Abstract
MicroRNAs (miRNAs) are emerging as the most potential regulator of neuronal development. Recent studies from our lab and elsewhere have demonstrated a direct role of miRNAs in regulating neuronal differentiation and synaptogenesis. MicroRNA-145, a miRNA identified to regulate pluripotency of stem cells, downregulates the protein levels of reprogramming transcription factors (RTFs) like OCT4, SOX2, and KLF4 (cell, 137,647–658,2009). Studies have shown that miR-145 is multifunctional and crucial for fate determination of neurons. In our recently published study, we have identified a set of miRNAs including miR-145 and miR-29b families differentially expressed in SH-SY5Y cells exposed sequentially with retinoic acid + brain-derived neurotrophic factor (RA+BDNF) for differentiation into mature neurons (Mol Neurobiol (2016) doi:https://doi.org/10.1007/s12035-016-0042-9). In the present study, we have identified the role of miR-29b in upregulation of miR-145, which is upregulated after exposure of RA+BDNF in a P53-dependent manner. In differentiating SH-SY5Y cells, expression of miR-29b downregulates expression of P85α, a P53 inhibitor, which results in upregulation of miR-145 and downregulation of RTF proteins. Ectopic expression of miR-145 and miR-29b in amounts equivalent to their endogenous expression has induced G1 phase cell cycle arrest. In conclusion, our studies have identified miR-29b as an upstream regulator of miR-145 and targets its RTF genes during differentiation of SH-SY5Y cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Generation of mature and functional neurons is a complex cellular process, achieved by timely synthesis of different proteins. The discovery of miRNAs as an extra layer of control in protein synthesis has opened new windows to understand the regulation of cellular processes [1,2,3]. Priming of proliferating neurons to differentiation requires molecular changes at transcriptional, post-transcriptional, and post-translational levels of gene expression. Cellular models of neuronal differentiation have helped heavily in identifying the key molecules of neural development. Identification of miRNAs involved in neuronal differentiation has further revealed the molecular mechanism of neural development [4].
Studies carried out by our group and elsewhere have identified dramatic alterations in global miRNA profile of neurons when they are differentiated into mature neurons [5,6,7]. However, only a limited number of target proteins are identified, which are directly regulated by miRNAs deregulated in differentiating neurons [5, 7,8,9]. Le et al. (2009) profiled expression of 175 miRNAs using microarray in SH-SY5Y, a human neuroblastoma, and observed upregulation in expression of 12 miRNAs (mainly miR-125b, miR-124a, miR-7, miR-199a, and miR-214) when cells are differentiated by exposure of RA+BDNF. Using real-time PCR-based TaqMan Low Density Arrays (TLDA), we have studied the effect of neuronal differentiation on global miRNA profile of SH-SY5Y (a human neuroblastoma) and PC12 (a rat pheocytochroma) cells and identified upregulation in expression of miRNA, as a more prominent change than downregulation [5, 6]. In PC12 cells, differentiation altered expression of 22 miRNAs (19 up- and 3 downregulated), while in SH-SY5Y, differentiation altered expression of 94 miRNAs (77 up- and 17 downregulated) [5, 6]. Upregulation observed in the expression of several miRNAs raised the possibility of involvement of transcription factors and other indirect factors in differentiating neurons. A master regulator of gene expression, TP53 (P53) is identified as one of the proteins which help cells choose their fate, proliferation and differentiation [10]. P53 proteins are multifunctional and are required by differentiating, dividing, and dying cells [11, 12].
In a recently published study, we have identified differentially expressed miRNAs in differentiated SH-SY5Y cells and demonstrated that upregulation of miR-145/143, miR-192, and miR-222 is P53 dependent [6]. miR-145 is well known to regulate the levels of OCT4, SOX2, and KLF4 (reprogramming transcription factors or RTFs) in human embryonic stem cells [13]. Interestingly, P53 did not play any role in upregulation of miR-29 family, which was upregulated by differentiation and known as brain-enriched miRNAs. Earlier studies have shown that knocking down miR-29 family induces apoptosis of neuronal cells and increased expression of miR-29 protects neurons [14,15,16]. In addition, age-dependent upregulation of miR-29 in fish suggested an adaptive mechanism to counteract the expression of aging-related phenotypes [16].
Keeping in mind the importance of miR-29 family in neural development and survival, we have further extended our studies to explore the role of the miR-29 family in neuronal differentiation. In the present study, we have demonstrated that miR-29 is an upstream regulator of P53-mediated upregulation of miR-145 expression and its target genes in differentiating neurons. Our findings have also supported earlier reports, that miR-145 targets reprogramming transcription factors and provided new evidence for the role of miR-29b and P53 as upstream regulator of miR-145 and its target genes.
