Abstract
Oligomerisation of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complexes is required for synaptic vesicle fusion and neurotransmitter release. How these regulate the release of pain peptides elicited by different stimuli from sensory neurons has not been established. Herein, K+ depolarization was found to induce multiple sodium dodecyl sulfate (SDS)-resistant SNARE complexes in sensory neurons exposed to botulinum neurotoxins (BoNTs), with molecular weights ranging from 104–288 k (large) to 38–104 k (small). Isoform 1 of vesicle-associated membrane protein 1 (VAMP 1) assembled into stable complexes upon depolarisation and was required for the participation of intact synaptosome-associated protein of relative molecular mass 25 k (SNAP-25) or BoNT/A-truncated form (SNAP-25A) in the large functional and small inactive SDS-resistant SNARE complexes. Cleaving VAMP 1 decreased SNAP-25A in the functional complexes to a much greater extent than the remaining intact SNAP-25. Syntaxin 1 proved essential for the incorporation of intact and SNAP-25A into the large complexes. Truncation of syntaxin 1 by BoNT/C1 caused /A- and/or /C1-truncated SNAP-25 to appear in non-functional complexes and blocked the release of calcitonin gene-related peptide (CGRP) elicited by capsaicin, ionomycin, thapsigargin or K+ depolarization. Only the latter two were susceptible to /A. Inhibition of CGRP release by BoNT/A was reversed by capsaicin and/or ionomycin, an effect overcome by BoNT/C1. Unlike BoNT/B, BoNT/D cleaved VAMP 1 in addition to 2 and 3 in rat sensory neurons and blocked both CGRP and substance P release. Thus, unlike SNAP-25, syntaxin 1 and VAMP 1 are more suitable targets to abolish functional SNARE complexes and pain peptide release evoked by any stimuli.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) drive the fusion of intracellular secretory vesicles with the pre-synaptic plasma membrane by forming a four-helix bundle which, in turn, causes neurotransmitter release that underlies synaptic transmission [1]. Synaptosome-associated membrane protein of relative molecular mass 25 k (SNAP-25) and syntaxin, the target (t) SNAREs, together with the vesicular (v) SNARE, vesicle-associated membrane protein (VAMP) are the key components of neuronal core SNARE complexes [2]. Of the four parallel α helices in the latter, one is contributed by syntaxin, another by VAMP and two by the pair of SNARE motifs in SNAP-25 [3–6]. Their assembly has been precisely dissected in vitro, using exogenously over-expressed fluorescently labelled SNAREs and cell-free reconstitution systems, and found to be resistant to sodium dodecyl sulfate (SDS) without boiling [1, 6–9]. The outcome from these approaches needs to be related to the situation in the native state because the fusion kinetics are strongly dependent on the experimental conditions. For example, even the weak interaction of syntaxin with VAMP can be sufficient to induce fusion of SNARE-containing liposomes [10]. Moreover, it is not possible to include in one assay all the numerous accessory proteins that are known to facilitate or inhibit exocytosis, by interacting with the SNAREs. Overcoming these hurdles requires a systematic investigation of SNARE complex assembly in situ. In this way, the relevance of cell-free studies can be highlighted. Furthermore, deciphering details of the functions/properties of endogenous SNAREs in different cells and vesicle secretory pathways are warranted, especially considering the subtleties of these natural processes [8]. In fact, a recent study confirmed that three oligomers of SNARE complexes are required for keeping the nanodisc pore open so as to efficiently release the vesicle contents [9]; a similar number had been observed using cracked PC-12 cell and recombinant SNAREs [11]. Oligomerisation of SNARE complexes seems to vary according to vesicle and cell types [12]. So far, there is no detailed characterization of SNARE oligomerisation in sensory neurons; hence, it is particularly important to investigate the functional relevance of such SNARE complexes for the exocytosis of pain peptides.
