Abstract
Glioblastoma multiforme (GBM) is the most common and aggressive brain tumor subtype. Despite that metastasis of GBM beyond the central nervous system (CNS) is rare, its malignancy is attributed to the highly infiltration trait, leading to the difficulty of complete surgical excision. Matrix gla protein (MGP) is a vitamin K-dependent small secretory protein, and functions as a calcification inhibitor. The involvement of MGP function in glioma cell dynamics remains to be clarified. The study showed that a low proliferative rat C6 glioma cell line named as C6-2 exhibited faster migratory and invasive capability compared to that observed in a high tumorigenic rat C6 glioma cell line (called as C6-1). Interestingly, C6-2 cells expressed higher levels of MGP molecules than C6-1 cells did. Lentivirus-mediated short hairpin RNA (shRNA) against MGP gene expression (MGP-KD) in C6-2 cells or lentivirus-mediated overexpression of MGP transcripts in C6-1 cells resulted in the morphological alteration of the two cell lines. Moreover, MGP-KD caused a decline in cell migration and invasion ability of C6-2 cells. In contrast, increased expression of MGP in C6-1 cells promoted their cell migration and invasion. The observations were further verified by the results from the implantation of C6-1 and C6-2 cells into ex vivo brain slice and in vivo rat brain. Thus, our results demonstrate that the manipulation of MGP expression in C6 glioma cells can mediate glioma cell migratory activity. Moreover, our findings indicate the possibility that high proliferative glioma cells expressing a high level of MGP may exist and contribute to tumor infiltration and recurrence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gliomas comprise more than 70% of all brain tumors and are the third leading cause of cancer death between the ages of 15 and 34 [1,2,3]. Glioblastoma multiforme (GBM) is the most aggressive glioma and classified as grade IV by histological classification of the World Health Organization (WHO) [4]. Despite advances in the diagnostic classification and therapeutic strategies, prognosis of GBM remains very poor and the median survival time of GBM patients is less than 2 years [5, 6], with only 0.05 to 4.7% of patients surviving 5 years past diagnosis [7, 8]. Unlike the peripheral tumor types, gliomas rarely metastasize outside the central nervous system (CNS). However, glioma tumor cells tend to extensively infiltrate into the surrounding brain tissue and result in rapid intracranial spreading [9, 10], which is a major obstacle to the effective treatment for GBM. The in vitro and in vivo evidence has postulated that the environment inducing glioma cell migration could suppress tumor cell proliferation, and vice versa [11,12,13]. GBM tumor cells can invade into the other brain regions via myelinated fibers of the white matter, and preferentially extend into the contralateral hemisphere through the corpus callosum [4, 13, 14]. Therefore, understanding GBM cell heterogeneity is crucial to design more effective remedy.
Matrix gla protein (MGP) is a highly conserved γ-carboxyglutamic acid-containing secretory protein with a high calcium-binding affinity [15, 16]. This protein was originally found in association with bovine bone matrix [17]. Later, MGP is known to be widely expressed in various vertebrate tissues including the kidney, lung, heart, arteries, and calcified atherosclerotic plaques [18,19,20]. Despite that the biological mechanism of MGP is not precisely known yet, its inhibitory role in soft tissue calcification has been recognized according to the findings from loss-of-function experiments [19]. In addition, the expression of MGP has been found to be associated with the progress of several cancer types, such as primary renal cell carcinomas, prostate carcinomas, testicular germ cell tumors, ovarian cancer, gastric cancer, and breast cancer [21,22,23,24]. Through the comparison of the transcriptional profiles between WHO grade II gliomas and recurrent high-grade (WHO grade III or IV) gliomas, it has been suggested that the elevated level of MGP expression is positively correlated to tumor progression [25]. Moreover, in GBM-derived tumor cells highly expressing inhibitor of differentiation 4 (Id4), smaller and less vascularized tumor was formed in the group with diminished MGP, suggesting Id-mediated increase in MGP expression and promotion of tumor angiogenesis [26]. Despite that reduced cell migration of human glioma cell lines can result from the transient interference of MGP messenger RNA (mRNA) expression [27], further evidence is needed to subsidize MGP-mediated regulation in the migratory ability of malignant glioma cells.
Rat C6 glioma cell line was originally generated from the brain tumor of Wistar-Furth rat after exposure to N,N′-nitroso-methylurea. The C6 cell line has been reported to form solid tumors in the rat brain with morphological similarity of human GBM, and has been widely applied in the study of neoplastic gliomas both in vitro and in vivo [28]. In our previous study using the two C6 glioma cell lines having the distinct in vitro and in vivo tumorigenicity, we have addressed that proliferation-associated genes, as well as tumor-promoting cytokines and chemokines, were abundantly expressed in the high tumorigenic C6 cells (C6-1) facilitating tumor formation in the rat cortex, compared to those detected in low tumorigenic C6 cells (C6-2) [29]. Examination of MGP expression through the comparative complementary DNA (cDNA) microarray analysis showed, however, that C6-2 cells exhibited much higher expression of MGP than C6-1 cells. Therefore, the aim of the study was to dissect the role of MGP in cell function using the two C6 cell lines. Our findings revealed that downregulation of MGP expression in C6-2 cells not only reduced their cell proliferation, but also attenuated their cell migratory and invasive ability. Meanwhile, elevated expression of MGP promoted C6-1 cell migration and invasion. The results are further confirmed by the observations from ex vivo brain slice experiments and in vivo glioma implantation. The implanted C6-1 cells with MGP overexpression migrated longer distances than the control C6-1 cells. Overall, we show substantial evidence to certify the regulatory role of MGP in glioma tumor cell infiltration and invasion.
