Abstract
In budding yeast, Saccharomyces cerevisiae, the phosphate signalling and response pathway, known as PHO pathway, monitors phosphate cytoplasmic levels by controlling genes involved in scavenging, uptake and utilization of phosphate. Recent attempts to understand the phosphate starvation response in other ascomycetes have suggested the existence of both common and novel components of the budding yeast PHO pathway in these ascomycetes. In this review, we discuss the components of PHO pathway, their roles in maintaining phosphate homeostasis in yeast and their conservation across ascomycetes. The role of high-affinity transporter, Pho84, in sensing and signalling of phosphate has also been discussed
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Role of phosphate
Phosphorus as inorganic phosphate (Pi) is an essential component of various biomolecules responsible for cellular structure and storage of cellular energy and is thus indispensable for survival of all living organisms. Hence, it is crucial for living organisms to maintain proper phosphate homeostasis, as any imbalance (hyper- or hypo-phosphatemia) will impact cellular processes, viz. cell differentiation and proliferation, metabolic functions, cytoskeletal organization (Dick et al. 2011). Non-motile microorganisms, which often encounter limiting conditions of essential nutrients, survive such adverse conditions through evolved complex signal transduction pathways usually involving a sensor, a transporter or a transceptor system. These pathways both by sensing or by limiting the uptake assess nutrient status in the ambient environment and initiate a signal transduction cascade, which leads to the activation of nutrient starvation responses. These responses help the cell in surviving the prevailing adverse conditions and to ensure that a wide variety of physiological activities properly adapt to the environmental conditions (Wykoff et al. 2007). However, if the starvation persists longer, the cells enter into a quiescence phase and can eventually die.
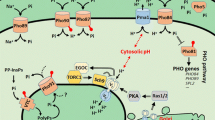
To enable survival in variable and scarce phosphate conditions, budding yeast, Saccharomyces cerevisiae, has evolved a phosphate responsive signalling and metabolism pathway, known as PHO pathway, which monitors cytoplasmic levels of phosphate. This pathway regulates genes which are required for maintaining proper phosphate homeostasis such as genes required for phosphate uptake from extracellular environment, mobilization of internal phosphate reserves (polyphosphates) during fluctuations in internal phosphate levels, involved in phosphate sensing and signalling and scavenging phosphate from extracellular sources (Mouillon and Persson 2006; Wykoff et al. 2007) (table 1). PHO pathway consists of three important components: (1) a membrane transporter system consisting of low- and high-affinity phosphate transporters, which enables survival over diverse phosphate concentration; (2) polyphosphates (polyP), which are phosphate reserves and are mobilized first during phosphate demands; and (3) the PHO regulation system, the core signalling cascade, which integrates signals from extra and intracellular environment and depending upon intracellular phosphate status, either up- or down-regulates the PHO responsive genes (Lenburg and O'Shea 1996; Oshima 1997; Persson et al. 2003; Mouillon and Persson 2006; Secco et al. 2012) (figures 1 and 2).
PHO pathway when environmental phosphate concentration is high: In high-Pi conditions, Pho4, transcriptional activator of PHO pathway, is hyper-phosphorylated by Pho80-Pho85 complex, which switches off PHO pathway. Low-affinity phosphate transporters, Pho87 and Pho90, uptake phosphate, excess of which is stored in form of polyP in vacuoles.
PHO pathway when environmental phosphate concentration is low: Under low-Pi conditions, Pho81 represses Pho80-Pho85 complex, which in turn leads to hypo-phosphorylation of Pho4. This Pho4 in turn activates PHO pathway genes mainly including high-affinity transporters (Pho84, Pho89) and secretory phosphatases (Pho5, Pho11 and Pho12).
2 Components of PHO pathway
2.1 Membrane transporter system
Plasma membrane of yeast cells is impermeable to phosphate because of its charged nature. Thus, two types of specialized transporters are required to facilitate entry of phosphate inside the cells, which are active over wide range of pH and environmental phosphate concentration (Persson et al. 1999).
2.1.1 Low-affinity transport system
This consists of two H+/Pi symporters: Pho87 and Pho90 (Roomans et al. 1977; Persson et al. 1999) (figure 1). These transporters have 12 transmembrane domains with a long N-terminal SPX domain (Syg1-Pho81-Xpr1), a domain that is common to many PHO genes. Earlier, Pho91 was also considered as a low-affinity transport system but it is localized onto the vacuoles and, thus, not involved in the uptake of phosphate from the extracellular environment (Wykoff and O'Shea 2001). These transporters, Pho87 and Pho90, have a Km of 1 mM and cater to the cellular phosphate requirement when yeast cells are grown in high-phosphate concentration. When the ambient levels of phosphate drops below their Km, there occurs PHO-pathway-mediated transporter switching and the high-affinity transporters replace the low-affinity ones. Under low-phosphate conditions, the low-affinity transporters undergo vacuolar degradation. Spl2, a Pho4 target, mediates vacuolar degradation of Pho87; however, the mechanism for Pho90 is not known. GFP-based localization experiments have suggested that for Pho90, the post-transcriptional regulation upon phosphate starvation is independent of both Spl2 and Pho4. However, it has been shown that SPX domain of Pho90 is required for this vacuolar degradation (Ghillebert et al. 2011). It appears that yeast cells use two distinct mechanisms for regulation of low-affinity transporters. This degradation of low-affinity transporters, upon phosphate scarcity, serves as a positive feedback loop for up-regulation of PHO pathway by further decreasing phosphate uptake by low-affinity transporters. However, the exact reason for this down-regulation of low-affinity transporters is not known. These low-affinity transporters have 12 transmembrane domains and, thus, occupy a large space on the membrane. Therefore, it has been proposed that the post-transcriptional down-regulation might create more space on the membrane for the high-affinity transporters.