Cleavage of any of the three key SNAREs by botulinum neurotoxins (BoNTs) disrupts vesicle fusion and, in turn, blocks neurotransmitter release [13]. There are seven serotypes of BoNTs, A to G. SNAP-25 is truncated by three serotypes at distinct peptide bonds: /A (Gln197-Arg198), /C1 (Arg198-Ala199) and /E (Arg180-Ile181) [14]. Syntaxins 1–3 are cleaved by /C1 at Lys253-Ala254 (syntaxin 1A) and Lys252-Ala253 (for syntaxin 1B) [15, 16], whereas /B, /D, /F and /G cleave VAMPs 1, 2 and 3 albeit at separate sites; notably, 1 is resistant to /B in rat [17–19]. Cleavage of the SNARE proteins at such sites inhibits their assembly into stable SDS-resistant complexes [20] because these scissile bonds reside inside the associating segments, the so-called SNARE motifs [1]. After the latter form four α-helical bundles, the core domains become protease-resistant [21, 22]. Also, spontaneously and loosely assembled SNAREs form trans-SNARE complexes that are not resistant to SDS, unlike the fully zippered/assembled cis-complexes [23]. Recently, it was shown that all of the three SNAREs contribute to docking of secretory vesicles [24], as well as fusion [23]. Despite these remarkable advances, it still remains unclear whether and how the formation of stable complexes by BoNT-modified SNAREs relate to their inhibition of neurotransmitter release, although divergent functions have been attributed to different SDS-resistant complexes [25].
Release of neuropeptides from large dense-core vesicles (LDCVs) is also inhibited by BoNTs, as well as numerous classic neurotransmitters from small synaptic clear vesicles. The former includes a pain mediator, calcitonin gene-related peptide (CGRP) [26], which is a major transmitter released from sensory trigeminal ganglionic neurons (TGNs) in the chronic phase of migraine [27]. CGRP levels are increased during a migraine attack and decreased upon recovery [28]. CGRP release from rat TGNs evoked by K+ depolarization is blocked by BoNT/A, /C1 and /D but not /B [26]. In contrast to depolarization-induced CGRP exocytosis, that elicited by the activation of the transient receptor potential vanilloid type 1 (TRPV1) with capsaicin proved insensitive to BoNT/A; the resultant truncated SNARE, SNAP-25A, can form SDS-resistant SNARE complexes and afford CGRP release during prolonged elevation of [Ca2+] [29]. Accordingly, this restricted inhibition of exocytosis by BoNT/A has been overcome by targeting BoNT/E protease as a chimera of /A into /E-insensitive TGNs, thereby blocking TRPV1-mediated CGRP release and alleviating inflammatory pain in an animal model [29, 30]. These effects are associated with /E-cleaved SNAP-25 being precluded from incorporating into stable SNARE complexes [29, 30]. It is known that BoNT/B does not affect CGRP exocytosis from rat sensory TGNs, but due to its cleavage of VAMP 1 in mouse neurons, transmitter release becomes blocked, suggesting VAMP 1 is involved in the release of this pain peptide [26]. However, support for this has not been sought from specific knockdown or knockout of this isoform. Moreover, the relative contributions of VAMP 1 and 2 to this exocytosis remain to be determined; likewise, there is a lack of evidence for syntaxin 1 being involved in CGRP release from sensory neurons.
Convincing evidence is presented herein for the involvement of VAMP 1 in CGRP release by knocking down its expression in TGNs. Also, the diversity and oligomerisation of the SNARE complexes (containing VAMP 1, syntaxin 1 and SNAP-25) in sensory neurons were examined in cells treated with different BoNT serotypes, to cleave either t- or v-SNAREs or both. Relating these modifications to the formation of SDS-resistant SNARE complexes and to the toxins’ differential blockade of CGRP release elicited by distinct stimuli proved informative.
Materials and Methods
Animals and Ethics Statement
Pups from rats (Sprague Dawley) or mice (Tyler’s Ordinary) were bred in an approved BioResource Unit at Dublin City University. The experiments, maintenance and care of the rodents complied with the European Communities (Amendment of Cruelty to Animals Act 1876) Regulations 2002 and 2005. Experimental procedures had been approved by the Research Ethics Committee of Dublin City University and licenced by the Irish authorities (permit number 100/3609).