Materials and Methods
Materials
Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12), antibiotics (penicillin/streptomycin), and puromycin were obtained from Invitrogen (Carlsbad, CA, USA). Fetal bovine serum (FBS) was purchased from HyClone Laboratories (Logan, UT, USA). The 8.0-μm pore size transwell inserts and 0.4-μm pore size Millicell-CM culture inserts were purchased from Millipore. 4′,6-diamidino-2-phenylindole (DAPI), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and proteinase inhibitors were purchased from Sigma-Aldrich (St. Louis, MO, USA). Vectastain ABC kit, FITC-avidin, and Cy3-avidin were purchased from Vector Laboratories (Burlingame, CA, USA). The information of antibodies used for the study is included in Table 1.
Cell Culture
The two C6 glioma cell lines were obtained from different laboratories, and named as C6-1 and C6-2 in this study. C6-1 cells were provided by Dr. Henrich Cheng (Taipei Veterans General Hospital) and purchased from the National Health Research Institute Cell Bank (Zhunan, Taiwan). C6-2 cells were originally obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The two C6 glioma cell lines have been characterized previously to display the different degrees of the tumorigenicity in the rat brain [29]. These C6 cells were cultured in DMEM/F12 containing 10% heat-inactivated FBS, and the media were changed every 2–3 days.
cDNA Microarray Assay
Total RNA preparation for cDNA microarray assay and data processing were as previously described [29].
Lentivirus-Mediated Gene Delivery
The short hairpin RNA (shRNA)-mediated knockdown of MGP was performed using shRNA lentiviral particles (Biosettia Inc.), which were designed to suppress the production of rat MGP in C6 glioma cells. The cultures that were transfected with shRNA lentiviral particles with non-effective scramble shRNA sequences (lenti-sh-ctrl) were used as the control group (scramble). The lentivirus vector constructs used for MGP gene knockdown (MGP-KD) are shown as follows: pLV-mU6-[sh-scramble]EF1a-GFP- puromycin (lenti-sh-ctrl), pLV-mU6-[sh-MGP]EF1a-GFP-puromycin (lenti-sh-MGP_58, lenti-sh-MGP_68, lenti-sh-MGP_122, lenti-sh-MGP_225). The shRNA sequences are shown in Table 2. Alternatively, the lentivirus vector construct used for the overexpression of MGP (NM_012862.1), pLV-EF1a-rnoMGP-mPGK-GFP-puromycin (lenti-MGP), was obtained from Biosettia Inc. The control lentivirus vector construct was pLV-EF1a-GFP-F2A-Blasticidin-S deaminase (Bsd)-mPGK-Puro (lenti-Bsd). Cell transfection was followed by the procedure as previously described [29]. After infection, the stable transfectants were selected using 3 mg/mL puromycin in the presence of 10% FBS for 48 h. The efficiency of MGP downregulation in C6-2 cells or MGP overexpression in C6-1 cells was confirmed by real-time polymerase chain reaction (RT-PCR), quantitative polymerase chain reaction (QPCR), and Western blotting. C6-2 cells that were infected by lenti-ctrl were referred as C6-2-scramble. C6-1 cells infected by control lentivirus (lenti-Bsd) were named as C6-1-mock.
Real-Time Polymerase Chain Reaction
Total RNA was isolated from the cultures, and cDNA was generated using MMLV reverse transcriptase (Thermo Fisher Scientific). Gel-based semiquantitative PCR (RT-PCR) reaction was performed using DNA polymerase (GoTaq Green Master Mix, Promega) to confirm the specificity of MGP-specific primers. The PCR products were separated on 1% agarose gels containing ethidium bromide. Quantitative PCR (QPCR) was performed on Roche LightCycler™ Real-time PCR system using a LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics). The results were normalized to internal control (GAPDH). The specific primer sequences are shown in Table 2.
Western Blot Analysis
The cells were gently homogenized in the lysis buffer consisting of PBS with 0.1% sodium dodecyl sulfate (SDS), 1 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM sodium orthovanadate, and proteinase inhibitor cocktail. Protein concentration was determined using a Bio-Rad DC kit. Subsequently, 100 μg of total protein was separated through 10–12.5% SDS polyacrylamide gel electrophoresis, and then transferred to the nitrocellulose membrane. Proteins were identified by incubating the membrane with primary antibodies (Table 1), and visualized by incubation in horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and the electrochemiluminescence (ECL) solution (PerkinElmer Life Sciences).
Immunofluorescence
The samples (cells and ex vivo tissue sections) were fixed in 4% paraformaldehyde (PFA) and permeabilized in PBS containing 0.1% Triton X-100 for 30 min. The samples were incubated with primary antibodies at the appropriate dilution at 4 °C overnight, biotin-conjugated secondary antibody for 1 h at room temperature, and fluorescein-avidin at room temperature for 45 min. Finally, the samples were subjected to DAPI nuclear staining. Immunostained cells were observed under a fluorescence microscope (Nikon E800) with a color digital camera.
F-Actin Staining
The cells (2 × 104 cells) were seeded on 12-mm coverslips placed into a 24-well tissue culture plate. After 24 h, the cultures were fixed in 4% PFA for 15 min. The cells were permeabilized in PBS containing 0.1% Triton X-100, and then incubated with Texas red-conjugated phalloidin (Thermo Fisher Scientific) for 1 h at room temperature. Nuclear staining was carried out using DAPI solution. F-actin staining was observed under an Olympus FLUOVIEW FV1000 confocal laser scanning microscope.
Cell Scratch Migration Assay
A monolayer scratch assay was performed as previously described [30]. Cells were grown in 10% FBS-containing medium and mechanically scratched by a sterile pipette tip. Cells were rinsed with PBS and grown in serum-free DMEM/F12 for various time periods. The cell motility, in terms of wound closure area, was determined by measurement of wound closure area using NIH ImageJ software. The degree of the wound closure is presented as the percentage of closure area over the scratch area at the initial time point.
