Abstract
Parkinson’s disease (PD) is a α-synucleinopathy in which intracellular aggregates of α-synuclein (α-syn) result in neurodegeneration and in the impairment of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex-mediated release of neurotransmitters. SNAP25 is a SNARE complex component: its concentration is increased in the cerebrospinal fluid of PD patients and this is related to the severity of cognitive and motor symptoms. Five SNAP25 single-nucleotide polymorphisms (SNPs) that modulate gene expression and were described to play a role in neurologic conditions (rs363050, rs363039, rs363043, rs3746544, and rs1051312) were analyzed in a cohort of 412 sporadic Italian PD patients and 1103 healthy controls (HC) in order to identify possible correlation with the disease. The SNAP25 rs1051312 C allele and CC genotype confer protection against PD onset, in particular in males (p = 0.003, OR(95%CI) = 0.67(0.51–0.88)) (pc = 0.008, OR(95%CI) = 0.28(0.10–0.70)). Co-segregation analyses revealed that the rs1051312 effect was reinforced when present within the rs363043 C-rs3746544 T-rs1051312 C haplotype (p = 3.3 × 10−4, OR = 0.47, 95%CI = 0.31–0.72), once again in males. Finally, rs363039 influenced age at onset (p = 0.02) and MMSE (Mini-Mental State Examination) scores (p = 0.01). The SNAP25 SNPs analyzed herein modulate gene expression at different levels as they are involved in binding miRNA and transcription factors; this suggests a possible synergistic effect of SNAP25 SNPs in the pathogenesis of PD. A replication in a larger and independent sample will help to further explore this hypothesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder after Alzheimer’s disease (AD) [1]; this disease results from the progressive loss of dopaminergic neurons in the substantia nigra pars compacta of the midbrain and is characterized by rigidity, resting tremor, and bradykinesia. Most PD patients eventually develop an impairment of cognitive domains, such as attention, executive and visuospatial functions, and memory, as well [2]. The etiopathogenesis of PD is still unclear but a genetic contribution to its onset is well established. Thus, for the familiar form of PD, mutations in 18 specific chromosomal regions with a causative effect have been described [3]. The majority of PD cases is nevertheless sporadic, and in this case, it is believed that a complex interplay between environmental and genetic factors triggers the pathology (double-hit hypothesis). In particular, the predominant sporadic variant of the disorder seems to be associated with a combination of common variants within several genes.
PD, together with dementia with Lewy body (DLB) and multiple system atrophy (MSA), is described as a α-synucleinopathy. The pathological hallmark of these disorders is indeed the formation of intracellular aggregates composed mainly of α-synuclein (α-syn) that result from protein misfolding [4]. α-Syn is localized in the presynaptic nerve terminals, but its physiological functions have yet to be defined. The mechanisms responsible for α-syn accumulation remain elusive but it is known that α-syn aggregates hamper soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex-mediated neurotransmitter release [5] and impair the size of synaptic vesicle pool [6]. The SNARE complex, which regulates the docking and fusion of synaptic vesicles to the presynaptic membrane, is formed by the interaction of vesicle-associated membrane protein (VAMP), present in the synaptic vesicles, with syntaxin and synaptosomal-associated protein of 25 kDa (SNAP25) in the presynaptic plasma membrane [7]. It was demonstrated that synaptic dysfunction is an initial event of the subsequent neurodegeneration in many neurodegenerative diseases of the CNS [8].
SNAP25 gene polymorphisms have been associated to distinct neuropsychiatric and neurological disorders such as autism [9], attention-deficit/hyperactivity disorder (ADHD) [10], schizophrenia [11], amyotrophic lateral sclerosis (ALS) [12], and AD [13]. A work by Garcia-Reitböck P also reported SNARE protein redistribution in an animal model of PD, and in a few cases of PD with peroxidase immunocytochemistry [14].