Material and Methods
Cells and Chemicals
SH-SY5Y cells were procured from ATCC, USA, and neural stem cells were procured from Stem Cell Technologies, Vancouver, British Columbia, Canada. Culture medium and all materials required for SH-SY5Y and neural stem cell (NSC) culture were procured from Thermo Scientific, CA, USA, and Stem Cell Technologies, respectively. Retinoic acid (RA) (R2625), BDNF (B3795), pifithrin-α (P4349), and propidium iodide (P4170) were procured from Sigma-Aldrich, Saint Louis, MO, USA. The transfection reagent DharmaFECT1 (T200102) was procured from Dharmacon, Pittsburgh, PA, USA. Mimics and inhibitors of miRNAs were obtained from Applied Biosystems, Foster City, CA, USA. Antibodies against β-actin (A2228), NANOG (SAB3500389), and OCT4 (SAB3500316) were procured from Sigma-Aldrich, Saint Louis, MO, USA. Antibodies for SOX2 (ab97959) and KLF4 (ab106629) were procured from Abcam, Cambridge, MA, USA. Antibody for P53 (134100) was procured from Life Technologies, Carlsbad, CA, USA. Antibody for P85α (4292) was procured from Cell Signaling Technology. Infrared-labeled secondary antibodies were obtained from LI-COR Biosciences, Lincoln City, NE, USA. Individual miRNA assays, reverse transcription (RT) kit, TaqMan universal master mixture, miRVana, and other reagents required for real-time PCR were obtained from Thermo Scientific, CA, USA. All other regular chemicals were purchased from Sigma-Aldrich.
Culture, Differentiation, Transfection of Cells, and Flow Cytometry Studies
SH-SY5Y and NSCs were grown, cultured, and differentiated as described in our earlier studies ([5, 6, 17]). In brief, SH-SY5Y cells were differentiated by exposing them with RA (10 μM) for 5 days, followed by 3-day BDNF (100 ng/ml) exposure. After 5 days of RA exposure, cells were washed with phosphate-buffered saline (PBS) and cell culture media were replaced with media containing BDNF (100 ng/ml) but no serum. Every alternate day, media of differentiating SHSY5Y cells were replaced with fresh media containing 10 μM RA (up to 5 days) or 100 ng/ml BDNF (3 days after RA). For NSC differentiation, they were seeded on PLL-coated surface and transferred in neurobasal medium containing N2 and B27 supplements. NSCs were procured from Stemcell Technologies and grown in NeuroCult NS-A basal medium (rat) in the presence of proliferation supplement provided by Stemcell Technologies and epidermal growth factor (20 ng/ml) and b-fibroblast growth factor (10 ng/ml).
All the transfections were carried out using the DharmaFECT1 transfection reagent as described by the manufacturer and described in earlier studies [5, 18]. Mimics and inhibitors of miR-145 and miR-29 were purchased from Qiagen, USA: Syn-miR-145 (Cat no. 4464066), Anti-miR-145 (Cat no. 4464084), Syn-miR-29 (Cat no. 4464084, MC 10103), and Anti-miR-29 (Cat no. 4464084 MC MH10103).
In brief, cell death in undifferentiated and differentiated SH-SY5Y cells was measured using FITC-labeled annexin-V as described in our earlier studies [17]. Cell cycle was measured as described in our earlier study [19]. In brief, transfected cells along with their respective controls were detached from the culture plate, washed twice with PBS, and fixed with 70% ethanol for 30 min at − 20 °C. After 30 min, cells were pelleted out, ethanol was removed, and the pellet was washed with PBS twice. Finally, cells were resuspended in resuspension buffer containing RNase A (10 mg/ml) and PI (1 mg/ml) for 30 min in the dark. Cell cycle experiments were assayed on BD Facs INFLUX.