In Vitro Transwell Cell Invasion Assay
The ability of in vitro cell invasion was determined by the method as described previously [31]. The cells were seeded at the density of 1 × 104 cells/well onto a PDL-precoated transwell insert (pore size, 8.0 μm), and placed a 24-well plate. The medium containing 10% FBS was added into the lower compartment of each well. After 6 h, the inserts were fixed in 4% PFA for 10 min, followed by staining with 0.05% crystal violet in PBS for 10 min. The upper surface of each transwell insert was carefully cleansed with cotton swabs. Cell invasion was determined by counting cells on another side of the transwell inserts from five randomly selected fields under a microscope. The ratio of migrated cells over the total seeded cells in each culture was measured. The results are presented as the fold of the data in experimental cultures over that in the relative control (scramble or mock).
MTT Cell Growth Assay
MTT assay was performed as previously described [29]. Briefly, cells were seeded at a density of 5 × 103 per well in 24-well plates using DMEM/F12 containing 10% FBS. After harvest, MTT solution (5 mg/mL; Sigma) was added to each well; 4 h later, the medium was removed and DMSO was added to dissolve formazan products. The absorbance was measured at 595 nm under an enzyme-linked immunosorbent assay reader.
Colony Formation Assay
The cells were seeded at the density of 200 cells per well in a six-well culture plate. The cultures were maintained in DMEM/F12 containing 10% FBS for 7 days, and then fixed with 4% PFA for 10 min, followed by staining using 0.05% crystal violet for 15 min. The number of cell colonies formed in each well was counted using NIH ImageJ analysis software.
Ex Vivo Brain Slice Culture and Glioma Cell Implantation
The brain slice cultures were prepared followed by the procedure as described with little modification [32, 33]. Animal surgery was approved by the Institutional Animal Care and Use Committees at the National Cheng Kung University (Tainan, Taiwan). Briefly, the brain tissues were removed from Sprague-Dawley rat pups at postnatal day 14, and then dissected coronally at a thickness of 300 μm using a Microslicer™ DTK-1000 vibratory tissue slicer. The tissue slices were then plated on Millicell-CM culture inserts and maintained on the surface of slice culture medium (50% MEM with Earle’s salts, 35% Earle’s balanced salt solution, 15% heat-inactivated horse serum, 1% GlutaMAX™) at 37 °C for 2 days. Two microliters of C6-2 (C6-2-scramble and C6-2-sh68) or C6-1 cell suspension (C6-1-mock and C6-1-MGP) at the density of 5 × 104 cells/slice was slowly injected into the corpus callosum of the brain slice using a P10 micropipettor. The cultures were maintained in the medium for another 4 days, and fixed in 4% PFA for 1 h. The injected cells were identified by GFP immunofluorescent staining, and observed under a Nikon E800 epifluorescence microscope equipped with a CCD camera.
Quantification of C6 Cell Migration in Brain Slices
Cell migration was evaluated using NIH ImageJ analysis software to quantify GFP intensity in the four rectangle areas with the size of 0.25 mm2 along the different distances from the injected site (0–500, 500–1000, 1000–1500, and 1500–2000 μm). Data were collected from five randomly sampled images captured from each culture using an epifluorescence microscope with a ×20 objective lens. The immunofluorescent intensity of GFP+ cells detected in a fixed rectangle area was measured using NIH ImageJ analysis software. The results are presented as the arbitrary units of GFP fluorescence intensity along the four distance intervals.
In Vivo Glioma Transplantation and Tumor Measurement
Adult male SD rats (250 ± 30 g; n = 12) were used according to the Institutional Animal Care and Use Committee (IACUC) guidelines of the National Cheng Kung University. After rats were anesthetized by intraperitoneal injection of chloral hydrate (50 mg/kg), a midline incision was cut and the underlying tissue was removed using a scalpel blade. A hole was made by a dentist drill fitted with a 0.9-mm-diameter carbide dental burr on the exposed skull at the position of 2 mm posterior to the bregma and 2 mm to the right of the sagittal suture. A Hamilton syringe with a 25-gauge needle was inserted into the brain at the depth of 3 mm above the corpus callosum. The fluid (5 μL) containing 5 × 105 C6-1 cells (C6-1-mock and C6-1-MGP) was slowly injected into the brain. After injection, the needle was maintained for additional 2 min to prevent leakage of solution. At 14-day post transplantation (dpi), the rat brains were removed, fixed by 4% PFA, and then cryoprotected in 30% (w/v) sucrose in PBS. The tissues were embedded in Tissue Tek OCT (Electron Microscopy Sciences), sectioned at 20-μm thickness, and then subjected to hematoxylin and eosin (H/E) staining and GFP immunofluorescence. Twenty H/E-stained coronal sections were captured by a scanner, and the tumor areas were measured using UTHSCSA Image tool for Windows (University of Texas Health Science Center at San Antonio) as described previously [29]. The volume (mm3) of the tumor was calculated by the tumor area (mm2) × section number × section thickness (20 μm).
Statistical Analysis
All data from each experiment were analyzed by two-tailed unpaired Student’s t test using Sigma-Plot 10 (Systat Software Inc.). The results are expressed as means ± SEM. The differences between control and compared culture are considered statistically significant at P < 0.05.
Results
Differential Expression of MGP in the Distinct Migratory Populations of Rat Glioma Cells
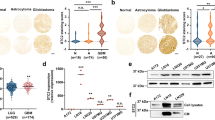
Through a comparative cDNA microarray analysis, we found that C6-1 cells with a higher tumorigenic behavior expressed a lower amount of MGP compared to C6-2 cells. MGP molecule is classified in many biology pathways most relevant to extracellular matrix, factor responses, and cation binding. The results from QPCR further verified that MGP mRNA expression was extremely lower in C6-1 cells than that of C6-2 cells (Fig. 1a). Since MGP was thought to be involved in the migratory ability of GBM tumor cells [27], the migration and invasion of C6-1 and C6-2 cells were examined. Through a cell scratch assay, we observed that the wound closure ability of C6-2 cells was better than that of C6-1 cells (Fig. 1b). Moreover, the results from the cell invasion examination showed that the number of C6-2 cells invading through the transwell insert was higher than C6-1 cells did (Fig. 1c). The observations reveal the positive correlation of MGP expression with the migratory and invasive ability of C6-2 cells versus C6-1 cells.