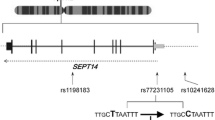
For these reasons, we analyzed five single-nucleotide polymorphisms (SNPs) within SNAP25 gene (rs363050, rs363039, rs363043, rs3746544, and rs1051312) in a cohort of sporadic Italian PD patients in order to identify possible correlation with the disease. The selection of SNPs was mainly based on the evidence from the existing literature. Rs363050, rs363039, and rs363043 are located in intron 1 of SNAP25 gene, in a region spanning about 13.8 kb, and are known to affect gene expression altering transcription factor binding [15, 16]; rs3746544 and rs1051312 lie instead in the 3′UTR region, which is also involved in regulatory aspects of gene expression.
Materials and Methods
Study Population
The study population included 412 consecutive patients with sporadic PD (185 females and 227 males), and 1103 age-matched healthy controls (HC) (626 females and 477 males) recruited among relatives of PD patients (mostly patients’ spouses with no maternal relationship for at least four generations). All patients enrolled in the study attended regular follow-up appointments with neurologists trained in movement disorders that evaluated their conditions and symptoms over time to confirm PD diagnosis. A complete anamnesis was collected to make sure that all the patients included in the study suffered from sporadic and not familiar PD. All the healthy controls were examined as well by neurologists that excluded the presence of any movement disorder and in particular PD. Subjects were recruited at three separate institutions: the unit of Parkinson’s disease and movement disorders of the IRCCS “C. Mondino” of Pavia, the Ospedale di Circolo and Fondazione Macchi in Varese, and the Neurologic Rehabilitation Unit of the Don C. Gnocchi Foundation, IRCCS in Milano. The Ethical Committees of the three institutions approved the study; all the participants gave informed consent. The following demographic and clinical variables were recorded in a part of the patient group: age, gender, age at onset of symptoms, current disability measured by the UPDRS-ME (Unified Parkinson Disease Rating Scale-Motor Examination), Hohen and Yahr stage [17] during ON and OFF periods and Schwab and England scale [18]. Cognitive performances were measured by the MMSE (Mini-Mental State Examination) [19] evaluated in ON phase. MMSE scores were age-, gender-, and education-corrected from the raw scores. All PD patients were undergoing dopaminergic treatment (levodopa and/or dopamine agonist agent). The presence of motor fluctuations was registered (Table 1).
Sample Collection and DNA Extraction
Whole blood was collected by venipuncture in EDTA-containing vacutainer tubes (Becton Dickinson Co., Rutherford, NJ); genomic DNA was extracted from peripheral blood mononuclear cells (PBMC) using standard phenol/chloroform procedure. DNA concentration for each sample was determined by measuring the optical density at 260 nm wavelengths using a spectrophotometer (SmartSpec Plus, Bio-rad, Irvine, CA, USA). DNA samples were stored at − 20 °C until use.
SNAP25 rs363050, rs363039, rs363043, rs3746544, and rs1051312 SNP Genotyping
Four out of five SNPs (rs363050, rs363039, rs363043, and rs3746544) within SNAP25 gene were analyzed by allelic discrimination real-time PCR using pre-designed TaqMan probes (Thermofisher scientific, USA) (C_329097_10, C_327976_10, C_2488346_10, and C_27494002_10 assays, respectively). PCR consisted of a hot start at 95 °C for 10 min followed by 40 cycles of 94 °C for 15 s and 60 °C for 1 min. Fluorescence detection took place at 60 °C. Assays were performed in 10-μl reactions, using TaqMan Genotyping Master Mix on 96-well plates using a CFX96 instrument (Bio-Rad Laboratories, USA). Control samples representing all possible genotypes and a negative control were included in each reaction. Results were analyzed using the Bio-Rad CFX Manager software (v. 3.1) (Bio-Rad Laboratories, USA).
Rs1051312 SNP genotypes were determined by restriction fragment length polymorphism (PCR-RFLP) assays as previously described [20].
