade SM, Livescu K, and Sridharan K 2009 Multi-view clustering via canonical correlation analysis; in Proceedings of the 26th Annual International Conference on Machine Learning, ICML '09 pp 129–136
Chen X, Xu X, Huang J and Ye Y 2013 TW-(k)-means: automated two-level variable weighting clustering algorithm for multiview data. IEEE Trans. Knowl. Data Eng. 25 932–944
Cho JH, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, Dakhlallah D, Price J, et al. 2011 Systems biology of interstitial lung diseases: integration of mrna and microrna expression changes. BMC Med. Genet. 4 8
Chuang HY, Rassenti L, Salcedo M, Licon K, Kohlmann A, Haferlach T, Foà R, Ideker T, et al. 2012 Subnetwork-based analysis of chronic lymphocytic leukemia identifies pathways that associate with disease progression. Blood 120 2639–2649
Codony C, Crespo M, Abrisqueta P, Montserrat E and Bosch F 2009 Gene expression profiling in chronic lymphocytic leukaemia. Best Pract. Res. Clin. Haematol. 22 211–222
Cottin V 2013 Interstitial lung disease. Eur. Respir. Rev. 22 26–32
Cui Y, Fern, XZ, Dy and JG 2007 Non-redundant multi-view clustering via orthogonalization. Proc. 7th IEEE International Conference on Data Mining (ICDM'07) pp 133–142
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA, et al. 2003 David: database for annotation, visualization, and integrated discovery. Genome Biol. 4 P3
Di W and Crawford MM 2012 View generation for multiview maximum disagreement based active learning for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 50 1942–1954
Ding CH 2003 Unsupervised feature selection via two-way ordering in gene expression analysis. Bioinformatics 19 1259–1266
Dudoit S, Yang YH, Callow MJ and Speed TP 2002 Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin. 12 111–140
Fält S, Merup M, Gahrton G, Lambert B and Wennborg A 2005 Identification of progression markers in b-cll by gene expression profiling. Exp. Hematol. 33 883–893
Fang G, Kuang R, Pandey G, Steinbach M, Myers CL and Kumar V 2010 Subspace differential coexpression analysis: problem definition and a general approach; in Pacific Symp Biocomput 15 145–56
Ghosh A, Dhara BC and De RK 2014 Selection of genes mediating certain cancers, using a neuro fuzzy approach. Neurocomputing 133 122–140
Gupta R, Rao N and Kumar V 2011 Discovery of error-tolerant biclusters from noisy gene expression data. BMC Bioinforma. 12 S1
Hong Y, Kwong S, Chang Y and Ren Q 2008 Consensus unsupervised feature ranking from multiple views. Pattern Recogn. Lett. 29 595–602
Huang DW, Sherman BT and Lempicki RA 2008 Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 4 44–57
Huang DW, Sherman BT and Lempicki RA 2009 Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13
Jaeger J, et al. 2003 Improved gene selection for classification of microarrays; in Pacific Symp. on Biocomputing pp 53–64
Ji J, Zhang A, Liu C, Quan X and Liu Z 2014 Survey: functional module detection from protein-protein interaction networks. IEEE Trans. Knowl. Data Eng. 26 261–277
Kim YM, Amini MR, Goutte C and Gallinari P 2010 Multi-view clustering of multilingual documents; in SIGIR (ACM) pp 821–822
Kohane IS, Butte AJ and Kho A 2002 Microarrays for an integrative genomics. MIT press
Li G, et al. 2008 A novel unsupervised feature selection method for bioinformatics data sets through feature clustering; in 2008 I.E. Int. Conf. on Granular Comput pp 41–47
Mitra P and Swarnkar T 2012 Graph based unsupervised feature selection for microarray data; in Proceedings of the 2012 I.E. International Conference on Bioinformatics and Biomedicine Workshops (BIBMW), IEEE Computer Society pp 750–751
Mitra P, Murthy CA and Pal SK 2002 Unsupervised feature selection using feature similarity. IEEE Trans. Pattern Anal. Mach. Intell. 24 301–312
Muller E, Gunnemann S, Farber I and Seidl T 2012 Discovering multiple clustering solutions: Grouping objects in different views of the data; in IEEE 28th International Conference on Data Engineering, IEEE pp 1207–1210
Pirim H, Ekiolu B, Perkins AD and Yüceer C 2012 Clustering of high throughput gene expression data. Comput. Oper. Res. 39 3046–3061
Safran M, Dalah I, Alexander J, Rosen N, Stein TI, Shmoish M, Nativ N, Bahir I, et al. 2010 GeneCards version 3: the human gene integrator. doi:10.1093/database/baq020
Sharma A, Imoto S and Miyano S 2012a A filter based feature selection algorithm using null space of covariance matrix for dna microarray gene expression data. Curr. Bioinforma 7 289–294
Sharma A, Imoto S and Miyano S 2012b A top-r feature selection algorithm for microarray gene expression data. IEEE/ACM Trans. Comput. Biol. Bioinformatics 9 754–764
Sun S 2013 A survey of multi-view machine learning. Neural Comput. Applic. 23 2031–2038
Swarnkar T, Simões SN, Martins-Jr DC, Anura A, Brentani H, Hashimoto RF and Mitra P 2014 Multiview clustering on ppi network for gene selection and enrichment from microarray data. In: IEEE International Conference on BioInformatics and BioEngineering
Tibshirani R and Witten D 2007 A comparison of fold-change and the t-statistic for microarray data analysis. Technical report, Stanford, CA: Stanford University
Varshavsky R, Gottlieb A, Linial M and Horn D 2006 Novel unsupervised feature filtering of biological data. Bioinformatics 22 e507–e513
Xiao Y, Hsiao TH, Suresh U, Chen HIH, Wu X, Wolf SE and Chen Y 2014 A novel significance score for gene selection and ranking. Bioinformatics 30 801–807
Xu C, Tao D and Xu C 2013 A survey on multi-view learning. arXiv preprint arXiv:1304.5634
Yang P, Hwa Yang Y, Zhou BB and Zomaya YA 2010 A review of ensemble methods in bioinformatics. Curr. Bioinforma. 5 296–308
Yu L and Liu H 2004 Efficient feature selection via analysis of relevance and redundancy. J. Mach. Learn. Res. 5 1205–1224
Yu S, Tranchevent LC, De Moor B and Moreau Y 2010 Gene prioritization and clustering by multi-view text mining. BMC Bioinf. 11 28
Author information
Authors and Affiliations
Corresponding author
Additional information
[Swarnkar T and Mitra P 2015 Graph-based unsupervised feature selection and multiview clustering for microarray data. J. Biosci.] DOI 10.1007/s12038-015-9559-8
Rights and permissions
About this article
Cite this article
Swarnkar, T., Mitra, P. Graph-based unsupervised feature selection and multiview clustering for microarray data. J Biosci 40, 755–767 (2015). https://doi.org/10.1007/s12038-015-9559-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-015-9559-8