Rat C6 glioma cells with abundant expression of MGP display high cell migratory capability. a Total RNA was isolated from C6-1 and C6-2 cells, and then subjected to QPCR for the measurement of MGP mRNA expression. The results confirmed the data from cDNA microarray analysis. b C6-1 and C6-2 cells reached confluence in 10% FBS-containing medium, and then subjected to cell scratch migration assay in serum-free medium. Four photographs were taken from each culture at each indicated time point. The area of wound closure was selected and measured by ImageJ software. The representative images shown in the upper panel were taken from the cultures at the initial time point and at 12 h after cell scratching. c C6-1 and C6-2 cells were plated onto the upper side of the transwell insert filters for cell invasion. After 6 h, the cells that migrated to the bottom side of the filter were stained (upper panel) and counted. Data are means ± SEM of at least three independent experiments. *p < 0.05, **p < 0.01, versus C6-1. Scale bar in b and c, 200 μm
Reduced Rat Glioma Cell Growth and Migration by MGP Downregulation
To determine the involvement of MGP in steering C6-2 cells to migrate, we performed a lentivirus-mediated shRNA delivery approach to downregulate MGP expression in C6-2 cells. The efficiency of lentivirus particles encoding shRNA against MGP (lent-sh-MGP_58, lenti-sh-MGP_68, lenti-sh-MGP_122, lenti-sh-MGP_225) in MGP gene knockdown (KD) was evaluated by QPCR and Western blot analysis. As shown in Fig. 2a, when compared to C6-2 cells treated by lenti-ctrl (scramble), MGP mRNA expression was significantly reduced in C6-2 cells infected by lent-sh-MGP_58 (sh-58), lenti-sh-MGP_68 (sh-68), lenti-sh-MGP_225 (sh-225), but not in C6-2 cells by lenti-sh-MGP_122 (sh-122). Moreover, a lower level of MGP production was observed in C6-2 cells infected by sh-68 (C6-2-sh68) when compared to that detected in C6-2-scramble and other transfectants (Fig. 2b). We also noticed that C6-2-sh68 cells were changed to a small round shape (Fig. S1A), whereas there was no significant change in the morphology of C6-2 cells transduced by sh-122 (C6-2-sh122). In accordance with the phase-contrast examination, F-actin staining showed the smaller cell size of C6-2-sh68 cells than that of C6-2-scramble cells (Fig. S1B). Moreover, lamellipodium was hardly found in C6-2-sh68 cells, whereas C6-2-scramble cells displayed an apparent lamellipodium structure (Fig. S1B).
Downregulation of MGP suppresses the cell growth and migration of rat glioma cells. a Total RNA isolated from C6-2-scramble (S) and C6-2 cells infected by lentivirus particles against MGP expression (sh58, sh68, sh122, sh225) were subjected to QPCR for the determination of MGP gene knockdown (KD). b Total proteins were extracted from C6-2-scramble (S) and C6-2 cells infected by lentivirus particles against MGP expression (sh58, sh68, sh122, sh225), and then subjected to Western blot analysis to examine the inhibition of MGP production. The equal level of GAPDH proteins is referred as a loading control. c The cell growth of C6-2-scramble (S), C6-2-sh68, and C6-2-sh122 in serum containing medium was examined by MTT assay at the indicated time points. d C6-2-scramble (S), C6-2-sh68, and C6-2-sh122 were seeded at the density of 200 cells/well onto a 6-well plate. The number of the colony formation in these cultures was counted at 7 day after seeding. e C6-2-scramble, C6-2-sh68, and C6-2-sh122 reached confluence in 10% FBS-containing medium, the cultures were subjected to cell scratch migration assay in serum-free medium (left hand panel). The representative images shown in the left hand panel were taken from the cultures at the initial time point, and 12 or 36 h after cell scratching. The quantification of the area of wound closure was conducted as described in the “Materials and Methods” section. f C6-2-scramble, C6-2-sh68, and C6-2-sh122 cells were plated onto the upper side of the transwell insert filters for cell invasion. After 6 h, the cells that migrated to the bottom side of the filter were counted (left hand panel). The quantification of the invaded cells was counted as described in the “Materials and Methods” section. Data are presented as mean ± SEM of at least three independent experiments. *p < 0.05, p*** < 0.001 versus C6-2-scramble. Scale bar in e and f, 200 μm
Furthermore, the results from MTT cell growth assay indicated that the growth of C6-2-sh68 was much slower than those detected in C6-2-scramble and C6-2-sh122 cells (Fig. 2c). Moreover, the colony formation ability of C6-2-sh68 cells was reduced when compared to that of C6-2-scramble or C6-2-sh122 cells (Fig. 2d). Further experiments were conducted to evaluate the migratory ability of C6-2 cells after MGP-KD using the cell scratch assay and transwell invasion analysis. As shown in Fig. 2e, C6-2-sh68 cells showed lower wound closure ability than C6-2-scramble and C6-2-sh122 had. Moreover, the cell invasion of C6-2 was reduced after MGP-KD (Fig. 2f). These results indicate that MGP-KD caused the inhibition of C6-2 cell dynamics, i.e., reduced cell proliferation and motility.