Statistical Analysis
The SPSS Software 24.0 (IBM) was used. Chi-square analysis was employed to exclude any deviation of SNP genotype distribution from Hardy-Weinberg equilibrium; p value was > 0.05 both in cases and in controls. Chi-squared statistics as appropriate were applied to 2 × N tables to compare case-control differences of SNP distributions. The association of each polymorphism with the disease was measured by the OR and its 95% confidence interval (CI). Bonferroni correction for multiple tests (p) was applied by multiplying the p value for the degrees of freedom (df) derived from the n-1 number of genotypes or alleles analyzed. P value Yate’s corrected (p) was calculated in 2 × 2 contingency tables. The co-segregation analysis was done using the SHEsisPlus software freely available at http://shesisplus.bio-x.cn/SHEsis.html. Age at onset and corrected MMSE evaluated in ON phase variable were not normally distributed after Kolmogorov-Smirnov and Shapiro-Wilk tests; mean ranks were calculated and Kruskal-Wallis test was applied for correlations with SNPs.
Results
Rs363050, rs363039, rs363043, rs3746544, and rs1051312 SNAP25 SNPs were analyzed in a cohort of 412 Italian sporadic PD patients; results were compared to those obtained in 1103 HC of the same geographic origin.
Allelic and genotypic frequencies of the 5 SNPs were in Hardy-Weinberg equilibrium (p > 0.05) in the two groups of subjects. Haplotype analysis of SNAP25 SNPs revealed a linkage disequilibrium between rs363050, rs363039, and rs363043 and between rs3746544 and rs1051312 (Fig. 1).
No differences between the genotypic and allelic distributions of rs363050, rs363039, rs363043, and rs3746544 SNPs were detected between PD and HC (Table 2). On the other hand, SNAP25 rs1051312 genotypes resulted to be differently distributed in PD patients and HC (p = 0.02, pc = 0.04). In particular, the CC genotype resulted to be a “protective” factor against PD onset as it was present in 7.4% of HC and only in 3.6% of patients (p = 0.005, pc = 0.01, OR(95%CI) = 0.47(0.26–0.81)). Notably, allelic distribution of the rs1051312 SNP was significantly skewed as well in PD compared to HC (p = 0.02, OR(95%CI) = 0.80(0.66–0.97 for C allele)) (Table 2).
When PD patients and HC were stratified according to gender, the differences in rs1051312 genotypes’ distributions were confirmed in males alone (p = 0.006, pc = 0.01). Notably, the SNAP25 rs1051312 CC genotype was confirmed as “protective”, being present in 7.3% of males HC and only in 2.2% of male PD patients (p = 0.004, pc = 0.008, OR(95%CI) = 0.28(0.10–0.70)). On the contrary, the opposite TT genotype resulted to be a risk factor for PD onset in males as it was present in 62.1% of PD patients and in 52.8% of HC subjects (p = 0.02, pc = 0.04, OR(95%CI) = 1.46(1.06–2.03)).
Allelic distribution was significantly different in PD compared to HC males as well (p = 0.003), with the C allele resulting in a protective effect (OR = 0.67 95%CI: 0.51–0.88), and, conversely, the T allele being a risk factor for PD (OR = 1.49 95%CI = 1.14–1.96) (Table 3). No differences were evident between PD and HC in the genotypic and allelic frequencies of the other SNAP25 polymorphisms (rs363050, rs363039, rs363043, and rs3746544) even when data were stratified according to gender.
Multinomial logistic regression analyses were performed next, considering the phenotype (PD or HC) as the dependent variable and gender and all the five SNAP25 SNPs as covariates. As expected, a significant difference of gender between PD and HC (p < 0.0001) emerged, this is due to the fact that twice as many men than women suffer from PD [21]. Regression’s results confirmed a role for rs1051312 SNP in PD pathogenesis, in particular rs1051312 CC genotype resulted once again “protective” (p = 0.017, OR = 0.48, 95%CI = 0.26–0.87).