Antibodies and Other Reagents
Cell culture reagents, cytosine-β-d-arabinofuranoside, capsaicin, ionomycin, thapsigargin, TrypZean, soybean trypsin inhibitor, monoclonal antibodies (mAbs) against syntaxins 1A and 1B (HPC1), CGRP or phosphorylated neurofilament subunit 200 kDa (NF-200), polyclonal rabbit antibody against CGRP, short hairpin RNA (shRNA) lentiviral transduction particles and non-targeted PLK0.1-puro control particles were purchased from Sigma (Arklow, Co Wicklow, Ireland). Rabbit polyclonal antibody against TRPV1 was bought from Alomone Labs (Jerusalem, Israel). mAbs against substance P (SP) was obtained from Abcam (Cambridge, UK). CGRP and SP enzyme immunoassay (EIA) kits were obtained from SPI-BIO (Montigny Le Bretonneux, France). Synaptic Systems GmbH (Goettingen, Germany) supplied rabbit VAMP 1, 2 and 3 polyclonal antibodies and VAMP 2 mAbs; rabbit SNAP-25 polyclonal IgG was from Novus antibody (Cambridge, UK), and goat anti-rabbit Alexa-594 and goat anti-mouse Alexa-488 were provided by Jackson ImmunoResearch (Hamburg, Germany). Precast 4–12 or 12 % NuPAGE Bis-Tris gels and nerve growth factor (7S) were bought from Bio-Sciences (Dun Laoghaire, Co. Dublin). Enhanced chemiluminescence reagents and pET29a were obtained from Merck Millipore (Carrigtwohill, Cork, Ireland). Talon metal affinity resin was purchased from Clontech Laboratories, Inc. (Mountain View, USA).
Production of Recombinant BoNTs
Protein engineering of recombinant neurotoxins was approved by the Biological Safety Committee of Dublin City University and the Environmental Protection Agency of Ireland and performed in accordance with European Union regulations. Genes encoding BoNT/B, /C1 and/D single chains (SCs) were codon optimised for expression in Escherichia coli and synthesised according to the published sequences (GenBank accession numbers M81186, AB200359 and X54254, respectively). After verifying the DNA sequences, the synthetic genes were individually cloned into pET29a vector between Nde I and Xho I sites. The resultant constructs were transformed into BL21.DE3 strain and the SC proteins expressed, using auto-induction medium [31]. Recombinant toxins were purified by immobilised metal affinity chromatography on TALON resin [32]. BoNT/B was further purified by cation exchange chromatography following a previously described protocol for a BoNT/BA chimera [33]. Conversion of BoNT/B, /C1 and /D SCs to di-chain (DC) forms was achieved by incubation of 1 mg of toxin with 8, 2 and 2 μg of TrypZean at 22 °C for 1 h before the addition of trypsin inhibitor (final concentration 100 μg/ml). Protein concentrations were quantified using Bradford reagent. Recombinant BoNT/A was prepared and characterised as before [30]. Specific neurotoxicities of the generated nicked recombinant toxins were measured using a mouse lethality assay [32]. Note that nicked recombinant toxins were used throughout this study.
Culturing of Primary Neurons
TGNs and dorsal root ganglionic neurons (DRGs) were isolated from rats or mice and cultured as before [26, 34]. Briefly, trigeminal or dorsal root ganglia were dissected from postnatal day 3–5 rats or mice, placed in ice-cold Dulbecco’s modified Eagle medium (DMEM), washed and centrifuged at 170×g for 1 min before incubating the pellets at 37 °C for 30 min with collagenase I (1 mg/ml). The suspension was then gently triturated through 10-ml Falcon pipettes before adding DNase I (1 mg/ml) for 15 min. Following centrifugation at 170×g for 5 min, the pellet was suspended, washed and cultured in medium [DMEM, 10 % (v/v) heat-inactivated foetal bovine serum, 100 IU/ml penicillin, 100 μg/ml streptomycin and nerve growth factor (50 ng/ml)]. Cells were seeded onto 24-well plates pre-coated with poly-l-lysine (0.1 mg/ml) plus laminin (20 μg/ml) and maintained in a CO2 incubator at 37 °C. After 24 h and every other day thereafter, the supernatant was replaced with medium containing the anti-mitotic agent, cytosine β-d-arabinofuranoside (10 μM). Rat cerebellar granule neurons (CGNs) were prepared as before [32].
Knockdown of VAMP 1 with shRNAs
At 7 days in culture, mouse TGNs were incubated in medium containing shRNA lentiviral particles that specifically target VAMP 1 or non-targeted PLK0.1-puro control particles (400 transducing units/well). After culturing as above for 7–10 days, the cells were stimulated for 30 min before EIA of the CGRP released and Western blot analysis of protein knockdown. The specific knockdown of VAMP 1 without off-target effects was established by Western blotting, using specific antibodies for the other SNAREs or VAMP 2. For calculating the percent knockdown, the band intensity for VAMP 1 in shRNA-treated neurons was subtracted from the value obtained for control cells; the resultant number was expressed relative to that of the control.
Targeted gene/protein ID is NM_009496 and sequence for shRNA lentiviral transduction particles is CCGGGCCATCATCGTGGTAGTGATTCTCGAGAATCACTACCACGATGATGGCTTTTTG.