Enhanced Rat Glioma Cell Migration by MGP Upregulation
Given the fact that C6-1 cells had the lower level of MGP expression (Fig. 1), the overexpression of MGP in C6-1 cells (C6-1-MGP) was conducted by lentivirus-mediated gene delivery. As shown in Fig. 3a, C6-1-MGP showed extensively high level of MGP gene expression, whereas MGP expression was scarce in C6-1 cells infected by lenti-ctrl (C6-1-mock). This was consistent with the results from Western blotting showing MGP was produced at a higher level in C6-1-MGP than that in C6-1-mock (Fig. 3b). A phase-contrast microscopy also indicated that C6-1-MGP cells showed extended branching cell processes (Fig. S1C, arrows). This was also consistent with the observations in F-actin staining (Fig. S1D, arrows). However, MGP overexpression in C6-1 cells did not affect the cell growth of C6-1 cells (Fig. 3c), as well as their colony formation ability (Fig. 3d). Yet, the results from the cell scratch analysis demonstrated that the wound closure ability of C6-1 cells was significantly enhanced after the upregulation of MGP expression when compared with that observed in C6-1-mock (Fig. 3e). The cell invasion of C6-1 was also increased by MGP overexpression when compared to that of C6-1-mock cells (Fig. 3f). These findings reveal that increased expression of MGP can promote the motility of high proliferative C6-1 cells.
Upregulation of MGP promotes the cell migration and invasion of rat glioma cells. a Total RNA isolated from C6-1 infected by lenti-bsd (C6-1-mock) and C6-1 infected by lentivirus particles containing MGP cDNA (C6-1-MGP) was subjected to QPCR for the determination of MGP expression. b Total proteins were extracted from C6-1-mock and C6-1-MGP, and subjected to Western blot analysis to examine MGP production. The equal level of GAPDH proteins is referred as a loading control. c The cell growth of C6-1-mock and C6-1-MGP cells in 10% FBS-containing medium was examined by MTT assay at the different time points after seeding. d C6-1-mock and C6-1-MGP cells were seeded at the density of 200 cells/well onto a 6-well plate. The number of the colony formation in the cultures was counted at 7 day after seeding. e C6-1-mock and C6-1-MGP cells reached confluence in 10% FBS-containing medium, the cultures were subjected to cell scratch migration assay in serum-free medium. The representative images shown in the upper panel were taken from the cultures at the initial time point, and 12 or 36 h after cell scratching. The quantification of the area of wound closure was conducted as described in the “Materials and Methods” section. f C6-1-mock and C6-1-MGP cells were plated onto the upper side of the transwell insert filters for cell invasion. After 6 h, the cells that migrated to the bottom side of the filter were counted (upper panel). The quantification of the invaded cells was counted as described in the “Materials and Methods” section. Data are presented as mean ± SEM of three independent experiments. *p < 0.05, ***p < 0.001 versus C6-1-mock. Scale bar in e and f, 200 μm
Promoted Migration of Rat Glioma Cells by MGP in the Brain
To validate the regulatory function of MGP in C6 glioma cell migration and invasion, an ex vivo organotypic brain slice culture was established. C6-2-scramble cells with GFP expression (GFP+-C6-2-scramble) and C6-2-sh68 cells tagged by GFP (GFP+-C6-2-sh68) were implanted into the brain slices at 2-day post brain slice preparation (Fig. 4a). After 4 days post injection, these implanted cells on the brain slices were traced by examining the location of GFP+ cells (Fig. 4b). The results showed that GFP+-C6-2-scramble and GFP+-C6-2-sh68 were observed at the areas within 2 mm from the injected site. However, the quantitation of GFP fluorescence intensity analyzed at the four migratory intervals between the injection site and the longest detectable region (0–2 mm) indicated that GFP+-C6-2-sh68 cells were largely accumulated at the injected region (Fig. 4b, arrows; 4C). GFP+-C6-2-sh68 cells located at the distance of 1.5–2 mm from the injected site were much less than GFP+-C6-2-scramble cells (Fig. 4c). Alternatively, we also conducted the experiments by implantation of GFP+-C6-1-mock and GFP+-C6-1-MGP cells into the brain slices and then examined their migration in the brain slice 4 days post injection (Fig. 4a). Similar to the findings as stated above, these injected GFP+-C6-1 cells were found at the injection site, and at the regions 2 mm away from the injection site (Fig. 4d, arrows). Interestingly, C6-1-MGP cells displayed a directional migratory pattern (Fig. 4d, arrowheads), whereas the migration of C6-1-mock cells was random. After a 4-day incubation, the farthest migration distance for the implanted C6 cells was about 2 mm (Fig. 4e). The amount of C6-1-MGP cells at the longer intervals (1.0–1.5 and 1.5–2.0 mm) was much higher than that of C6-1-mock cells (Fig. 4e). In addition, C6-1-MGP cells remaining at the injection site were less than C6-1-mock cells. These results further reveal that the migration of C6-1-MGP cells was much faster than that observed in C6-1-mock cells.