Co-segregation analyses of SNAP25 gene SNPs with the SHEsisPlus software (http://shesisplus.bio-x.cn/SHEsis.html) were performed next. Results showed a difference in rs3746544-rs1051312 haplotype distribution, and in particular in the T-C haplotype, between PD and HC males (p = 0.01); the “protective” OR (OR = 0.67, 95%CI: (0.51–0.88)) nevertheless did not differ from the one conferred by rs1051312 C allele alone (OR = 0.67 95%CI:0.51–0.88) (Table 4). The major difference in haplotype distributions between PD and HC males emerged instead for the rs363043-rs3746544-rs1051312 SNPs haplotypes (p = 0.01) (Table 5). In particular, we detected a cumulative protective effect given by the co-presence of the rs363043 C-rs3746544 T-rs1051312 C alleles (C-T-C haplotype), a haplotype that was significantly more present in HC than in PD males (p = 3.3 × 10−4, OR = 0.47, 95%CI = 0.31–0.72). Thus, the C-T-C haplotype was observed to confer a stronger protective effect against PD onset than the one given by the rs1051312 C allele alone or by the rs3746544 T-rs1051312 C haplotype (Tables 4 and 5). Again, no differences emerged when female HC and PD patients were analyzed.
Possible correlations between SNAP25 SNPs and some PD clinical features, in particular: age at onset, corrected MMSE, and the presence of fluctuations and of hallucinations were finally analyzed. Age at disease onset was available for 355 patients and was not normally distributed (p < 0.0001). Notably, rs363039 was significantly associated with PD age at onset, with the AA genotype being associated with a lower age at onset and, on the contrary, the GG genotype being associated with a higher age at onset passing through the intermediate values of the heterozygous (AG) subjects (p = 0.02). MMSE scores were available for 283 patients (151 males) and were also not normally distributed (p < 0.0001). None of SNAP25 SNPs resulted to influence MMSE scores in the overall group of PD patients; nevertheless, gender stratification once again showed a role for rs363039. In particular, in males, the rs363039 GG genotype was associated with a lower MMSE score and, on the contrary, AA genotype correlated with a better MMSE score passing through the intermediate values of the heterozygous (AG) patients (p = 0.01) (Table 6) in male PD patients. Finally, SNAP25 SNPs were not associated with either fluctuations or hallucinations.
Discussion
We analyzed five SNPs (rs363050, rs363039, rs363043, rs3746544, and rs1051312) of SNAP25 gene in order to evaluate their possible involvement in sporadic PD onset risk. These SNAP25 SNPs were selected because of their involvement in distinct neuropsychiatric and neurological disorders both of the childhood (autism and ADHD) [9, 10] and of the adulthood (schizophrenia, ALS, and AD) [11,12,13] and because they can influence the regulation of SNAP25 synthesis [15, 16]. Because (1) PD is a α-synucleinopathy, characterized by the presence of neurotoxic Lewy bodies (LBs) which are composed of α-syn aggregates [22], (2) α-syn influences SNARE-dependent membrane fusion by reducing vesicle docking in vitro [23], and (3) SNAP25 is one of the SNARE complex components, it seemed possible that SNAP25 polymorphisms could be associated with PD. Notably, this possibility was reinforced by recent results showing that SNAP25 concentration is increased in the cerebrospinal fluid of PD patients and such increase is related to cognitive and motor symptom severity and, on the other hand, post-mortem brain analyses indicated that presynaptic (including SNAP25) and postsynaptic proteins are depleted in PD dementia [24].
Results herein show that both SNAP25 rs1051312 C allele and CC genotype confer protection against PD onset in the investigated cohort. Result gained power when PD patients were stratified by gender, with a major effect being observed in males.