2.1.2 High-affinity transport system
This system consists of two transporters: Pho84, a H+/Pi- symporter and Pho89, a Na+/Pi- symporter with a Km of 10 μM (figure 2). These transporters also have 12 transmembrane domains and have different pH optima: pH 4.5 and 9.5 for Pho84 and Pho89, respectively. This difference in pH optima has been proposed to ensure phosphate uptake over wide pH range. Both these transporters are under tight regulation of PHO pathway and are expressed only in phosphate limiting conditions. However, their maximum activity at their pH optima is quite different, as the activity of Pho84 is 100-fold higher than that of Pho89, making Pho84 the main high-affinity phosphate transporter (Ogawa et al. 2000; Persson et al. 2003; Mouillon and Persson 2006; Secco et al. 2012).
2.2 Polyphosphate metabolism
Polyphosphates (PolyP) are linear phosphate polymers in which individual phosphates are linked together by high-energy phosphoranhydride bonds. The chain length varies from a few to hundreds of phosphate residues (Kornberg et al. 1999). These polyP are present in almost all organisms ranging from bacteria, fungi, plants and animals (Kulaev and Kulakovskaya 2000). In yeast, almost all (around 99%) of polyP is present in vacuoles (Kornberg et al. 1999) and it is this fraction that is mobilized during phosphate limiting conditions (figures 1 and 2). The remaining fraction is present in other subcellular locations like cytosol, internal cell wall layer, mitochondria and nucleus (Secco et al. 2012). When a yeast cell is grown in phosphate rich environment, phosphate is taken inside the cells by the low-affinity transporters and is utilized for various cellular functions. The excess of phosphate is stored in the form of polyP and helps the cell meet increased phosphate demands during cell growth and division.
Genome-wide expression analysis, followed by detection of polyP levels by PAGE and enzymatic analysis (Ault-Riché et al. 1998), identified genes involved in polyP metabolism which had one or more Pho4 binding sites, CACGTG or CACGTT (Oshima 1997), within the 500 bp promoter region. The synthesis of polyP is mediated by PHM1, PHM2, PHM3 and PHM4, as the mutants of these genes showed significant reduction in polyP accumulation (Ogawa et al. 2000) (figure 1). There was negligible polyP accumulation in case of phm3Δ, phm4Δ and phm1Δphm2Δ double deletion. Data on measurement of rate of phosphate uptake suggested that polyP levels also help in maintaining the rate at which the cell takes up phosphate. Yeast cells that are defective in polyP synthesis (phm2Δ, phm1Δphm2Δ, phm3Δ and phm4Δ) showed a delay in phosphate uptake profile as compared to the wild type (Ogawa et al. 2000). It has been also been proposed that polyP synthesis is required to maintain a check on the intracellular Pi levels (Ogawa et al. 2000).
In contrast, not much is known about the genes involved in polyP degradation. Till date, two polyphosphatases are known: PPX1, an exopolyphosphatase (Wurst et al. 1995), which acts at the terminals of polyP chains and PHM5 or PPN1, an endopolyphosphatase, which acts in between the polyP chains (Ogawa et al. 2000; Sethuraman et al. 2001) (figure 2). However, in a double deletion mutant of these two genes (ppx1Δphm5Δ), polyP utilization is delayed but still occurs, suggesting the presence of other polyphosphatases (Lichko et al. 2008). In addition to this, phm5 mutant also shows growth defect and is unable to survive in minimal medium in the stationary phase. It has been suggested that the excess accumulation of long polyP chains might be the reason for loss of viability of phm5 mutant (Ogawa et al. 2000; Sethuraman et al. 2001).
PolyP reserves get utilized first upon a phosphate limitation event and are mobilized immediately, possibly by activation of polyphosphatases independent of PHO pathway. However, if these phosphate reserves are not enough to meet the phosphate requirement, i.e. the starvation is either severe or persists for longer time, yeast cells up-regulate the PHO responsive genes via the PHO regulatory system.
2.3 PHO regulation system
The PHO regulation system consists of the core PHO genes including cyclin-dependent kinase inhibitor, Pho81, cyclin-cyclin-dependent kinase complex, Pho80-Pho85, the transcription factor, Pho4 and the PHO responsive genes involved in acquisition, storage and uptake of phosphate (Lenburg and O'Shea 1996; Oshima 1997; Persson et al. 2003; Mouillon and Persson 2006; Secco et al. 2012) (figure 2). The activity of this system is dependent on the availability of phosphate and is mediated by the basic helix loop helix transcription factor, Pho4 (Berben et al. 1990), which binds as a homodimer (Shao 1998) in association with a co-transcription factor, Pho2 (Vogel et al. 1989). Pho4 binds to a specific motif, CACGTG/CACGTT, found in the promoter region of the PHO-responsive genes (Oshima 1997). The activity of Pho4 is regulated by Pho80-Pho85 mediated phosphorylation of its five-serine proline (SP) dipeptides SP1, SP2, SP3, SP4 and SP6, which regulates its subcellular localization – nuclear or cytoplasmic (Komeili 1999). Phosphorylation at sites SP2 and SP3 regulate the nuclear export of Pho4p, at SP4 inhibits the nuclear import and at SP6 blocks the interaction of Pho4p with Pho2p (Komeili 1999). However, the role of phosphorylation at SP1 is not known. The activity of Pho2p is also under the regulation of phosphorylation by Cdc2/Cdc28-type kinase at the SPIK site (amino acid residues 230–233). This phosphorylation improves both the transcriptional activation activity and interaction between Pho4 and Pho2 (Liu 2000). This whole signalling cascade is under the control of CDK inhibitor, Pho81, which negatively regulates the activity of Pho80-Pho85 complex. Pho81 is constitutively localized in the nucleus independent of the Pi levels, where it interacts with Pho80 under low- and high-Pi conditions. This leads to the formation of a ternary complex Pho81-Pho80-Pho85 (Ogawa et al. 2000; Mouillon and Persson 2006; Secco et al. 2012).