Upregulation of MGP promotes cell migration of rat glioma cells on ex vivo brain slices. a C6-2 (C6-2-scramble or C6-2-sh68) or C6-1 (C6-1-mock or C6-1-MGP) cells expressing GFP were injected into the region nearby the corpus callosum (CC) of rat brain slices (a, upper panel). The migratory cells were analyzed in the four indicated regions (500 μm × 500 μm at each interval) from the injected site. The diagram containing the procedure of ex vivo experiments illustrates that cells were implanted into the brain slices at 2 days after slice preparation, and then maintained in the culture for another 4 days. b At 6-day post slice preparation, the brain slices were fixed and then subjected to GFP immunofluorescent staining for the examination of C6-2 (C6-2-scramble or C6-2-sh68) cells (green) under a fluorescence microscope. c The quantification of GFP immunofluorescent intensity in each indicated region was conducted as described in the “Materials and Methods” section. d The implanted GFP+ C6-1-mock and GFP+ C6-1-MGP cells (green) were investigated under a fluorescence microscope. e The quantification of GFP immunofluorescent intensity in each indicated region was conducted. The representative images shown in b and d were taken from one of three repeated brain slices. Arrows in b and d indicate the cells migrated away from the implanted size (open arrows). Arrowheads in b and d show the track of the directional migration. The results in c and e are represented as the arbitrary units of GFP fluorescence intensity along the four distance intervals. Data are presented as mean ± SEM of three independent brain slices. #,*p < 0.05, p*** < 0.001 versus C6-2-scramble or C6-1-mock. Scale bar in b and d, 50 μm
Furthermore, the implantation of C6-1-mock and C6-1-MGP cells into the adult rat brain was performed. The implanted C6-1-mock and C6-1-MGP cells around the injected site were then examined by observing GFP+ cells at 7-dpi (Fig. 5a). Interestingly, the tumor size formed by implanted C6-1-MGP cells at the injected site was smaller than the one detected in C6-1-mock-injected group, although the difference was not biostatistical significant (Fig. 5b). C6-1-mock cells were found scantly at the periphery of the tumor (Fig. 5c (a), arrows), corpus callosum (Fig. 5c (b), arrows), and subventricular zone (Fig. 5c (c), arrows). However, C6-1-MGP cells were highly spread over the boundary of the tumor site (Fig. 5c (d, e), arrows), migrated into the corpus callosum (Fig. 5c (e), arrows), and invaded into the subventricular zone (Fig. 5c (f), arrows). Notably, accumulative C6-1-MGP cells were observed along the ventricle (Fig. 5c (f), arrowheads). Thus, the results demonstrate that the increased invasion and infiltrative ability of C6-1 cells by MGP overexpression might raise the risk of diffusive tumor generation.
Tumor formation and cell invasion of C6-1-mock and C6-1-MGP cells after implantation into rat brains. a C6-1-mock and C6-1-MGP cells were implanted into adult rats as described in the “Materials and Methods” section. b Brain tissue sections collected at 7-dpi were subjected to H/E histological staining for the measurement of tumor volume. The tumor volumes generated by C6-1-mock or C6-1-MGP cells at 7-dpi were 8.66 ± 1.75 or 5.60 ± 2.58 mm3, respectively (n = 6 rats in each animal group). Results are expressed as means ± SEM. p = 0.35 versus C6-1-mock group. c The brain tissue sections were also subjected to GFP immunofluorescence to examine the location of C6-1-mock and C6-1-MGP cells. These GFP+ C6-1 cells in brain regions denoted in a (a–f) are indicated by arrows. The GFP+ C6-1 clumps along the ventricle are indicated by arrowheads. cc corpus callosum, ctx cortex, T tumor, V ventricle. Scale bar, 50 μm
Discussion
In this study, we show evidence that rat glioma cells containing abundant expression of MGP (C6-2) displayed a faster migratory activity than that observed in the high proliferative glioma cells (C6-1) with a lower expression level of MGP. The results from in vitro experiments demonstrate that the downregulation of MGP expression reduced the cell motility of C6-2 cells, whereas increased expression of MGP improved C6-1 cell migration and invasion. The observations using an ex vivo brain slice model further confirm that the upregulation of MGP expression promoted the migration of C6-1 cells under three-dimensional microenvironment, whereas the downregulation of MGP expression reduced C6-2 cell invasion in brain slice.
Altered Expression of MGP in Glioma Cell Proliferation
Despite that the association of MGP upregulation in the poor prognosis of glioma and breast cancer has been reported [24, 27], its involvement in tumor cell dynamics (i.e., cell proliferation, migration, and survival) remains obscure. The recent study has indicated that microRNA-mediated repression of MGP expression in MCF-7 breast tumor cell line induced MCF-7 cell proliferation [34]. In contrast, our findings demonstrate that the declined cell growth of C6-2 cells was caused by MGP downregulation through the shRNA-mediated approach. Thus, it is very likely that MGP effect on the regulation of tumor cell proliferation is dependent on cancer types. Nevertheless, we found that MGP overexpression in high proliferative C6-1 did not affect their cell growth. In comparison with genes expressed in C6-2 cells, C6-1 cells not only highly express molecules associated with cell proliferation, but also produce cytokines and chemokines abundantly [29]. Besides, it is noted that MGP overexpression in C6-1 caused no significant change in the expression of IL-6, IL-33, CXCL12, and TGFβ2 (Fig. S2). These cytokines and chemokines abovementioned have been reported to enhance glioma progression [29, 35, 36]. Thus, based on the discrepancy of cell proliferation between C6-1 and C6-2 cells, MGP effect on the promotion of glioma cell proliferation remains arguable. Moreover, given that GBM tumor consists of heterogeneous subpopulations with dissimilar morphologies, self-renewal, and proliferative capacities [37], evidence provided in this study indicates that the deregulation of MGP expression, in general, could be insufficient to reduce glioma cell proliferation.