RNA Isolation and Real-time PCR Assays
The total RNA, including small RNAs, was isolated using mirVana miRNA isolation kits as per manufacturer’s protocol. Real-time PCR of SOX2, OCT4, KLF4, NANOG, and hypoxanthine-guanine phosphoribosyltransferase (HPRT) endogenous control was carried out using probes labeled with FAM (5′) and TAMRA (3′) and TaqMan master mixes. Before real-time PCR, reverse transcription (RT) was carried out using high capacity cDNA kit of Life Technologies, USA. The RT reaction mixture contains 1X RT buffer, 8 mM dNTP mix, 1X RT random primers, and 0.25 μl each of MultiScribe reverse transcriptase and RNase inhibitor in a total volume of 5 μl which also includes 1 μg total RNA. The thermal cycling conditions used for RT reactions were 10 min at 25 °C, 120 min at 37 °C, and 5 s at 85 °C. Sequences of primers and probes used for real-time PCR are as follows:
-
SOX2: FP: 5′-TGCGAGCGCTGCACAT-3′, RP: 5′-GCAGCGTGTACTTATCCTTCTTCTTC-3′, Probe: 5′-CCGGCGGAAAACCAAGACGCT-3′
-
OCT4: FP: 5′-ACCCACACTGCAGCAGATCA3′, RP: 5′-CACACTCGGACCACATCCTTCT-3′, Probe: 5′-CCACATCGCCCAGCAGCTTGG-3′
-
KLF4: FP: 5′-ACCTACACAAAGAGTTCCCAT-3′, RP: 5′-TGTGTTTACGGTAGTGTGCCTG-3′, Probe: 5′-TCATCTGAGCGGGCGAATTTCCA-3′
-
NANOG: FP: 3′-ACAACTGGCCGAAGAATAGCA-3′, RP: 5′-GGTTCCCAGTCGGGTTCAC-3′, Probe: 5′-TGACGCAGAAGGCCTCAGCACCT-3′,
-
HPRT: FP: 5′-TGCTGAGGATTTGGAAAGGG-3′, RP: 5′-ACAGAGGGCTACAATGTGATG-3′, Probe: 5′-AGGACTGAACGTCTTGCTCGAGATG-3′
Real-time PCRs of individual miRNAs of miR-29b, miR-145, and miR-143 were carried out using individual TaqMan miRNA assays of Thermo Fisher Scientific, USA. miRNA RT was carried out by using high-capacity microRNA reverse transcription kit Thermo Fisher Scientific, USA, as described by the manufacturer. The RT reaction mixture for miRNAs contains 1X RT buffer, 8 mM dNTP mix, 1X individual miRNA RT primer, and 1 μl each of MultiScribe reverse transcriptase and RNase inhibitor in a total volume of 7.5 μl which includes 350 ng total RNA. The thermal cycling conditions used for miRNA RT reactions were 30 min at 16 °C, 30 min at 42 °C, and 5 min at 85 °C.
Western Blotting
The total cell lysates were prepared using CelLytic M cell lysis reagent of Sigma-Aldrich, USA, supplemented with a protease inhibitor cocktail and Dithiothreitol. Western blotting was performed as described in our earlier paper [17]. For detection of target proteins, IR-labeled secondary antibodies were used, and membranes were scanned using ODYSSEY Clx system, LI-COR, as described in our earlier studies [5, 20].
In Silico Analysis
Gene ontology pathways for miRNA targets were identified using GO process tool of MetaCore platform of Thomson Reuters, London, UK, a data mining and online platform.
Statistical Analysis
All the experiments were carried out in triplicate. Either Student’s t test or ANOVA was employed to calculate pairwise comparison and statistical significance. P ≤ 0.05 in Student’s t test was considered as significant.
Results
Differentiation Upregulates Expression of miR-29 and miR-145/143 Families in SH-SY5Y and NSCs
SH-SY5Y cells were differentiated by exposure of RA or RA+BDNF. Exposure of RA induced formation and elongation of neurites and additional exposure of BDNF in RA exposed SH-SY5Y cells serving to form interconnections and synaptic junctions between dendrites or neurites (Fig. 1a,b). Differentiation of SH-SY5Y cells by exposure of RA or RA+BDNF significantly upregulated the expression of miR-29 and miR-145/143 families (Fig. 1a,b). In SH-SY5Y cells, maximum increase was observed in miR-29b and miR-145 after RA or RA+BDNF exposure (Fig. 1a,b). Interestingly, miR-29 family is expressed more after RA+BDNF exposure, while miR-145/143 is expressed more in RA-exposed SH-SY5Y cells (Fig. 1c,d). Interestingly, differentiation of NSCs also upregulated the expression of miR-29 and miR-145/143 families; however, fold changes are lesser in comparison to those of SH-SY5Y cells (Fig. 2a,b).
Effect of neuronal differentiation on expression of miR-29 and miR-145 families. a Phase contrast images of undifferentiated (Control), 5 days exposed with retinoic acid (RA) or 5 days exposed with RA and 3 days with BDNF (RA+BDNF). b Immunofluorescence of βIII tubulin and Map2, markers of differentiated neurons in RA+BDNF-exposed SH-SY5Y cells. c, d Real-time PCR of miR-29 and miR-145 families with TaqMan primers specific for mature miRNAs in Control, RA, or RA+BDNF differentiated SH-SY5Y cells. SH-SY-5Y cells were differentiated using our previously published protocol [6]. In brief, SH-SY5Y cells were differentiated by the exposure of RA and BDNF for 8 days. Before real-time PCR, an equal amount of total RNA (350 ng) was used for cDNA synthesis and further equal amount of cDNA (0.5 μl) was used in even PCR reactions. Relative quantification was carried out considering respected control. U6 snRNA was used for normalization. RQ, relative quantification. *P ≤ 0.05. RA 10 μM; BDNF 100 ng/ml
Differentiation of NSCs induced expression of miR-29 and miR-145 and NSCs. Real-time PCR of miR-29 and miR-145 families with TaqMan primers specific for mature miRNAs in a undifferentiated (0 Day) and b differentiated (8 Day) NSCs. NSCs were differentiated using our previously published protocol [5]. Before real-time PCR, an equal amount of total RNA (350 ng) was used for cDNA synthesis and further equal amount of cDNA (0.5 μl) was used in even PCR reactions. Relative quantification was carried out considering respected controls. U6 snRNA was used for normalization. RQ, relative quantification. *P ≤ 0.05
Increased Expression of miR-145 Induced G1 Arrest and Apoptosis in Naive SH-SY5Y Cells
SH-SY5Y cells were transfected with miR-145 mimics (Syn-miR-145), which increases the expression of miR-145 equal to their amount (25 nM) in differentiated SH-SY5Y cells (Supple I).Transfection of miR-145 mimics in SH-SY5Y cells significantly increases the number of cells lying in G1 stage of cell cycle, similar to RA-exposed cells. However, G1 arrest observed in miR-145 mimic transfected cells was lower in comparison to that in RA-exposed cells. Annexin-PI-based analysis of cell death in naive and RA-exposed cells transfected with mimics of miR-145 was performed to understand the cell stage-specific role of miRNA. In naive SH-SY5Y cells, transfection of miR-145 mimics induced G1 phase arrest as observed with cells exposed to RA (Fig. 3a). However, transfection of miR-145 mimics did not alter the cell cycle of RA exposed SH-SY5Y cells (Fig. 3a). Transfection of miR-145 mimics also induced apoptosis in naive SH-SY5Y cells, while no significant alterations were observed in RA-exposed SH-SY5Y cells (Fig. 3b and Supple 2).