2.3.1 Yeast cells grown under low-phosphate conditions
Studies have shown that when the ambient level of phosphate drops below the Km of low-affinity transporters, yeast cells immediately respond by mobilizing their polyP reserves. However, if a starvation event persists longer or is severe in nature, a complex metabolic network mediated by inositol pyrophosphates is activated and helps the cell in sensing the low-phosphate environment and also in Pho81 mediated inhibition of Pho80-Pho85 kinase activity. Biochemical experiments (involving use of culture extracts from cells grown in high Pi and low Pi and chromatography) performed to identify regulators of Pho80-Pho85-Pho81 activity suggest the role of an evolutionary conserved metabolite, inositol heptakisphosphate (IP7), in this regulation (Lee et al. 2007). IP7 is produced by sequential phosphorylation reaction of inositol 1,4,5-trisphosphate (IP3) by inositol polyphosphate multikinase Arg82. This results in the formation of inositol tetrakisphosphate (IP4) and inositol pentakisphosphate (IP5), which further undergoes phosphorylation by Ipk1 to form inositol hexakisphosphate (IP6). IP6 then undergo phosphorylation by Kcs1 and Vip1 to form inositol pyrophosphates (IP7) and double pyrophosphorylated form of IP8, bis-diphosphoinositol tetrakisphosphate (PP-IP4-PP), respectively (Odom et al. 2000; Saiardi et al. 2001; Mulugu et al. 2007; Saito 2010). Since upon phosphate starvation, the intracellular levels of IP7 increases, yeast mutants defective in production of IP7 show failure of inhibition of Pho80-Pho85, even in phosphate-starved conditions (Lee et al. 2007). The increased level of IP7 allosterically modulates Pho81 such that it leads to a conformational change in the ternary complex Pho81-Pho80-Pho85. This conformational change prevents access to the kinase active site by Pho4, which leads to lack of Pho4 phosphorylation (Lee et al. 2009; Lee et al. 2011). Hypo-phosphorylated Pho4 remains localized to the nucleus where it interacts with Pho2 to activate phosphate responsive genes, including the high-affinity transporters, PHO84 and PHO89, secretory phosphatase, PHO5, and a negative regulator of low-affinity transporter, SPL2 (figure 2). The high-affinity transporters, as discussed above, have a very low Km and thus can uptake phosphate even under very low extracellular phosphate concentrations. On the other hand, the secretory phosphatases help in phosphate scavenging by cleaving phosphate from different phosphorylated substrates like nucleic acids, phospho-sugars, phospholipids, etc. (Ogawa et al. 2000; Persson et al. 2003; Mouillon and Persson 2006; Secco et al. 2012) (figure 2).
2.3.2 Yeast cells grown under high- or normal-phosphate conditions
When the ambient phosphate concentration is either high or normal, phosphate is taken in the cell by low-affinity transporters and is utilized for various cellular processes. The excess of cytoplasmic phosphate is stored in the vacuoles in the form of polyP. During such conditions, there is no accumulation of IP7. This lack of IP7 prevents the activation of Pho81 and thus the conformational change in the ternary complex (Lee et al. 2009, Lee et al. 2011). As a result, the kinase domain of Pho85 remains available to the transcription factor Pho4 (figure 1). The hyper-phosphorylated Pho4 is a preferred substrate for the nuclear export receptor, Msn5 and is exported to the cytoplasm (Kaffman et al. 1998) (figure 1). The absence of hypo-phosphorylated nuclear Pho4, thus, prevents the up-regulation of phosphate responsive genes under high-phosphate conditions (Ogawa et al. 2000; Persson et al. 2003; Mouillon and Persson 2006; Secco et al. 2012) (figure 1).
A complex feedback mechanism exists, which enables the yeast cells to switch from the low- to high-affinity transporter system and vice versa, depending upon the environmental Pi levels (Wykoff et al. 2007). During high-phosphate conditions, cells use low-affinity transport system to cater the phosphate requirements. However, as the phosphate conditions become limiting, PHO pathway gets activated and it causes the up-regulation of high-affinity transporters and consequently, down-regulation of the low-affinity transporters. A recent study highlighted the advantages of having a dual transport system consisting of low- and high-affinity transporters (Levy et al. 2011). In case of a single transport system, when the nutrients in the ambient environment start depleting initially, there is no effect on cellular nutrient pool as the transporters uptake nutrients. However, as the external nutrient level approaches Km of the transporters, the uptake decreases and a starvation response is initiated. Soon after the starvation response, the cell growth is limited. Thus, a single transporter system gives cells less time to prepare for future starvation events (Levy et al. 2011). In contrast, it has been seen that the dual transport system helps the cell in prolonging the time period between starvation response and growth limitation. In this case, the two low- and high-affinity transporters have different dissociation constants. When the ambient nutrient levels are high, low-affinity transporters transport nutrients inside the cell. As the levels of the nutrient drop below Km of the low-affinity transporters, the uptake of nutrients via these transporters decreases and a starvation response is initiated, which includes the up-regulation of high-affinity transporters. These high-affinity transporters then uptake nutrients until the nutrient levels reach Km of high-affinity transporters, following which the cell growth is inhibited. Thus, the existence of PHO dual transport system increases the time period between the initiation of starvation response and limitation of growth and this foreseeing of a possible future PHO limitation might help the cell to adapt better (Levy et al. 2011).