MGP Function in Glioma Cell Migration
Similar to the findings that human astrocytoma cells with high motility displayed a lower proliferative capability [11], we show here that MGP-enriched C6-2 cells exhibiting less proliferative and tumorigenic activity have higher migratory and invasion ability. Alike the role of MGP in tumor cell proliferation, MGP function in the cell migration and invasion of peripheral tumors is debated. For instance, the inverse correlation of MGP expression to tumor size, lymph-node metastasis, and tumor grade has been reported in the renal cell carcinomas [21]. Moreover, it has been suggested that the loss of MGP expression is associated with prostatic carcinoma progression and metastasis [21]. The recent findings have shown that silence of MGP expression in breast tumor cell line MCF-7 can increase MCF-7 cell invasion [34]. However, in a mouse model, MGP has been indicated to alter endothelial adhesion and trans-endothelial migration in vitro, as well as promoted in vivo osteosarcoma metastasis into the lung [38]. The observations regarding MGP expression in human GBM specimens have evidently indicated that MGP expression is positively correlated to GBM grades [25, 27]. Although the transient inhibition of MGP expression in human glioma cell lines (U373fast, H4, and U343MG) can reduce the migration and invasion of these cell lines [27], it is not clear if decreased glioma cell behavior can be prolonged by stably declined expression of MGP. The distinct proliferative and migratory capabilities of rat C6-1 and C6-2 glioma cells provide us advantageous useful cell models to further dissect the function of MGP in the regulation of glioma cell migration. The findings shown here that stably MGP-KD in C6-2 cells suppressed their cell motility, whereas MGP overexpression in C6-1 cells promoted their cell migration and invasion. The structure of filopodia and lamellipodia was rarely formed in C6-2 cells with MGP-KD, reflecting the observations showing lower cell migration and invasion after MGP downregulation in C6-2 cells. Alternatively, the elongation feature of C6-1 cells receiving forced MGP overexpression might be a cause of promoted cell migration and invasion. In addition, the study using ex vivo brain slice has shown that C6-1 cells with MGP overexpression displayed in directed linear migration path and moved toward longer distance than control cells did. We present here that implantation of C6-1 cells with MGP overexpression can form a smaller tumor in the rat brain at 7-day post implantation than the control group, although it has a non-biostatistical difference. The in vitro findings demonstrate again that the deregulation of MGP expression did not alter the proliferation of C6-1 glioma cells as discussed above. However, our in vitro, ex vivo, and in vivo studies that clearly pointed to the positive control of MGP in rat glioma cell migration and invasion, suggest that the upregulation of MGP in high proliferative glioma cell population may enhance intracranial migration and cause poor prognosis. Thus, the identification of this glioma cell subpopulation in human GBM tissues can allow us to precisely determine if MGP is a potential promising target for anti-glioma progression and recurrence.
Factors in the Regulation of MGP Expression
MGP expression has been detected in human GBM-derived tumor cell population with enriched expression of inhibitor of differentiation/DNA binding 4 (Id4) that was highly produced in human GBM and promoted angiogenesis in glioma xenografts [26]. It has been also addressed that elevation of MGP expression was involved in Id4-mediated glioma cell proliferation and Id4-induced angiogenesis [26]. Those results show the positive correlation of Id4 expression with MGP production, and point to its regulatory roles in glioma progression by promoted angiogenesis. Yet, based on our observation that Id4 expression in C6-2 cells is at a much lower level than C6-1 cells (Fig. S3A), the regulation of MGP expression in C6-2 cells is Id4-irrelevant. Others have shown that MGP can be upregulated by bone morphogenetic protein 4 (BMP-4) in bovine aortic endothelial cells (BAEC) in TGFβ-dependent pathway [39]. Since TGFβ and BMP-4 gene expression was significantly higher in C6-1 cells than that of C6-2 cells (Fig. S3B), TGFβ and BMP-4 control mechanism for the regulation of MGP expression cannot explain the fact that abundant expression of MGP was detected in C6-2 cells, but not in C6-1 cells. Thus, the discrepancy of MGP expression levels in C6-1 and C6-2 cells might be due to the complex regulatory pathways induced by the distinct multiple intrinsic factors in these two cell lines. In addition, molecular mechanisms responsible for MGP expression in rodent glioma cells might be very different from the ones defined in human cells or human GBM-derived tumor cells as described above. Nevertheless, further evidence is needed to identify the regulatory roles of the abovementioned factors in glioma MGP expression.
Conclusions
Our study shows evidently that MGP expressing in rat glioma cells contribute to the positive control of tumor cell migratory ability. In conjunction with clinical relevance of increased MGP expression within higher grade glioma, our study suggests that the upregulated level of MGP in the highly proliferative rat glioma cell model can enhance glioma cell migration both in vitro and in vivo. This finding sheds light on the importance of the characterization of MGPhigh/proliferativehigh glioma subpopulations in GBM specimens, which could be as a novel therapeutic target for anti-glioma infiltration to gain more effective tumor eradication.
References
Ohgaki H (2009) Epidemiology of brain tumors. Methods Mol Biol 472:323–342
Robins HI, Peterson CG, Mehta MP (2003) Combined modality treatment for central nervous system malignancies. Semin Oncol 30(4 Suppl 9):11–22
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M (2006) Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2(9):494–503 quiz 491 p following 516
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114(2):97–109
Cloughesy TF, Cavenee WK, Mischel PS (2014) Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9:1–25
Parker NR, Khong P, Parkinson JF, Howell VM, Wheeler HR (2015) Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol 5:55