Cytochemical Staining
For immuno-staining, cells on coverslips (IBIDI, Martinsried, Germany) were washed, fixed, permeabilised and blocked as before [26]. NF-200 mAbs (dilution 1:500) and rabbit antibody against CGRP (1:1,000), SP mAbs (1:1,000) and rabbit antibody against CGRP (1:1,000), CGRP mAbs (1:1,000) and rabbit polyclonal TRPV1 antibody (1:100) or rabbit VAMP 1 polyclonal antibody (1:1,000) and mouse mAbs VAMP 2 (1:1,000) were applied in the blocking solution for 1 h at 22 °C and, after washing, fluorescent secondary antibodies (goat anti-rabbit Alexa 568 or 488 and goat anti-mouse Alexa 488 or 568) were added for 1 h. Bright field and fluorescent pictures were taken with an inverted confocal microscope (Zeiss LSM 710). Omission of primary antibody from the fluorescence staining gave the background for secondary antibody; the signal intensity above this was taken as positive reactivity.
Drug Treatments
At 7 days in culture, medium was gently aspirated from the TGNs; control or cells treated with BoNTs were incubated with basal buffer (low concentration of potassium (LK), mM: 3.5 KCl, 22.5 hydroxyethyl piperazineethanesulfonic acid (HEPES), 78.5 NaCl, 1 MgCl2, 2.5 CaCl2, 3.3 glucose and 0.1 % bovine serum albumin, pH 7.4) or stimulation solutions (high concentration of potassium (HK), mM: 22.5 HEPES, pH 7.4, 22 NaCl, 60 KCl, 1 MgCl2, 2.5 CaCl2, 3.3 glucose and 0.1 % bovine serum albumin] or 1 μM capsaicin in LK). Release buffer containing 5 μM ionomycin or 0.05 % dimethyl sulfoxide (DMSO) vehicle was employed for Ca2+ reversibility studies. In the cases where ionomycin (12.5 μM) or thapsigargin (15 μM) were used for inducing CGRP release, these were diluted in basal buffer before adding to the cells, as described in the figure legends.
Quantification of CGRP and SP Release
In the case of TGNs treated with or without toxins, 0.5 ml of basal release buffer (LK or LK plus vehicles) or stimulation buffers (HK or LK plus thapsigargin, capsaicin and/or ionomycin) was added into each well, followed by a 30-min incubation at 37 °C [26]. To determine the amounts of CGRP or SP released, 0.1 or 0.05 ml of sample, respectively, was added to 96-well plates coated with mAbs against CGRP or antiserum against SP and EIA performed following instructions for the kits.
Assay of the Endocytosis of BoNTs
TGNs were pre-treated with 50 nM BoNT/B or /D for 24 h in culture medium and washed before applying 50 nM BoNT/A for 10 min in HK. After removal of the unbound toxins by washing, cells were cultured for 24 h before quantifying SNAP-25 cleavage by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting [30]. Endocytosis of BoNT/A into neurons was based on their cleavage intracellularly of SNAP-25, by densitometric scanning of the blots and analysis by ImageJ (National Institutes of Health, Bethesda, MD). For BoNT/A, the percent of SNAP-25 remaining was calculated relative to the total [i.e. BoNT/A-cleaved (SNAP-25A) and BoNT/A-uncleaved); in the case of BoNT/B or /D, intact VAMP 1 or 2 was normalised according to a loading control (syntaxin 1) before analysis relative to the untreated control cells.
Treatment of TGNs with BoNTs: Monitoring Effects on CGRP Release and SNARE Cleavage
After 7 days in culture, fresh medium or that containing 100 nM BoNT/A, /B, /C1 or /D were applied individually or sequentially to TGNs for 24 h at 37 °C. After removal of unbound toxin and measurement of CGRP release, the cells in each well were lysed by 0.2 ml of 2× SDS sample buffer, heated for 5 min at 95 °C and separated by SDS electrophoresis, using precast NuPAGE 12 % Bis-Tris gels [26]. Each SNARE was detected with specific IgGs and anti-species secondary antibodies. After development in enhanced chemiluminescence reagent, the lanes were analysed using the gel documentation system (G BOX Chemi-16) and intensities quantified with ImageJ software. The ratios calculated for intact VAMP or syntaxin and the requisite internal standard (i.e. a SNARE not susceptible to the toxin in use) in BoNT-treated samples were expressed relative to those for controls; in the case of SNAP-25, the proportions of full-length and truncated forms were calculated.