3 Comparative analysis of PHO pathway in other ascomycetes
3.1 PHO pathway in Schizosaccharomyces pombe
Recent attempts to understand the phosphate starvation response in other ascomycetes have suggested the existence of both common and novel components of the budding yeast PHO pathway in other ascomycetes (Henry et al. 2011). Studies in Schizosaccharomyces pombe (Sp) have suggested the occurrence of a phosphate starvation response consisting of high-affinity transporter SpPHO84 and acid phosphatase SpPHO1 (orthologues of Saccharomyces cerevisiae (Sc): PHO84 and PHO5, respectively) (Maundrell et al. 1985; Schwaninger et al. 1990; Schweingruber et al. 1992; Henry et al. 2011; Carter-O'Connell et al. 2012) (figure 3b). However, homology searches have failed to identify the orthologues of the core PHO regulon (ScPHO4, ScPHO2 and ScPHO81) (Henry et al. 2011). Further, acid phosphatase activity in low-phosphate conditions did not confirm roles of putative orthologues of cyclin-CDK complex SpPHO80-SpPHO85 in phosphate starvation response (Henry et al. 2011). However, for upstream pathways like IP7 and polyphosphate metabolism, similar mutant phenotypes were seen in both the yeasts. It is, therefore, possible that these two fungi have evolved a similar phosphate starvation response by two different mechanisms, suggesting the existence of common ancestry (Henry et al. 2011). Role for a zinc transcription factor, SpPHO7, and few upstream proteins like IP7 and SpCsk1 in regulation of phosphate transcriptional stress response has been identified. Like ScPHO4, SpPHO7 also binds to an upstream region of SpPHO1 (orthologue of ScPHO5). However, unlike ScPHO4, which is tightly under the control of phosphate status of the cell and binds to PHO pathway genes only during low-phosphate conditions, SpPHO7 is more of a general stress response transcription factor (involved in iron, copper, osmotic and alternative carbon utilization stress) and is always bound to its targets. In the phosphate-enriched condition, SpCsk1 prevents the full activation of the starvation response. During low-phosphate conditions, an unknown mechanism prevents this repression leading to the full activation of phosphate starvation response ( Henry et al. 2011; Carter-O'Connell et al. 2012).
Schematic representation of comparison of PHO pathway in S. cerevisiae, S. pombe, C. glabrata and N. crassa: (a) In S. cerevisiae, levels of IP7 increase in response to limited phosphate levels. IP7 allosterically modulates Pho81 and thus the ternary complex Pho81-Pho80 and Pho85. This in turn leads to hypo-phosphorylated Pho4 and thus activation of phosphate starvation response. (b) In S. pombe, phosphate depletion has been proposed to increase levels of IP7, which in turn directly or indirectly by an unknown mechanism, regulates the transcription factor Pho7 that is usually under Csk1 repression. However, during phosphate starvation the Csk1 mediated repression of Pho7 is removed and phosphate starvation response is induced (adapted from Henry et al. 2011). (c) In C. glabrata, a phosphate starvation response similar to S. cerevisiae is present. Low phosphate levels cause increase in IP7 concentration, which in turn inactivates the ternary complex Pho81-Pho80-Pho85. This leads to the activation of C. glabrata homologue of transcription factor Pho4. However, unlike S. cerevisiae, Pho2 is not required for up-regulation of phosphate starvation response (adapted from Kerwin and Wykoff 2009). (d) In N. crassa, PHO pathway is similar to S. cerevisiae. The core pathway consists of NUC-2, MAK-2, PREG, PGOV and NUC-1 (Gras et al. 2013). The dotted line indicate probable interactions.
3.2 PHO pathway in Candida species
In case of Candida glabrata (Cg), a phosphate starvation response similar to S. cerevisiae has been proposed (figure 3c). Comparative genomic analysis between S. cerevisiae (figure 3a) and C. glabrata (figure 3c) for PHO pathway genes has identified orthologues of most of the genes. Analyses of deletion mutants of these orthologues for acid phosphatase activity show similar phenotypes in C. glabrata as S. cerevisiae, suggesting similar functions in phosphate starvation response (Kerwin and Wykoff 2009). Upon phosphate starvation, Candida up-regulates a starvation response including secretion of phosphatases and up-regulation of high-affinity transporter (ScPHO84). However, homology searches have failed to identify orthologues of acid phosphatases (ScPHO5, ScPHO11, ScPHO12) and the high-affinity transporter ScPHO89 in Candida species (Kerwin and Wykoff 2009). Instead, CgPMU2, a gene containing a phosphomutase-like domain, identified by complementation of Scpho5Δ strain with C. glabrata genomic DNA library, encodes for phosphatases secreted in the periplasmic space during low-phosphate conditions (Orkwis et al. 2010). This starvation response is expected to be under the control of CgPHO4, which is regulated by its nuclear or cytoplasmic localization. CgPHO4 is regulated by cyclin/CDK/CDK-inhibitor complex, ScPHO80-ScPHO85-ScPHO81. However, CgPHO2 (a co-activator of CgPHO4) is not required for up-regulation of phosphate starvation response in Candida (Kerwin and Wykoff 2009; 2012). Further, the motif CACGTG/CACGTT found in the promoter region of all ScPHO4-regulated genes (Oshima 1997) is absent in C. glabrata (Kerwin and Wykoff 2009). The inability to identify SPL2 orthologues indicates that either the positive feedback loop of PHO pathway is absent or C. glabrata has other analogs. Therefore, although the general response to phosphate starvation seems similar in the two ascomycetes, there are some differences in the manner the pathway is regulated which might have arisen due to different ecological niches and the type of the phosphate resources available to them (Kerwin and Wykoff 2009) (see figure 3).