Overexpression of miR-145 selectively induces cell cycle arrest and apoptosis in undifferentiated SH-SY5Y. Effect of transient transfection of miR-145 on a cell cycle and b apoptosis of SH-SY5Y cells transfected with mimics of miR-145 in undifferentiated or differentiated stage (all experiments for flow cytometric studies were performed in triplicates; only representative images are shown here)
Ectopic Expression of miR-29b Induces P53 Protein and Decreases P85α in SH-SY5Y Cells
SH-SY5Y cells are transfected with 25 nM of miR-29b mimics, which induces expression of miR-29b similar to their amounts found in differentiated SH-SY5Y cells (Supple 3). Differentiation of SH-SY5Y cells with RA or RA+BDNF significantly downregulates P85α levels; however, levels of P53 are induced by differentiation in SH-SY5Y cells (Fig. 4a) [6]. Transfection of miR-29b mimics (25 nM) significantly reduced the P85α levels and induced P53 levels (Fig. 4b,c). Analysis of 3′-UTR of P85α revealed the presence of targeting sites for miR-29, which is conserved among different mammals (Supple 4).
miR-29b regulates P85α levels in differentiating SH-SY5Y cells. a, b Western blot and densitometry of P85α in whole cell lysates prepared from control (undifferentiated), RA, or RA+BDNF exposed SH-SY5Y cells or SH-SY5Y cells transfected with either NTC or mimics and inhibitors of miR-29b. c Western blot and densitometry of P53 in whole cell lysate prepared from SH-SY5Y cells transfected with either NTC or mimics or inhibitors of miR-29b. All the transfections were made by DharmaFECT transfection reagent using 25 nM siRNA. NTC, non-target control; ddCt, double delta cycle threshold; IDV, integrated density value. *P ≤ 0.05, densitometry of western blots were carried out by ImageJ software, Syn-mimic; Anti-inhibitor
Ectopic Expression of miR-145 Regulates Expression of miR-29b Through P53
Transfection of miR-29b mimics (25 nM) to SH-SY5Y cells induced a fivefold change in the expression of miR-145/143, while no substantial changes were observed in expression of miR-29b, when cells are transfected with either mimics or inhibitors of miR-145 (Fig. 5a,b). Pre-exposure of SH-SY5Y cells with P53 inhibitors (PFTα) significantly prevented the miR-29b-mediated induction of miR-145/143 in SH-SY5Y cells (Fig. 5c).
miR-29b increased expression of miR-145 in P53-dependent fashion. a Real-time PCR of miR-145/143 in total RNA isolated from SH-SY5Y cells transfected with mimics (Syn-miR-29b) or inhibitors (Anti-miR-29b) of miR-29b and b expression of miR-29b in SH-SY5Y cells transfected with mimic (Syn-miR-145) or inhibitors (Anti-miR-145) of miR-145 along with NTC. c Real-time PCR of miR-145/143 in total RNA isolated from SH-SY5Y cells transfected either with NTC or exposed with PFTα or transfected with mimics of miR-29b (Syn-miR-29b) or transfected with mimics of miR-29b and exposed with PFTα (Syn-miR-29b+PFTα). All the transfections were made by DharmaFECT transfection reagent using 25 nM siRNA. NTC, non-target control; ddCt, double delta cycle threshold; IDV, integrated density value. *P ≤ 0.05. PFTα, pifithrin-α 10 μM for 24 h
Differentiation of SH-SY5Y and NSCs Downregulates RTF Protein Levels
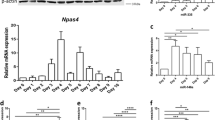
Immunoblotting of RTF proteins in SH-SY5Y cells significantly downregulated RTF proteins in differentiated SH-SY5Y or NSCs (Fig. 6a,b). In both SH-SY5Y and NSCs, differentiation produced maximum downregulation in KLF4 followed by SOX2 proteins. Further, transfection of miR-145 or miR-29b mimics (in amounts required to reach that in differentiated cells) significantly downregulated the levels of all the RTF proteins (Fig. 7a,b). Interesting to note was downregulation of NANOG, after SH-SY5Y and NSCs transfected with mimics of miR-145 or miR-29b, which does not have a binding site for miR-145 (Fig. 7a,b).