2-D Gel Electrophoresis
A 2-D SDS-PAGE method [35] was used to investigate the SDS-resistant SNARE complexes containing SNAP-25, VAMP 1 and syntaxin 1 present in control and BoNT-treated TGNs. After stimulation with K+ depolarization for 5 min at 37 °C, the cells were solubilized immediately in SDS sample buffer without boiling. Proteins were separated by SDS-PAGE on 4–12 % precast Bis-Tris gel, and each sample lane was cut into strips corresponding to different distances of migration. Small pieces of the chopped gels were boiled for 10 min in SDS sample buffer, left overnight at room temperature and boiled again for 5 min before loading the extracted proteins onto a second 12 % precast Bis-Tris gel. After re-electrophoresis and transfer to polyvinylidene difluoride membrane, SNAREs released from the complexes were detected by Western blotting.
Statistical Analysis
Probability values were determined with the use of Student’s two-tailed t test (non-significant (N.S.); p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001).
Results
Rat TGNs Display Characteristics of Sensory Neurons—Nociceptive Marker and Pain Mediators: Selective Truncation of SNAP-25 and/or Syntaxin 1 by BoNT/A or /C1 Blocks Release of Pain Peptides Whereas Cleavage of VAMP 2 by /B Failed to Inhibit This Release, Unlike/D that Additionally Truncated VAMP 1
As a means for affirming their neuronal identity, rat TGNs at 7 days in culture were labelled with a monoclonal antibody for NF-200, a conventional marker for neurons. Confocal microscopy revealed that this stained both the cell soma and processes; all these were, also, shown to contain CGRP, a major pain-mediating peptide (Fig. 1a). Moreover, a sensory neuron marker, TRPV1, was found in the CGRP-positive neurons by labelling with a selective antibody (Fig. 1b). Notably, another pain peptide, SP, co-occurred with CGRP in those neurons (Fig. 1c). Focusing at higher resolution on the cell body and fine fibres unveiled a striking punctate labelling of SP which co-localised well with CGRP (Fig. 1d). In conclusion, these cultured TGNs exhibit characteristics of nociceptive sensory neurons.
Representative confocal micrographs demonstrating that cultured TGNs are NF-200 positive, contain CGRP plus SP and express TRPV1. Rat TGNs were grown on IBIDI chambers for 7 days, fixed and permeabilized prior to labelling with a NF-200 mAb and rabbit CGRP polyclonal antibody, followed by goat anti-mouse Alexa 488 and anti-rabbit Alexa 568, b rabbit anti-TRPV1 and CGRP mAb followed by goat anti-rabbit Alexa 488 and anti-mouse Alexa 568 and c, d SP mAb and rabbit CGRP polyclonal antibody, followed by goat anti-mouse Alexa 488 and anti-rabbit Alexa 568. a, c Images of bright field (left panel) and its combination with fluorescent staining (right panel). Notably, a high-resolution confocal image (d) shows SP and CGRP are co-localised in punctate spots on the cell body and neurites (arrow). Scale bars in a–c, 20 μm
Recombinant BoNT/A-/D were purified from E. coli lysate (see “Materials and Methods”) and successfully nicked to a disulphide linked DC forms (Fig. 2a). The specific neurotoxicity for BoNT/A, /B, /C1 and /D are 2.0 ×, 4.0 ×, 0.6 × and 5 × 108 mLD50/mg, respectively.