Early observation in C. albicans pathogenic isolates by Cassone et al. (1983) gave indirect evidence of relationship between external Pi and hyphal growth, which was reduced in absence of external Pi. Several candidate PHO pathway genes have been shown to influence hyphal formation and filamentation (Nobile et al. 2012). However, it is surprising that even with 24 genes associated with PHO pathway in C. albicans genome, not much is known about the function of most genes and their role in fugal pathogenicity. Phosphatase activity, which is a phosphate limitation response, was previously shown to be very low or delayed in C. albicans (Smith et al. 1973). However, recently Romanowski et al. (2012) showed that the phosphate limitation had a dramatic effect on phosphatase activity, which corroborates with S. cerevisiae results. This phosphate limitation leads to extensive filamentation observed in C. albicans and could represent a mechanism by which it invades tissues to obtain phosphate. This extensive filamentation was found to be dependent on CaPHO4 (Romanowski et al. 2012). CaPHO4, the homologue of ScPHO4, is required for growth in phosphate-depleted medium (Homann et al. 2009). Up-regulation of CaPHO4 in response to phosphate limitation lead to an increased expression of the secreted phosphatase (Romanowski et al. 2012). Negative regulator of PHO pathway, CaPHO85, the homologue of ScPHO85, was shown to complement Scpho85Δ (Miyakawa 2000) and was involved in temperature-dependent filamentation under control of CaHsp90 (Shapiro et al. 2012). In contrast, CaGRF1, the homologue of ScPHO2, is not required for phosphate limitation response (Homann et al. 2009). Thus, it is possible that pathogenicity and hyper-filamentation (a fungal virulence factor) in C. albicans is PHO pathway dependent.
3.3 PHO pathway in Neurospora crassa
The regulation of phosphate homeostasis in Neurospora crassa is similar to the regulation in S. cerevisiae (figure 3d). Neurospora also has a core phosphate regulatory unit consisting of NUC-1, PREG, PGOV and NUC-2, which have homology to ScPho4, ScPho80, ScPho85 and ScPho81, respectively (Metzenberg 1979; Gras et al. 2007) (figure 3d). NUC-1 is a basic helix-loop-helix transcriptional regulator whose activity is under the control of PREG, a cyclin-like protein kinase and PGOV, which is a cyclin-dependent protein kinase (Peleg and Metzenberg 1994; Kang and Metzenberg 1990; Peleg et al. 1996). The PREG-PGOV complex is in turn regulated by the ankyrin repeat protein, NUC-2 (Poleg et al. 1996). When Neurospora is grown under adequate supply of phosphate, PREG-PGOV complex is active and it antagonizes the function of NUC-1, preventing the up-regulation of phosphate starvation response genes (Poleg et al. 1996). An immunofluorescence study to understand the effect of ambient phosphate concentration on posttranslational regulation of NUC-1, suggests that under such high-Pi conditions, NUC-1 predominantly localizes to the cytoplasm (Peleg et al. 1996). When the ambient phosphate levels becomes limiting, NUC-2 inhibits the PREG-PGOV complex. Under such conditions, there occurs a 5-fold increase in the levels of NUC-1 and it accumulates in the nucleus (Peleg et al. 1996), where it binds to a 6 bp motif CACGTG in the promoter region of phosphate responsive genes. This includes genes encoding for phosphate permeases and repressible alkaline phosphatase that help the cell to survive in the prevailing low-phosphate environment (Metzenberg 1979; Lenburg and O'Shea 1996; Gras et al. 2007; Leal et al. 2007). In order to provide insight into molecular events involved in phosphate sensing, a suppression subtractive hybridization study was performed by Gras et al. (2007). This study identified genes that might play a role in phosphate sensing and signalling. Interestingly, as compared to a wild-type strain, in a nuc-2 deletion strain, genes involved in initiation of mRNA translation were found to be significantly up-regulated. The other genes that showed a differential expression covered diverse cellular processes like amino acid metabolism, protein synthesis, protein fate, extracellular matrix protein precursor signal transduction mechanisms (MAP kinase) and transport facilitation (Gras et al. 2007). In another recent study by Gras et al. (2013), the role of MAP kinase MAK-2 in phosphate signalling pathway has been further explored. Comparison of transcriptional profile of a mak-2 deletion strain, grown in low phosphate with a wild-type grown in low- and high-phosphate conditions revealed over 900 differentially expressed genes. Detailed analysis of the differentially expressed gene set showed the inability of the mak-2 deletion strain to sense the ambient phosphate conditions. Irrespective of the external Pi concentration, the mak-2 deletion strain showed a profile similar to wild-type strain grown in high-Pi conditions. Several genes like those encoding for acetamidase, chitinase, acid and alkaline phosphatases, carboxipeptidase S1, glucan 1,3-β-glucosidase and NUC-2 and PHO-4 proteins were down-regulated in a mak-2 deletion mutant when grown both in low- and high-Pi conditions. Interestingly, NUC-2 and NUC-1 mutants also show a similar phenotypic profile. In silico protein interactions have suggested that MAK-2 interacts with PREG and PGOV and not with MUC-2 and MUC. Based on all these results, it has been proposed that under adequate supply of phosphate, MAK-2 is inactive. In such a case, PREG-PGOV complex interacts with the transcriptional regulator NUC-1, preventing its nuclear localization and thereby prevent up-regulation of phosphate starvation response genes. However, when the phosphate concentration becomes limiting, MAK-2 inhibits the PREG-PGOV complex ultimately leading to nuclear localization of NUC-1 and up-regulation of phosphate responsive genes (Gras et al. 2013).