The rs1051312 and the rs3746544 SNPs are located in the 3′UTR region of SNAP25 gene: they result to be in linkage disequilibrium; in silico simulation analyses suggest that rs1051312 and rs3746544 are miRSNPs, thus they can alter specific miRNAs binding sites and, as a consequence, SNAP25 gene expression [25]. Functional SNPs occurring in miRNA sequences or in binding sites of miRNA are reported to be associated with susceptibility to various diseases [26]. In particular, in PD, a research identified numerous miRNA-binding site variants associated with the disorder [27]. Particularly, rs1051312 and rs3746544 are believed to influence the function of miR-510 and miR-641 molecules and both rs1051312 and rs3746544 are included in the sequence of miR-641 binding site. The interaction of miR-510 and the 3′UTR of SNAP25 d is instead influenced by rs1051312 alone (Fig. 2). Hence, both rs1051312 and rs3745544 might contribute to regulate SNAP25 synthesis. For these reasons we conducted co-segregation analyses of the five SNAP25 SNPs selected for this study. Results evidenced that “C-T-C haplotype” (co-presence of rs363043 C, rs3746544 T, and rs1051312 C alleles) confers protection against PD onset in male subjects, a protection stronger than the one conferred by rs1051312 C allele alone or by “T-C haplotype” (co-presence of rs3746544 T and rs1051312 C alleles).
Representation of the putative effect of SNAP25 rs3746544 and rs1051312 SNPs on miRNA binding. The four panels show the four haplotypes, the allelic variants of the SNPs are shown by the white letters in black circles in the sequence of SNAP25 3′ UTR. Shifted circles of the miRNAs depict the mismatches
Rs363043, rs363050, and rs363039 are located in intron 1; they all produce transcription factor binding site (TFBS) changes (gain/loss of TFBS) [15]; therefore, the “C-T-C haplotype” might contribute to modulate SNAP25 synthesis by miRNAs and transcription factors’ binding. Analyses of possible correlations between SNAP25 SNPs and PD clinical features reinforced the hypothesis that these SNPs do play a role in the disease. Thus, the rs363039 SNP modulated age at onset with a negative effect (lower age at onset) given by AA genotype. The GG genotype, on the contrary, correlated with a higher age at onset with an intermediate effect of the heterozygous genotype AG. Importantly, the same SNAP25 rs363039 SNP influenced MMSE scores as well in male patients alone. In this case, the AA genotype was “positive” conferring a higher MMSE, whereas the GG genotype was correlated with lower MMSE. Also, in this case, the heterozygous genotype AG resulted in intermediate MMSE scores. Rs36039 is located in intron 1 of the SNAP25 gene and has been previously associated with both cognitive performance and structural maturation of gray matter in healthy children [28]. In particular, individuals homozygous for the A allele showed increased cognitive performances. Rs363039 SNP along with other polymorphisms on the SNAP25 gene has also been previously associated with intelligence within a normal population of Dutch children, adolescents, and adults [15]. These analyses, however, showed that the rs363039 G allele was associated with higher Wechsler Intelligence Scale verbal and performance subtests scores [15]. Our results are pointing in the opposite direction as those of Gosso et al. but confirm findings by Soderqvist et al. [28]. Within this context, it seems important to underline that: (1) the rs363039 A, together with the rs363050 G allele, was shown to predict improvements in behavioral parameters after a multidimensional rehabilitative approach in AD patients [13]; and (2) the rs363050 A and rs363043 T alleles, which segregate together with rs363039 G allele because of their linkage disequilibrium, correlate with impaired cognitive scores and functional MRI parameters in AD [29].
A limitation of this study is the size of the PD cohort; thus, a replication in a larger and independent cohort, which will also need to be more balanced in terms of gender, is needed to confirm the possibility that SNAP25 SNPs play a role in the pathogenesis of PD. Additionally, in future studies, it will be interesting to evaluate if there is a relationship between SNAP25 SNPs, the efficacy of the levodopa or dopamine agonist therapy and tobacco smoking as nicotine has been reported to modulate dopamine release [30].
References
Schneider SA, Obeso JA (2015) Clinical and pathological features of Parkinson's disease. Curr Top Behav Neurosci 22:205–220. https://doi.org/10.1007/7854_2014_317
Aarsland D (2016) Cognitive impairment in Parkinson's disease and dementia with Lewy bodies. Parkinsonism Relat Disord 22(Suppl 1):S144–S148. https://doi.org/10.1016/j.parkreldis.2015.09.034.