Effects of BoNTs on SNARE cleavage and inhibition of CGRP and SP release. a Nicked recombinant BoNT/A-/D were subjected to SDS-PAGE in the presence or absence of dithiothreitol (DTT) followed by Coomassie staining. Note that heavy chain (HC) and light chain (LC) were separated only in the presence of DTT, confirming that the inter-chain disulphide bond had been formed in each serotype. b–e Rat TGNs cultured for 7 days were incubated with various doses of each BoNT for 24 h in culture medium. Ca2+-dependent CGRP release evoked by 60 mM KCl was assayed [26]. The cleavage of respective SNAREs by BoNTs was revealed by Western blotting of cell lysate with the indicated antibodies. The proportions of intact substrate remaining were calculated relative to an uncleaved internal control, using digital images of the gels. Concentrations of BoNT used in the representative blots in panels b–e were 100, 10, 1, 0.1, 0.01 and 0 nM; from right to left. f K+-evoked SP release is inhibited by BoNT/A, /C1 or /D but not /B. Data plotted are means ± SEM, n ≥ 3; n.s., not significant, **p < 0.01, ***p < 0.001 versus toxin-free control
In order to test the purified BoNTs (Fig. 2a) on the blockade of CGRP exocytosis due to the selective cleavage of certain SNARE protein at their distinct scissile bonds, rat cultured TGNs were incubated 24 h at 37 °C with BoNT/A, /C1, /B or /D at varying concentrations. CGRP release evoked by K+ depolarization from toxin treated or non-treated neurons over 30 min was measured using EIA, and the same cells were solubilised and subjected to SDS-PAGE followed by Western blotting to detect the cleavage of respective SNAREs. Notably, K+-evoked CGRP release was inhibited by BoNT/A with a concentration dependence identical to that for SNAP-25 cleavage (Fig. 2b). BoNT/C1 cleaved syntaxin 1 and, additionally, removed eight residues from C-terminal of SNAP-25 rather than nine by /A resulting in the blockade of CGRP release (Fig. 2c); however, the contribution of syntaxin 1 to CGRP release could not be determined because SNAP-25 also got truncated. Interestingly, 100 nM recombinant /B did not significantly affect CGRP release though VAMP 2 (Fig. 2d) and 3 (data not shown) got extensively cleaved. Notably, additional cleavage of VAMP 1 by /D gave concentration-dependent blockade of CGRP exocytosis (Fig. 2e). Likewise, K+-evoked CGRP release from rat DRGs was inhibited by BoNT/D but not /B (Fig. S1). In contrast, both toxins were found to inhibit CGRP release from mouse DRGs (Fig. S1). These data suggest that VAMP 1 may be also essential for CGRP release from other sources of sensory neurons.
Considering the striking co-localization of CGRP and SP in TGNs (Fig. 1d) and reported co-release of both peptides from LDCVs [36], it was necessary to establish whether SP release also requires VAMP 1. BoNT/B (100 nM) did not significantly inhibit SP release from rat TGNs, but the latter was blocked by BoNT/D, /A or /C1 (Fig. 2f). This provides evidence for the contribution of VAMP 1 to the exocytosis of these pain peptides.
Knockdown of VAMP 1 Expression in Rat TGNs Significantly Reduced K+-Evoked CGRP Release Demonstrating a Pivotal Role in Exocytosis; Its Cleavage by BoNT/D Inhibited the Stimulation-Dependent Endocytosis of BoNT/A
VAMP 1 expression in rat TGNs was significantly knocked down in neurites (Fig. 3a) and cell bodies (data not shown), by its specific shRNA lentiviral particles, compared to that in control cells treated with non-targeted virus; VAMP 2 content remained unaffected, as revealed by immunofluorescence staining with isoform-specific antibodies (Fig. 3a). Western blotting indicated that the extent of knockdown of VAMP 1 was ∼95 %, without any alteration in VAMP 2 (Fig. 3b). This, in turn, reduced the K+-evoked Ca2+-dependent CGRP release by up to ∼78 % (Fig. 3c). Such an essential role of VAMP 1 in mediating this exocytosis was confirmed by its cleavage and that of VAMP 2 by treatment with 50 nM BoNT/D for 24 h and associated inhibition to (∼80 %) of K+-evoked CGRP release (Fig. 3d, right panel). Although /B cleaved VAMP 2 (∼80 %), but not the resistant isoform 1 (see “Introduction”), CGRP release remained unaffected (Fig. 3d, right panel). This indicates that VAMP 1, rather than 2, serves an important function in CGRP exocytosis. In this context, it is pertinent that VAMP 1 was detected exclusively in sensory TGNs and not in CGNs (Fig. S2a).