4 Role of Pho84 in phosphate signalling, sensing and transport
Pho84, a high-affinity transporter, is up-regulated upon a phosphate starvation event. A pho84Δ deletion strain shows a number of phenotypes: arsenate resistance, poor growth on low-phosphate medium, constitutive up-regulation of PHO pathway as measured by Pho5 activity and negligible levels of polyP (Wykoff et al. 2007). These all phenotypes are based on the existence of a feedback mechanism in PHO pathway. It has been proposed that because of low polyP levels, a pho84Δ deletion strain lacks the phosphate buffering system (Wykoff et al. 2007; Ogawa et al. 2000). As a consequence, yeast cells up-regulate their PHO pathway every time the cellular phosphate demand increases, e.g. during cell growth and division. Due to this up-regulation, the low-affinity transporters get down-regulated, further lowering the phosphate uptake. Since the major high-affinity transporter, Pho84 is deleted, this cell enters a state in which PHO pathway is constitutively on but the phosphate uptake is negligible (Wykoff et al. 2007). While the reason for the depleted polyP reserves in a pho84Δ strain (Ogawa et al. 2000) is not known, it is possible that there might be some alternate pathway regulating this step.
In the common lab strain S288c, PHO pathway has been found to be constitutively up-regulated with 15 of 32 PHO pathway genes up-regulated (Gagneur et al. 2009). Since, pho84Δ leads to constitutive up-regulation of PHO pathway even in a phosphate-rich environment and S288c shows such an up-regulation (even in high Pi), it has been hypothesized that Pho84 might be non-functional in S288c. Comparison of protein sequence of Pho84 in 26 yeast strains ( http://www.yeastgenome.org/cgi-bin/FUNGI/alignment.pl?locus=YML123C ) showed that a non-conservative amino acid substitution, P259L, is a rare mutation found only in lab strains including S288c and isogenic strains like BY4741 and BY4742. It has been suggested that this substitution of proline by leucine in Pho84 transmembrane domain may prevent kinks, which reduces protein flexibility and therefore, might affect its transporter function (Cordes et al. 2002).
Studies done to understand the effect of phosphate on growth, metabolism and other regulatory functions in S. cerevisiae have identified links between phosphate signalling and Protein Kinase A (PKA) pathway. Re-addition of phosphate to a phosphate-starved yeast cell, in the presence of glucose, has been shown to up-regulate PKA pathway and causes trehalase activation, mobilization of trehalose, repression of STRE genes, etc. Analysis of yeast strains containing strong constitutive overexpression of phosphate transporters (Pho87, Pho90, Pho84, Pho89 and Pho91) revealed the contribution of each of the phosphate transporters in transport and signalling property. Yeast strains containing constitutive overexpression of Pho87 and Pho84 were able to sustain phosphate induced trehalase activation, whereas the others could not. Also, this phosphate-induced trehalase activation in phosphate-starved cells was maximum in strain containing a constitutive expression of Pho84, suggesting a key role of Pho84 in PKA signalling as well as phosphate transport (Giots et al. 2003). The existence of these two features in Pho84: a transporter and a receptor, identified it as a ‘transceptor‘. Furthermore, it has also been shown that this phosphate-mediated induction of PKA pathway in phosphate-limited cells can cause down-regulation of Pho84 via a negative feedback loop. This down-regulation is caused by phosphorylation of Pho84 followed by ubiquitination which marks the protein for endocytosis and eventually causes degradation in vacuoles (Lundh et al. 2009). Both phosphorylation and ubiquitination occur at amino acid residues in the loop connecting the two helical transmembrane domains, as the Pho84 central loop mutant (with 304-372 amino acid deletion) shows no ubiquitination and also reduced phosphorylation. Interestingly, non-transported organic phosphate esters like glycerol-3-phosphate can trigger PKA signalling (Popova et al. 2010). The existence of this signalling function without any transport strongly suggests the role of Pho84 as a receptor. Furthermore, other competitive inhibitors of phosphate transport via Pho84 like phosphonoacetic acid could not trigger PKA signalling. This suggests that simple binding of a substrate at the binding site is not enough to trigger signalling function of Pho84. Binding of only specific compounds at these sites trigger conformational changes, which in turn activates the signalling cascade, and the complete transport cycle is not required to trigger. It has been seen that same binding sites of Pho84 are responsible for both signalling and transport. Using Substituted Cysteine Accessibility Method (SCAM), Popova et al. (2010) identified F160 in transmembrane-IV and V392 in transmembrane-VIII as residues whose side chains are exposed in the phosphate binding site of Pho84. Covalent modification of these residues to cysteine, coupled with addition of MTSEA (a thiol-reactive reagent for labeling proteins at cysteine residues), causes inhibition of both signalling and transport function, suggesting that same residues might be responsible for both the functions. In a later study Samyn et al. (2012), based on 3-dimensional structure of Pho84 (generated using bacterial GlpT permease as a template and using multiple sequence alignment of phosphate transporters from plants, bacteria and fungi), two amino acid residues (D358 in transmembrane-VII and K492 in transmembrane-XI) were identified to play an important role in phosphate transport.