Regulation of reprogramming transcription factors (RTFs) during differentiation of SH-SY5Y cells and neural stem cells. Western blot and densitometry of RTFs (SOX2, OCT4, KLF4, NANOG) in a total cell lysates prepared from undifferentiated (control) or RA or RA+BDNF exposed differentiated SH-SY5Y cells and b from undifferentiated (0 Day) or differentiated (8 Day) rat cerebral cortex NSCs. IDV, integrated density value; RQ, relative quantification. *P ≤ 0.05, RA 10 μM; BDNF 100 ng/ml
Ectopic expression of miR-145 and miR-29b regulate the level of reprogramming transcription factors (RTFs) in differentiated SH-SY5Y cells. Western blotting and densitometry of RTFs (SOX2, OCT4, KLF4, NANOG) in whole cell lysates prepared a from SH-SY5Y cells transfected with either NTC or mimics (Syn-miR-145) or inhibitors (Anti-miR-145) of miR-145 and b from SH-SY5Y cells transfected with mimics (Syn-miR-29b) or inhibitors (Anti-miR-29b) of miR-29b. All the transfections were made by DharmaFECT transfection reagent using 25 nM siRNA. NTC, non-target control; IDV, integrated density value; RQ, relative quantification, *P ≤ 0.05
Inhibition of P53 Prevents miR-29b Induced G1 Arrest in SH-SY5Y Cells
Transient transfection of miR-29b mimics (25 nM) induced proportion of SH-SY5Y cells at G1 phase of cell cycle (Fig. 8a,b) from 53.5 to 63.6% of total cells as studied by PI staining and flow cytometry detection. Interestingly, pre-exposure of PFTα substantially prevented the cells moving to G1 phase of cell cycle (Fig. 8c,d).
Effect of overexpression of miR-145 and miR-29b on G1 phase arrest in SH-SY5Y cells. Flow cytometry-based PI uptake assay for cell cycle analysis of a SH-SY5Y cells transfected with NTC, b NTC exposed with pifithrin-α (PFTα), c mimics of miR-29b (Syn-miR-29b), or d transfected with mimics of miR-145 or e transfected with mimics of miR-29b and exposed with pifithrin-α (Syn-miR-29b+PFTα). f Bar diagram of G1, S, and G2 phase percentages of SH-SY5Y cells. All values are the mean of three individual experiments. Significant changes are calculated by Student’s t test. Pifithrin-α 10 μM for 24 h
The Similarity in Cellular Processes Targeted by miR-29b and miR-145 miRNA Target Proteins
Out of the top 10 most significantly targeted biological pathways, 8 are the same among miR-29b and miR-145 (Fig. 9a,b) targeted pathways. GO process analyses of different processes targeted by in silico predicted targets of miR-145 and miR-29b have identified anatomical structure development and system development on the top most significant processes for miR-145 and miR-29b, respectively. Analyses of the top 10 significantly targeted GO processes of miR-145 and miR-29b have identified 8 processes, which are the same between miR-145 and miR-29b targets (Figs. 9a,b).
Discussion
The present study demonstrates the role of miR-29b in upregulation of miR-145 and its target RTF genes in SH-SY5Y cells differentiated with sequential exposure of RA+BDNF. Moreover, our studies have identified P53 proteins as the connecting link between upregulation of miR-29b and miR-145 during neuronal differentiation. Upregulation of miRNA and its target proteins by another miRNA during neuronal maturation supports our earlier findings that upregulation in miRNA expression is more prominent than downregulation during neuronal differentiation [5, 6]. Increased transcription of several genes and decreased translation of most of mRNAs are general phenomena observed in stressed cells [21,22,23]. A recently published study has shown that during stress, phase separation takes place in stressed bodies, which releases 5′ UTR resulting in higher protein translation. Evidences are emerging from several different types of studies which support the hypothesis that protein translation is regulated locally by different mechanisms in the cells [24]. Our studies have shown that in differentiating SH-SY5Y cells, upregulation of miR-29b controls the upregulation of miR-145 by downregulatingP85α (a, which results in increased levels of P53 proteins). Our earlier studies have identified miR-29b and miR-145 as two of the dramatically upregulated miRNAs in SH-SY5Y cells, differentiated with sequential exposure of RA and BDNF [6]. On exploring the role of P53 protein in differentiation of SH-SY5Y cells, we have observed that upregulation of miR-145 is P53 dependent, while upregulation of miR-29b is P53 independent [6]. The present study demonstrates that miR-29b acts as an upstream regulator of miR-145 and its target genes in differentiating SH-SY5Y cells and identified the P53 protein as the connecting link between miR-29b and miR-145 expression (Fig. 10).