VAMP 1 is essential for CGRP exocytosis and activity-dependent endocytosis of BoNT/A in rat TGNs. a Representative immunofluorescence images, prepared using VAMP 1 and 2 specific antibodies, show that infection of the cells with VAMP 1 shRNA lentivirus reduced its expression in neurites. TGNs at 7 days in culture were treated with VAMP 1 shRNA lentiviral particles for 10 days before washing, fixation and permeabilization. Note that VAMP 1 (red) displayed minimal levels of signals in neurites of the treated cells compared to the control, in contrast to the striking unaltered staining (green) observed for VAMP 2. Scale bars, 10 μm. b Duplicate Western blots demonstrate a significant loss of VAMP 1 in the shRNA-treated cells but not the control; note that there was no change in VAMP 2. c EIA data shows a substantial reduction of K+-evoked Ca2+-dependent CGRP release from the treated cells only. d Representative Western blots revealing truncation of VAMP 2 by BoNT/B which proved ineffective in lowering the stimulated endocytosis of BoNT/A, as reflected by SNAP-25 cleavage. On the other hand, BoNT/D cleaved VAMP 1 and 2 and blocked the uptake of BoNT/A. This arises from /D (but not /B) blocking evoked exocytosis of CGRP (right panel). e Estimation of the extents of cleavage of VAMP 1, 2 and SNAP-25 in d; exocytosis-coupled endocytosis of BoNT/A was blocked by pre-treating TGNs with BoNT/D, whereas /B failed to cause significant change. Data plotted are means ± SEM; n = 3; n.s., not significant, ***p < 0.001
As reported, BoNT/A enters neurons via synaptic vesicle recycling [37, 38] which is coupled to stimulated exocytosis; therefore, inhibition of exocytosis should block stimulated uptake of BoNT/A. To test this, rat TGNs were exposed to BoNT/D for 24 h before applying 50 nM BoNT/A in high [K+] for 10 min, washing and post-incubating for 24 h in fresh culture medium. This reduced the subsequent depolarization-induced internalisation of BoNT/A (Fig. 3d), as revealed by a significant decrease in SNAP-25 cleavage compared to that in cells treated with BoNT/A alone (Fig. 3d, e). In stark contrast, though BoNT/B cleaved VAMP 2, it failed to block CGRP exocytosis (Fig. 3d, e) and reduce stimulated BoNT/A uptake (Fig. 3d, e). Thus, it is concluded that VAMP 1 is essential for CGRP release and activity-dependent endocytosis by TGNs.
SNAP-25A Detected in SDS-Resistant SNARE Complexes: Additional Cleavage of VAMP 1 by BoNT/D Resulted in Preferential Incorporation of Intact Rather than Truncated SNAP-25 into Stable SNARE Complexes
Cultured TGNs were stimulated briefly with high [K+] buffer before being harvested in SDS sample buffer. Cell lysates were subjected to first dimensional gel electrophoresis without boiling. Then, gel strips chopped, according to the migration of the proteins, were boiled at 100 °C and kept at room temperature for 24 h before each sample was subjected to electrophoresis in the second dimension to resolve the SNARE complexes. Notably, most of syntaxin 1, SNAP-25 and VAMP 1 were present in rat and mouse control TGNs in the free forms before boiling, except for some detected in several SDS-resistant SNARE complexes, with predominant molecular weights of 104–288 k (Fig. 4a(I), b(I)); each contained the three SNAREs. Although VAMP 3 and negligible 2 were also detected in SDS-resistant complexes (Fig. S3), their contribution to neuropeptide release is excluded (see above and “Discussion”). Depolarisation of rat TGNs induced incorporation of VAMP 1 into large SDS-resistant SNARE complexes (Fig. S2b). When rat TGNs were incubated for 24 h in culture medium with 100 nM BoNT/A before stimulation, ∼70 % of the SNAP-25 got cleaved (Fig. 4a, lysate II); this SNAP-25A as well as the remaining full-length protein together with syntaxin 1 and VAMP 1 appeared in the 104–288 k complexes with a similar pattern to the untreated samples (Fig. 4a (II versus I)). Sequential treatment with /A followed by /B caused no change in the proportions of full-length SNAP-25 and SNAP-25A in complexes migrating at 104–288 k (Fig. 4a (IV)), although VAMP 2 and 3 got proteolysed (not shown); presumably, this outcome occurred because VAMP 1 is insensitive to /B in rat (Fig. 4a, lysate IV). Consistently, BoNT/B did not reduce the stable complexes in TGNs (Fig. 4a (III)). In striking contrast, BoNT/B cleaved VAMP 1 in mouse TGNs resulting in the reduction of SNARE complexes (Fig. 4b (III)). Exposure of /A-treated mouse TGNs to /B significantly decreased (by ∼70 %) the fraction of SNAP-25A present in the 104–288 k complexes but to a much lesser extent than the remaining SNAP-25 (Fig. 4b (IV versus II)). Notably, these patterns of SNARE complex formation accord with those observed in cells sequentially treated with /A followed by /D both in rat (Fig. 4a (V)) and mouse (Fig. 4b (V)). These results indicate the importance of VAMP 1 for enabling SNAP-25A to incorporate into SDS-resistant SNARE complexes in sensory neurons. Moreover, the noted occurrence of SNAP-25A in SDS-resistant SNARE complexes required VAMP 1 and syntaxin 1 (see below) with intact SNAP-25 being preferable to SNAP-25A.