Phosphate metabolism is an important pathway to understand the sensing and signalling of essential nutrient, phosphate. Since phosphate is an essential component, an accurate and robust sensing of the nutrient is critical for an organism. Thus, a dual-affinity transporter system, consisting of low- and high-affinity transporters, gives an evolutionary advantage to the organism to respond to a wider concentration of phosphate in the environment and time to adapt to changes in phosphate availability. Within ascomycetes, there are several molecular components that are conserved in sequence and function, while other components have diversified functions and responses. Thus, comparative analysis of PHO pathway will help in understanding the diversification of this pathway and its relationship with specific environmental responses and evolutionary history of a species especially fungal species.
References
Ault-Riché D, Fraley CD, Tzeng CM and Kornberg A 1998 Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180 1841–1847
Berben G, Legrain M, Gilliquet V and Hilger F 1990 The yeast regulatory gene PHO4 encodes a helix-loop-helix motif. Yeast 6 451–454
Carter-O'Connell I, Peel MT, Wykoff DD and O'Shea EK 2012 Genome-wide characterization of the phosphate starvation response in Schizosaccharomyces pombe. BMC Genomics 13 697
Cassone A, Carpinelli G, Angiolella L, Maddaluno G and Podo F 1983 31P nuclear magnetic resonance study of growth and dimorphic transition in Candida albicans. J. Gen. Microbiol. 129 1569–1575
Cordes FS, Bright JN and Sansom MSP 2002 Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323 951–960
Dick CF, Dos-Santos ALA and Meyer-Fernandes JR 2011 Inorganic phosphate as an important regulator of phosphatases. Enzyme Res. 2011 103980
Gagneur J, Sinha H, Perocchi F, Bourgon R, Huber W and Steinmetz LM 2009 Genome-wide allele- and strand-specific expression profiling. Mol. Syst. Biol. 5 274
Ghillebert R, Swinnen E, De Snijder P, Smets B and Winderickx J 2011 Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem. J. 434 243–251
Giots F, Donaton MCV and Thevelein JM 2003 Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47 1163–1181
Gras DE, Persinoti GF, Peres NTA, Martinez-Rossi NM, Tahira AC, Reis EM, Prade RA and Rossi A 2013 Fungal Genetics and Biology. Fungal Genet. Biol. 60 140–149
Gras DE, Silveira HCS, Martinez-Rossi NM and Rossi A 2007 Identification of genes displaying differential expression in the nuc-2 mutant strain of the mold Neurospora crassa grown under phosphate starvation. FEMS Microbiol. Lett. 269 196–200
Henry TC, Power JE, Kerwin CL, Mohammed A, Weissman JS, Cameron DM and Wykoff DD 2011 Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal transduction pathway in yeasts. Eukaryot. Cell 10 198–206
Homann OR, Dea J, Noble SM and Johnson AD 2009 A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5 e1000783
Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M and Sherlock G 2012 The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nuc. Acid. Res. 40 D667–674
Kaffman A, Rank NM, O'Neill EM, Huang LS and O'Shea EK 1998 The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396 482–486
Kang S and Metzenberg RL 1990 Molecular analysis of nuc-1+, a gene controlling phosphorus acquisition in Neurospora crassa. Mol. Cell Biol. 10 5839–5848
Kerwin CL and Wykoff DD 2009 Candida glabrata PHO4 is necessary and sufficient for Pho2-independent transcription of phosphate starvation genes. Genetics 182 471–479
Kerwin CL and Wykoff DD 2012 De novo generation of a phosphate starvation-regulated promoter in Candida glabrata. FEMS Yeast Res. 12 980–989
Komeili A 1999 Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284 977–980
Kornberg A, Rao NN and Ault-Riché D 1999 Inorganic polyphosphate a molecule of many functions. Annu. Rev. Biochem. 68 89–125
Kulaev I and Kulakovskaya T 2000 Polyphosphate and phosphate pump. Annu. Rev. Microbiol. 54 709–734
Leal J, Squina FM, Martinez-Rossi NM and Rossi A 2007 The transcription of the gene for iso-orotate decarboxylase IDCase, an enzyme of the thymidine salvage pathway, is downregulated in the pregc mutant strain of Neurospora crassa grown under phosphate starvation. Can. J. Microbiol. 53 1011–1015
Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ and Gasch AP 2011 A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7 514
Lee PS, Greenwell PW, Dominska M, Gawel M, Hamilton M and Petes TD 2009 A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5 e1000410
Lee YS, Mulugu S, York JD and O'Shea EK 2007 Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316 109–112
Lenburg M and O'Shea E 1996 Signaling phosphate starvation. Trends Biochem. Sci. 21 383–387
Levy S, Kafri M, Carmi M and Barkai N 2011 The competitive advantage of a dual-transporter system. Science 334 1408–1412
Lichko LP, Kulakovskaya TV, Kulakovskaya EV and Kulaev IS 2008 Inactivation of PPX1 and PPN1 genes encoding exopolyphosphatases of Saccharomyces cerevisiae does not prevent utilization of polyphosphates as phosphate reserve. Biochemistry (Mosc.) 73 985–989
Liu C 2000 Regulation of the yeast transcriptional factor PHO2 activity by phosphorylation. J. Biol. Chem. 275 31972–31978
Lundh F, Mouillon J-M, Samyn D, Stadler K, Popova Y, Lagerstedt JO, Thevelein JM and Persson BL 2009 Molecular mechanisms controlling phosphate-induced downregulation of the yeast Pho84 phosphate transporter. Biochemistry 48 4497–4505
Maundrell K, Nurse P, Schönholzer F and Schweingruber ME 1985 Cloning and characterization of two genes restoring acid phosphatase activity in pho1− mutants of Schizosaccharomyces pombe. Gene 39 223–230