Studies carried out in recent past have identified direct and indirect roles of miR-29 family in brain development, brain aging, neuronal maturation, and neurodegeneration [12, 13, 16]. Earlier studies from our and others’ labs have identified upregulation in miR-29b expression in differentiated neurons [6, 25, 26]. Maximum upregulation observed in miR-29b, among different members of miR-29 family (miR-29a/b/c) in differentiating SH-SY5Y cells, indicates that miR-29b is the most important among the miR-29 family. Similar to SH-SY5Y cells, members of miR-29 family are also upregulated in differentiating NSCs, which indicates that differentiation-mediated upregulation of miR-29 family is common among different types of neurons. In our earlier studies, we have shown that differentiation induced levels of P53 proteins in SH-SY5Y cells, when expression of several miRNAs including miR-29 family and miR-145 are upregulated. Induction of P53 proteins in cells transfected with mimics of miR-29b indicates that miR-29b has a role in induction of P53 protein in differentiating neurons. Downregulation of P85α observed in cells transfected with inhibitors of miR-29b indicates that miR-29b induces P53 proteins by downregulating its repressor in differentiating neurons. Earlier studies of Park et al. (2008) have also shown the role of miR-29b in induction of P53 proteins by downregulating P85α in HeLa cells [27]. Members of miR-29 family have been shown to target multiple targets based on the nature of cells and tissues; however, their role in regulating P53 levels and associated functions seems to be common. Induction of apoptosis and cell cycle arrest in differentiated SH-SY5Y cells, transfected with mimics of miR-145, indicates different roles of miR-145 in undifferentiated and differentiated SH-SY5Y cells. Transfection of miR-145 mimics selectively induced G1 arrest and apoptosis in undifferentiated cells indicates toward the selective role of miR-145 in helping undifferentiated cells undergo differentiation.
Levels of P53 have been reported to regulate expression of neurogenesis-regulating genes, proliferation, and differentiation of the neural stem cell in the developing brain [28,29,30,31]. During post-natal brain development, increased P53 levels coordinate with cyclin-dependent kinases to control cell cycle and differentiation of neurons [32]. Interestingly, studies of Suzuki et al. (2009) have demonstrated the ability of P53 to induce the expression of growth-suppressive miRNAs (miR-16-1, miR-143, and miR-145) in a transcription-independent manner [33]. Similar to the study of Park et al., we have also found induction in P53 levels in SH-SY5Y cells overexpressed with miR-29b is mediated by downregulation of P85α [27].
Studies of Xu et al. (2009) have identified the crucial role of miR-145 in regulating pluripotency by targeting the expression of RTFs or Yamanaka factors like OCT4, SOX2, and KLF4 in human embryonic stem cells [11]. Later on, several independent studies have supported the findings that miR-145 regulates expression of RTFs or Yamanaka factors in different types of cells [34,35,36,37,38]. Similar to recent studies of Morgado et al. (2016), we have found that increased expression of miR-145 in RA+BDNF differentiated SH-SY5Y cells downregulates Yamanaka factors, which helps differentiated cells maintain their non-proliferating state [38]. Similar to earlier reports, an inverse relation has been observed between expression of miR-145 and RTF protein levels in differentiated SH-SY5Y and neural stem cells. Moreover, transfection of SH-SY5Y cells with the amount of miR-29 mimics equal to their levels in differentiated SH-SY5Y cells substantially downregulated levels of RTF (SOX2, KLF4, OCT4, and NANOG) further confirmed that miR-29b acts upstream to miR-145 in regulating the protein levels of RTFs. G1 phase arrest, observed in cells transfected with amount of miR-145 mimics equivalent to amount of miR-145 expressed in differentiated SH-SY5Y cells, indicates that increased expression of miR-145 is a prerequisite for neuronal differentiation. Earlier studies have also shown that transcription of a miR-29 family is regulated by MYC proteins, which promote neuronal differentiation by controlling division of progenitor cells [39, 40].
Members of miR-29 family, which are upregulated by both RA or RA+BDNF in a P53-independent manner, are reported to express in higher amounts in mature neurons while downregulated in AD patients [16, 26]. Expression of miR-29b is regulated in a P53-independent manner; however, overexpression of miR-29 significantly induced P53 levels and miR-145/143 expression in SH-SY5Y cells, which indicates that miR-29b acts upstream to P53 and miR-145/143. No significant changes observed in expression of miR-29b by miR-145 or miR-143 in SH-SY5Y cells further confirmed that miR-29b acts upstream to P53 and miR-145 in differentiating SH-SY5Y cells. Interestingly, studies of Park et al. (2009) have reported that miR-29 activates P53 by targeting P85α and CDC42 in cancer cells. However, further studies are required to confirm the same in neurons [27]. Accumulation of cells at G1 phase after transfection with mimics of miR-29b or miR-145 indicates that expression of these miRNAs helps differentiated cells to maintain their post-mitotic state. Reversal of cell cycle arrest in SH-SY5Y cells, which are pre-exposed with PFTα and transfected with miR-29b mimics, confirms that P53 acts as upstream to miR-145 and helps in regulating cell cycle arrest of differentiating neurons. Similar to our findings, earlier studies have shown that increased expression of miR-29 protects matured neurons from apoptosis, while a loss of miR-29 induces neuronal death [6, 12, 14, 26].