Omuro A, DeAngelis LM (2013) Glioblastoma and other malignant gliomas: a clinical review. JAMA 310(17):1842–1850
Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA et al (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro-Oncology 16(7):896–913
Claes A, Idema AJ, Wesseling P (2007) Diffuse glioma growth: a guerilla war. Acta Neuropathol 114(5):443–458
Suzuki SO, Iwaki T (2005) Dynamic analysis of glioma cells: looking into “movement phenotypes”. Neuropathology 25(3):254–262
Giese A, Loo MA, Tran N, Haskett D, Coons SW, Berens ME (1996) Dichotomy of astrocytoma migration and proliferation. Int J Cancer 67(2):275–282
Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P (2006) Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia 53(8):799–808
Giese A, Bjerkvig R, Berens ME, Westphal M (2003) Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol 21(8):1624–1636
Price SJ, Pena A, Burnet NG, Pickard JD, Gillard JH (2004) Detecting glioma invasion of the corpus callosum using diffusion tensor imaging. Br J Neurosurg 18(4):391–395
Price PA, Rice JS, Williamson MK (1994) Conserved phosphorylation of serines in the Ser-X-Glu/Ser(P) sequences of the vitamin K-dependent matrix Gla protein from shark, lamb, rat, cow, and human. Protein Sci 3(5):822–830
Price PA, Williamson MK (1985) Primary structure of bovine matrix Gla protein, a new vitamin K-dependent bone protein. J Biol Chem 260(28):14971–14975
Price PA, Urist MR, Otawara Y (1983) Matrix Gla protein, a new gamma-carboxyglutamic acid-containing protein which is associated with the organic matrix of bone. Biochem Biophys Res Commun 117(3):765–771
Fraser JD, Price PA (1988) Lung, heart, and kidney express high levels of mRNA for the vitamin K-dependent matrix Gla protein. Implications for the possible functions of matrix Gla protein and for the tissue distribution of the gamma-carboxylase. J Biol Chem 263(23):11033–11036
Hale JE, Fraser JD, Price PA (1988) The identification of matrix Gla protein in cartilage. J Biol Chem 263(12):5820–5824
Shanahan CM, Cary NR, Metcalfe JC, Weissberg PL (1994) High expression of genes for calcification-regulating proteins in human atherosclerotic plaques. J Clin Invest 93(6):2393–2402
Levedakou EN, Strohmeyer TG, Effert PJ, Liu ET (1992) Expression of the matrix Gla protein in urogenital malignancies. Int J Cancer 52(4):534–537
Januchowski R, Zawierucha P, Rucinski M, Nowicki M, Zabel M (2014) Extracellular matrix proteins expression profiling in chemoresistant variants of the A2780 ovarian cancer cell line. Biomed Res Int 2014:365867
Bosma AJ, Weigelt B, Lambrechts AC, Verhagen OJ, Pruntel R, Hart AA, Rodenhuis S, van ’t Veer LJ (2002) Detection of circulating breast tumor cells by differential expression of marker genes. Clin Cancer Res 8(6):1871–1877
Yoshimura K, Takeuchi K, Nagasaki K, Ogishima S, Tanaka H, Iwase T, Akiyama F, Kuroda Y et al (2009) Prognostic value of matrix Gla protein in breast cancer. Mol Med Rep 2(4):549–553
van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G et al (2003) Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol 163(3):1033–1043
Kuzontkoski PM, Mulligan-Kehoe MJ, Harris BT, Israel MA (2010) Inhibitor of DNA binding-4 promotes angiogenesis and growth of glioblastoma multiforme by elevating matrix GLA levels. Oncogene 29(26):3793–3802
Mertsch S, Schurgers LJ, Weber K, Paulus W, Senner V (2009) Matrix gla protein (MGP): an overexpressed and migration-promoting mesenchymal component in glioblastoma. BMC Cancer 9:302
Grobben B, De Deyn PP, Slegers H (2002) Rat C6 glioma as experimental model system for the study of glioblastoma growth and invasion. Cell Tissue Res 310(3):257–270
Fang KM, Yang CS, Lin TC, Chan TC, Tzeng SF (2014) Induced interleukin-33 expression enhances the tumorigenic activity of rat glioma cells. Neuro-Oncology 16(4):552–566
Fang KM, Lin TC, Chan TC, Ma SZ, Tzou BC, Chang WR, Liu JJ, Chiou SH et al (2013) Enhanced cell growth and tumorigenicity of rat glioma cells by stable expression of human CD133 through multiple molecular actions. Glia 61(9):1402–1417
Chai KM, Wang CY, Liaw HJ, Fang KM, Yang CS, Tzeng SF (2014) Downregulation of BRCA1-BRCA2-containing complex subunit 3 sensitizes glioma cells to temozolomide. Oncotarget 5(21):10901–10915
Sliwa M, Markovic D, Gabrusiewicz K, Synowitz M, Glass R, Zawadzka M, Wesolowska A, Kettenmann H et al (2007) The invasion promoting effect of microglia on glioblastoma cells is inhibited by cyclosporin A. Brain 130(Pt 2):476–489
Humpel C (2015) Organotypic brain slice cultures: a review. Neuroscience 305:86–98
Tiago DM, Conceicao N, Caiado H, Laize V, Cancela ML (2016) Matrix Gla protein repression by miR-155 promotes oncogenic signals in breast cancer MCF-7 cells. FEBS Lett 590(8):1234–1241
Hau P, Jachimczak P, Schlaier J, Bogdahn U (2011) TGF-beta2 signaling in high-grade gliomas. Curr Pharm Biotechnol 12(12):2150–2157
Weissenberger J, Loeffler S, Kappeler A, Kopf M, Lukes A, Afanasieva TA, Aguzzi A, Weis J (2004) IL-6 is required for glioma development in a mouse model. Oncogene 23(19):3308–3316
Soeda A, Hara A, Kunisada T, Yoshimura S, Iwama T, Park DM (2015) The evidence of glioblastoma heterogeneity. Sci Rep 5:7979
Zandueta C, Ormazabal C, Perurena N, Martinez-Canarias S, Zalacain M, San Julian M, Grigoriadis AE, Valencia K et al (2016) Matrix-Gla protein promotes osteosarcoma lung metastasis and associates with poor prognosis. J Pathol 239:438-449
Yao Y, Zebboudj AF, Shao E, Perez M, Bostrom K (2006) Regulation of bone morphogenetic protein-4 by matrix GLA protein in vascular endothelial cells involves activin-like kinase receptor 1. J Biol Chem 281(45):33921–33930
Acknowledgements
The authors are grateful for the technical assistance provided by Ms. Chia-Hsin Ho. This study was also supported by the Ministry of Science and Technology (MOST 103-2314-B-006-007-MY3), Taiwan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Animal surgery was approved by the Institutional Animal Care and Use Committees at the National Cheng Kung University (Tainan, Taiwan).
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Fu, MH., Wang, CY., Hsieh, YT. et al. Functional Role of Matrix gla Protein in Glioma Cell Migration. Mol Neurobiol 55, 4624–4636 (2018). https://doi.org/10.1007/s12035-017-0677-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-017-0677-1