Klein C, Westenberger A (2012) Genetics of Parkinson's disease. Cold Spring Harb Perspect Med 2(1):a008888. https://doi.org/10.1101/cshperspect.a008888
Yasuda T, Nakata Y, Mochizuki H (2013) α-Synuclein and neuronal cell death. Mol Neurobiol 47(2):466–483. https://doi.org/10.1007/s12035-012-8327-0
Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC (2010) Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329(5999):1663–1667. https://doi.org/10.1126/science.1195227.
Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK, Chaudhry FA, Nicoll RA et al (2010) Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65(1):66–79
Gundersen CB (2017) The structure of the synaptic vesicle-plasma membrane interface constrains SNARE models of rapid, synchronous exocytosis at nerve terminals. Front Mol Neurosci 10:48. https://doi.org/10.3389/fnmol.2017.00048
Burgoyne RD, Morgan A (2011) Chaperoning the SNAREs: A role in preventing neurodegeneration? Nat Cell Biol 13(1):8–9. https://doi.org/10.1038/ncb0111-8
Guerini FR, Bolognesi E, Chiappedi M, Manca S, Ghezzo A, Agliardi C, Sotgiu S, Usai S et al (2011) SNAP-25 single nucleotide polymorphisms are associated with hyperactivity in autism spectrum disorders. Pharmacol Res 64(3):283–288. https://doi.org/10.1016/j.phrs.2011.03.015
Gao Q, Liu L, Chen Y, Li H, Yang L, Wang Y, Qian Q (2015) Synaptosome-related (SNARE) genes and their interactions contribute to the susceptibility and working memory of attention-deficit/hyperactivity disorder in males. Prog Neuro-Psychopharmacol Biol Psychiatry 57:132–139. https://doi.org/10.1016/j.pnpbp.2014.11.001
Honer WG, Young CE (2004) Presynaptic proteins and schizophrenia. Int Rev Neurobiol 59:175–199. https://doi.org/10.1016/S0074-7742(04)59007-4
Ikemoto A, Nakamura S, Akiguchi I, Hirano A (2002) Differential expression between synaptic vesicle proteins and presynaptic plasma membrane proteins in the anterior horn of amyotrophic lateral sclerosis. Acta Neuropathol 103(2):179–187. https://doi.org/10.1007/s004010100449
Guerini FR, Farina E, Costa AS, Baglio F, Saibene FL, Margaritella N, Calabrese E, Zanzottera M et al (2016) ApoE and SNAP-25 polymorphisms predict the outcome of multidimensional stimulation therapy rehabilitation in Alzheimer's disease. Neurorehabil Neural Repair 30(9):883–893. https://doi.org/10.1177/1545968316642523
Garcia-Reitböck P, Anichtchik O, Bellucci A, Iovino M, Ballini C, Fineberg E, Ghetti B, Della Corte L et al (2010) SNARE protein redistribution and synaptic failure in a transgenic mouse model of Parkinson's disease. Brain 133(7):2032–2044. https://doi.org/10.1093/brain/awq132
Gosso MF, de Geus EJ, Polderman TJ, Boomsma DI, Heutink P, Posthuma D (2008) Common variants underlying cognitive ability: Further evidence for association between the SNAP-25 gene and cognition using a family-based study in two independent Dutch cohorts. Genes Brain Behav 7(3):355–364. https://doi.org/10.1038/sj.mp.4001868
Braida D, Guerini FR, Ponzoni L, Corradini I, De Astis S, Pattini L, Bolognesi E, Benfante R et al (2015) Association between SNAP-25 gene polymorphisms and cognition in autism: Functional consequences and potential therapeutic strategies. Transl Psychiatry 5:e500. https://doi.org/10.1038/tp.2014.136
Hoehn MM, Yahr MD (1967) Parkinsonism: onset, progression and mortality. Neurology 17:427–442
Schwab RS, jr EAC (1969) Amantadine HCL (Symmetrel) and its relation to Levo-Dopa in the treatment of Parkinson's disease. Trans Am Neurol Assoc 94:85–90