In summary, our study is the first to demonstrate upstream role of miR-29b in regulating expression of miR-145 and its target RTF proteins during neuronal differentiation. Our studies have also shown that P53 protein acts as the connecting link between upregulation of miR-29b and miR-145. In differentiated SH-SY5Y cells, miR-29 upregulates the P53 levels by downregulating P85α, which results in upregulation of miR-145, a miRNA known to downregulate RTF proteins. In conclusion, coordinated action of miR-29b and miR-145 regulates protein levels of RTF genes in differentiated SH-SY5Y cells and helps the neuronal cells to maintain their non-proliferating stage.
Change history
03 July 2019
The original version of this article unfortunately contained an error. In Figure 8, the image under section a) NTC, and b) NTC���+���PFT�� are copied by mistake.
03 July 2019
The original version of this article unfortunately contained an error. In Figure 8, the image under section a) NTC, and b) NTC���+���PFT�� are copied by mistake.
Abbreviations
- BDNF:
-
brain-derived neurotrophic factor
- DTT:
-
Dithiothreitol
- HPRT:
-
hypoxanthine-guanine phosphoribosyltransferase
- IDV:
-
integrated density value
- miRNA:
-
microRNA
- mRNA:
-
messenger RNA
- PCR:
-
polymerase chain reaction
- RA:
-
retinoic acid
- RTF:
-
reprogramming transcription factor
- RQ:
-
relative quantification
- RT:
-
reverse transcription
- IR:
-
infrared
- PBS:
-
phosphate buffer saline
- PI:
-
propidium iodide
- PLL:
-
poly-l-lysine
- PVDF:
-
polyvinylidene fluoride
References
Coolen M, Bally-Cuif L (2009) MicroRNAs in brain development and physiology. Curr Opin Neurobiol 19(5):461–470
Petri R, Malmevik J, Fasching L, Åkerblom M, Jakobsson J (2014) miRNAs in brain development. Exp Cell Res 321(1):84–89
Singh T, Jauhari A, Pandey A, Singh P, B Pant A, Parmar D, Yadav S (2014) Regulatory triangle of neurodegeneration, adult neurogenesis and microRNAs. CNS Neurol Disord Drug Targets 13(1):96–103
Stiles J, Jernigan TL (2010) The basics of brain development. Neuropsychol Rev 20(4):327–348
Pandey A, Singh P, Jauhari A, Singh T, Khan F, Pant AB, Parmar D, Yadav S (2015) Critical role of the miR-200 family in regulating differentiation and proliferation of neurons. J Neurochem 133(5):640–652
Jauhari A, Singh T, Pandey A, Singh P, Singh N, Srivastava AK, Pant AB, Parmar D et al (2016) Differentiation induces dramatic changes in miRNA profile, where loss of dicer diverts differentiating SH-SY5Y cells toward senescence. Mol Neurobiol:1–10
Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H et al (2009) MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol 29(19):5290–5305
Makeyev EV, Zhang J, Carrasco MA, Maniatis T (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27(3):435–448
Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G (2012) p53 dynamics control cell fate. Science 336(6087):1440–1444
Quadrato G, Di Giovanni S (2012) Gatekeeper between quiescence and differentiation: p53 in axonal outgrowth and neurogenesis. Int Rev Neurobiol 105:71–89
Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS (2009) MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137(4):647–658
Roshan R, Shridhar S, Sarangdhar MA, Banik A, Chawla M, Garg M, Singh VP, Pillai B (2014) Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA 20(8):1287–1297
Yang G, Song Y, Zhou X, Deng Y, Liu T, Weng G, Yu D, Pan S (2015) MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep 12(2):3081–3088
Ripa R, Dolfi L, Terrigno M, Pandolfini L, Savino A, Arcucci V, Groth M, Tozzini ET et al (2017) MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging. BMC Biol 15(1):9
Ouyang YB, Xu L, Lu Y, Sun X, Yue S, Xiong XX, Giffard RG (2013) Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia 61(11):1784–1794
Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A et al (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc Natl Acad Sci 105(17):6415–6420
Yadav S,Pandey A,Shukla A, Talwelkar SS,Kumar A, Pant AB,Parmar D, (2011) miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J Biol Chem 286(43):37347–37357
Jin HY, Gonzalez-Martin A, Miletic AV, Lai M, Knight S, Sabouri-Ghomi M, Head SR, Macauley MS et al (2015) Transfection of microRNA mimics should be used with caution. Front Genet 6:340
Jauhari A,Singh T, Singh P, Parmar D, Yadav S, (2018) Regulation of miR-34 family in neuronal development. Mol Neurobiol 55(2):936–945