Metzenberg RL 1979 Implications of some genetic control mechanisms in Neurospora. Microbiol. Rev. 43 361–383
Miyakawa Y 2000 Identification of a Candida albicans homologue of the PHO85 gene, a negative regulator of the PHO system in Saccharomyces cerevisiae. Yeast 16 1045–1051
Mouillon J-M and Persson BL 2006 New aspects on phosphate sensing and signalling in Saccharomyces cerevisiae. FEMS Yeast Res. 6 171–176
Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA and York JD 2007 A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316 106–109
Neurospora crassa Sequencing Project 2014 Broad Institute of Harvard and MIT ( http://www.broadinstitute.org/ )
Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR and Johnson AD 2012 A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148 126–138
Odom AR, Stahlberg A, Wente SR and York JD 2000 A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287 2026–2029
Ogawa N, DeRisi J and Brown PO 2000 New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11 4309–4321
Orkwis BR, Davies DL, Kerwin CL, Sanglard D and Wykoff DD 2010 Novel acid phosphatase in Candida glabrata suggests selective pressure and niche specialization in the phosphate signal transduction pathway. Genetics 186 885–895
Oshima Y 1997 The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72 323–334
Peleg Y and Metzenberg RL 1994 Analysis of the DNA-binding and dimerization activities of Neurospora crassa transcription factor NUC-1. Mol. Cell Biol. 14 7816–7826
Peleg Y, Addison R, Aramayo R and Metzenberg RL 1996 Translocation of Neurospora crassa transcription factor NUC-1 into the nucleus is induced by phosphorus limitation. Fungal Genet. Biol. 20 185–191
Persson BL, Lagerstedt JO, Pratt JR, Pattison-Granberg J, Lundh K, Shokrollahzadeh S and Lundh F 2003 Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43 225–244
Persson BL, Petersson J, Fristedt U, Weinander R, Berhe A and Pattison J 1999 Phosphate permeases of Saccharomyces cerevisiae structure, function and regulation. Biochim. Biophys. Acta 1422 255–272
Poleg Y, Aramayo R, Kang S, Hall JG and Metzenberg RL 1996 NUC-2, a component of the phosphate-regulated signal transduction pathway in Neurospora crassa, is an ankyrin repeat protein. Mol. Gen. Genet. 252 709–716
Popova Y, Thayumanavan P, Lonati E, Agrochão M and Thevelein JM 2010 Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc. Natl. Acad. Sci. USA 107 2890–2895
Romanowski K, Zaborin A, Valuckaite V, Rolfes RJ, Babrowski T, Bethel C, Olivas A, Zaborina O and Alverdy JC 2012 Candida albicans isolates from the gut of critically ill patients respond to phosphate limitation by expressing filaments and a lethal phenotype. PLoS ONE 7 e30119
Roomans GM, Blasco F and Borst-Pauwels GW 1977 Cotransport of phosphate and sodium by yeast. Biochim. Biophys. Acta 467 65–71
Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM and Snyder SH 2001 Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc. Natl. Acad. Sci. USA 98 2306–2311
Saito H 2010 Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 13 677–683
Samyn DR, Ruiz-Pávon L, Andersson MR, Popova Y, Thevelein JM and Persson BL 2012 Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H+ transceptor and its effect on signalling to the PKA and PHO pathways. Biochem. J. 445 413–422
Schwaninger R, Dumermuth E and Schweingruber ME 1990 Effects of seven different mutations in the pho1 gene on enzymatic activity, glycosylation and secretion of acid phosphatase in Schizosaccharomyces pombe. Mol. Gen. Genet. 221 403–410
Schweingruber ME, Edenharter E, Zurlinden A and Stockmaier KM 1992 Regulation of pho1-encoded acid phosphatase of Schizosaccharomyces pombe by adenine and phosphate. Curr. Genet. 22 289–292
Secco D, Wang C, Shou H and Whelan J 2012 Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586 289–295
Sethuraman A, Rao NN and Kornberg A 2001 The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98 8542–8547
Shao D 1998 A cysteine residue in helixII of the bHLH domain is essential for homodimerization of the yeast transcription factor Pho4p. Nuc. Acid. Res. 26 710–714
Shapiro RS, Sellam A, Tebbji F, Whiteway M, Nantel A and Cowen LE 2012 Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr. Biol. 22 461–470
Smith RF, Blasi D and Dayton SL 1973 Phosphatase activity among Candida species and other yeasts isolated from clinical material. Appl. Microbiol. 26 364–367
Vogel K, Hörz W and Hinnen A 1989 The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell Biol. 9 2050–2057
Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ and Oliver SG 2012 PomBase: a comprehensive online resource for fission yeast. Nuc. Acid. Res. 40 D695–699
Wurst H, Shiba T and Kornberg A 1995 The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 177 898–906
Wykoff DD and O'Shea EK 2001 Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159 1491–1499
Wykoff DD, Rizvi AH, Raser JM, Margolin B and O'Shea EK 2007 Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol. Cell 27 1005–1013
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Luis M Corrochano
[Tomar P and Sinha H 2014 Conservation of PHO pathway in ascomycetes and the role of Pho84. J. Biosci. 39 1–12] DOI 10.1007/s12038-014-9435-y
Rights and permissions
About this article
Cite this article
Tomar, P., Sinha, H. Conservation of PHO pathway in ascomycetes and the role of Pho84. J Biosci 39, 525–536 (2014). https://doi.org/10.1007/s12038-014-9435-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-014-9435-y