Folstein MF, Folstein SE, McHugh PR (1975) ʽʽMini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12:189–198
Öner Ö, Akın A, Herken H, Erdal ME, Çiftçi K, Ay ME, Bicer D, Öncü B et al (2011) Association among SNAP-25 gene DdeI and MnlI polymorphisms and hemodynamic changes during methylphenidate use: A functional near-infrared spectroscopy study. J Atten Disord 15(8):628–637. https://doi.org/10.1177/1087054710374597
Elbaz A, Bower JH, Maraganore DM, McDonnell SK, Peterson BJ, Ahlskog JE, Schaid DJ, Rocca WA (2002) Risk tables for parkinsonism and Parkinson's disease. J Clin Epidemiol 55(1):25–31
Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M (1998) Alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc Natl Acad Sci U S A 95:6469–6473
Lai Y, Kim S, Varkey J, Lou X, Song JK, Diao J, Langen R, Shin YK (2014) Nonaggregated α-synuclein influences SNARE-dependent vesicle docking via membrane binding. Biochemistry 53(24):3889–3896. https://doi.org/10.1021/bi5002536
Bereczki E, Bogstedt A, Höglund K, Tsitsi P, Brodin L, Ballard C, Svenningsson P, Aarsland D (2017) Synaptic proteins in CSF relate to Parkinson's disease stage markers. NPJ Parkinsons Dis 3(7):1149–1158. https://doi.org/10.1016/j.jalz.2016.04.005.
Ye C, Hu Z, Wu E, Yang X, Buford UJ, Guo Z, Saveanu RV (2016) Two SNAP-25 genetic variants in the binding site of multiple microRNAs and susceptibility of ADHD: A meta-analysis. J Psychiatr Res 81:56–62. https://doi.org/10.1016/j.jpsychires.2016.06.007
Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W (2008) Polymorphisms in microRNA targets: A gold mine for molecular epidemiology. Carcinogenesis 29(7):1306–1311. https://doi.org/10.1093/carcin/bgn116
Ghanbari M, Darweesh SK, de Looper HW, van Luijn MM, Hofman A, Ikram MA, Franco OH, Erkeland SJ (2016) Genetic variants in microRNAs and their binding sites are associated with the risk of Parkinson disease. Hum Mutat 37(3):292–300. https://doi.org/10.1002/humu.22943
Söderqvist S, McNab F, Peyrard-Janvid M, Matsson H, Humphreys K, Kere J, Klingberg T (2010) The SNAP25 gene is linked to working memory capacity and maturation of the posterior cingulate cortex during childhood. Biol Psychiatry 68(12):1120–1125. https://doi.org/10.1016/j.biopsych.2010.07.036
Guerini FR, Agliardi C, Sironi M, Arosio B, Calabrese E, Zanzottera M, Bolognesi E, Ricci C (2014) Possible association between SNAP-25 single nucleotide polymorphisms and alterations of categorical fluency and functional MRI parameters in Alzheimer's disease. J Alzheimers Dis 42(3):1015–1028. https://doi.org/10.3233/JAD-140057
Ma C, Liu Y, Neumann S, Gao X (2017) Nicotine from cigarette smoking and diet and Parkinson disease: A review. Transl Neurodegener 6:18. https://doi.org/10.1186/s40035-017-0090-8
Funding
This work was supported by the National Institutes of Health [Ricerca Corrente 2015].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The Ethical Committees of the three institutions approved the study; all the participants gave informed consent.
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Agliardi, C., Guerini, F.R., Zanzottera, M. et al. SNAP25 Gene Polymorphisms Protect Against Parkinson’s Disease and Modulate Disease Severity in Patients. Mol Neurobiol 56, 4455–4463 (2019). https://doi.org/10.1007/s12035-018-1386-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-018-1386-0