Pandey A, Jauhari A, Singh T, Singh P, Singh N, Srivastava AK, Khan F, Pant AB, Parmar D, Yadav S (2015) Transactivation of P53 by cypermethrin induced miR-200 and apoptosis in neuronal cells. Toxicol Res 4(6):1578–1586
Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172(6):803–808
Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23(24):2452–2462
Farny NG, Kedersha NL, Silver PA (2009) Metazoan stress granule assembly is mediated by P-eIF2α-dependent and independent mechanisms. RNA 15(10):1814–1821
Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA (2017) Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168(6):1028–1040. e1019
Fenn AM, Smith KM, Lovett-Racke AE, Guerau-de-Arellano M, Whitacre CC, Godbout JP (2013) Increased micro-RNA 29b in the aged brain correlates with the reduction of insulin-like growth factor-1 and fractalkine ligand. Neurobiol Aging 34(12):2748–2758
Kole AJ, Swahari V, Hammond SM, Deshmukh M (2011) miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev 25(2):125–130
Park S-Y, Lee JH, Ha M, Nam J-W, Kim VN (2009) miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat Struct Mol Biol 16(1):23–29
Gil-Perotin S, Haines JD, Kaur J, Marin-Husstege M, Spinetta MJ, Kim KH, Duran-Moreno M, Schallert T et al (2011) Roles of p53 and p27 Kip1 in the regulation of neurogenesis in the murine adult subventricular zone. Eur J Neurosci 34(7):1040–1052
Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen A-J, Perry SR, Tonon G et al (2008) p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455(7216):1129–1133
Meletis K, Wirta V, Hede S-M, Nistér M, Lundeberg J, Frisén J (2006) p53 suppresses the self-renewal of adult neural stem cells. Development 133(2):363–369
Armesilla-Diaz A, Bragado P, Del Valle I, Cuevas E, Lazaro I, Martin C, Cigudosa J, Silva A (2009) p53 regulates the self-renewal and differentiation of neural precursors. Neuroscience 158(4):1378–1389
Lookeren Campagne MV, Gill R (1998) Tumor-suppressor p53 is expressed in proliferating and newly formed neurons of the embryonic and postnatal rat brain: Comparison with expression of the cell cycle regulators p21Waf1/Cip1, p27Kip1, p57Kip2, p16Ink4a, cyclin G1, and the proto-oncogene bax. J Comp Neurol 397(2):181–198
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K (2009) Modulation of microRNA processing by p53. Nature 460(7254):529–533
Barros R, Pereira D, Callé C, Camilo V, Cunha AI, David L, Almeida R, Dias-Pereira A et al (2016) Dynamics of SOX2 and CDX2 expression in Barrett’s mucosa. Dis Markers 2016:1–7
Zou G, Liu T, Guo L, Huang Y, Feng Y, Huang Q, Duan T (2016) miR-145 modulates lncRNA-ROR and Sox2 expression to maintain human amniotic epithelial stem cell pluripotency and β islet-like cell differentiation efficiency. Gene 591(1):48–57
Morgado AL, Rodrigues CM, Solá S (2016) MicroRNA-145 regulates neural stem cell differentiation through the Sox2–Lin28/let-7 signaling pathway. Stem Cells 34(5):1386–1395
Ozen M, Karatas OF, Gulluoglu S, Bayrak OF, Sevli S, Guzel E, Ekici ID, Caskurlu T et al (2015) Overexpression of miR-145–5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. Cancer Investig 33(6):251–258
Liu X, Huang J, Chen T, Wang Y, Xin S, Li J, Pei G, Kang J (2008) Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res 18(12):1177–1189
Nolan K, Mitchem MR, Jimenez-Mateos EM, Henshall DC, Concannon CG, Prehn JH (2014) Increased expression of microRNA-29a in ALS mice: functional analysis of its inhibition. J Mol Neurosci 53(2):231–241
Annis RP, Swahari V, Nakamura A, Xie AX, Hammond SM, Deshmukh M (2016) Mature neurons dynamically restrict apoptosis via redundant pre-mitochondrial brakes. FEBS J 283:4569–4582
Acknowledgments
Mr. Abhishek Jauhari is grateful to UGC, New Delhi, and Ms. Tanisha Singh is grateful to DST, New Delhi, for providing research fellowships. The technical assistance of Mr. B S Pandey and Mr. Puneet Khare is also gratefully acknowledged. The CSIR-IITR communication reference number is 3493.
Funding
Funding for the work carried out in the present study had been provided by the CSIR network project (miND).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Jauhari, A., Singh, T. & Yadav, S. Expression of miR-145 and Its Target Proteins Are Regulated by miR-29b in Differentiated Neurons. Mol Neurobiol 55, 8978–8990 (2018). https://doi.org/10.1007/s12035-018-1009-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-018-1009-9