VAMP 1 forms SDS-resistant SNARE complexes with syntaxin 1 and intact or SNAP-25A in TGNs; cleaving VAMP 1 decreased the amount of large SNAP-25A-containing complexes but to a much lesser extent than those containing intact SNAP-25. Rat (a) and mouse (b) TGNs were incubated for 24 h with 100 nM BoNT/A, /B or /A followed by /B or /D. After toxin removal, cells were stimulated with HK for 5 min, before lysis in SDS sample buffer for 2-D gel electrophoresis and analysis by Western blotting (see “Materials and Methods”). Syntaxin 1, SNAP-25 and VAMP 1 were detected in the large SNARE complexes (104–288 k) (a (I) and b (I)). The incorporation of SNAP-25A into large SNARE complexes requires intact VAMP 1; these were reduced by BoNT/D (V versus II) but not /B in rat (a IV versus a II) because it does not cleave VAMP 1 (a (III)). Treatment of mouse TGNs with either BoNT/B or /D decreased the content of SNAP-25A in the SNARE complexes (b (IV), b (V) versus b (II)). Representative blots from three independent experiments are shown; Asterisk indicates that 20 % of sample was used in these lanes
Cleavage of Syntaxin 1 by BoNT/C1 in TGNs Reduced the Occurrence of /C1-Truncated SNAP-25 (SNAP-25C1) in the Large Complexes Without Affecting the Presence of the Intact Form
As /C1 cleaves off eight C-terminal residues from SNAP-25, rather than the nine removed by /A, in addition to proteolysing syntaxin 1 (Fig. 5a (lysate II), b), its effects on the oligomerisation of SDS-resistant SNARE complexes were investigated. In contrast to the appearance of SNAP-25A in 104–288 k complexes (cf. Fig. 4a (II)), SNAP-25C1 predominantly occurred in the lower size range of 38–104 k with VAMP 1 (Figs. 5a (II) and S4), which were found to lack the ability to mediate CGRP release (see below). Only a minute amount of SNAP-25C1 resided in the 104–288 k complexes (Fig. 5a (II)), in direct contrast to SNAP-25A (cf. Fig. 4a (II)) that formed larger complexes. Unlike SNAP-25C1, the residual full-length form was detected predominantly in the larger 104–288 k complexes along with the intact syntaxin remaining (Fig. 5a (II)). Thus, SNAP-25, but not SNAP-25C1, can incorporate into the 104–288 k complexes after proteolysis (∼80 %) of syntaxin 1 (Fig. 5a (II)). Apparently, the residual intact syntaxin 1 preferentially binds to intact SNAP-25 and VAMP 1, with greater affinity than for SNAP-25C1.
Syntaxin 1 and VAMP 1 in rat TGNs are essential for retaining intact, BoNT/A- and /C1-truncated SNAP-25 in large SNARE complexes. a After cleavage of syntaxin 1 and SNAP-25 with /C1, the residual intact syntaxin and SNAP-25 still got incorporated into large complexes (II) unlike the BoNT/A- and /C1-truncated SNAP-25 (II, V, VI) that predominately ended up in the small forms. Subsequent cleavage of VAMP 1 by /D lowered the content of both complexes (IV). Consistently, BoNT/D alone reduced the level of large SNARE complexes (III). Incorporation of BoNT/A (V) or /C1-truncated (VI) SNAP-25 into the small SNARE complexes was independent of the sequence of toxins applied. Notably, SNARE complexes were virtually abolished by sequential treatment with BoNT/A, /C1 and /D (VII). Asterisk indicates that 20 % of sample was used in these lanes. b Schematic of the cleavage sites on SNAP-25 for the BoNT/A and /C1; /A cleaves one more residue than /C1 which additionally cleaves syntaxin 1, whereas BoNT/D and /B cleave VAMP 1 at distinct sites
Intact Syntaxin 1 and VAMP 1 Are Required for Creating Large Oligomers Containing SNAP-25: When Syntaxin 1 Was Cleaved SNAP-25A and SNAP-25C1 Occurred in Smaller Forms with VAMP 1
Treatment of rat TGNs with BoNT/D cleaved VAMP 1 resulting in significant reduction of syntaxin 1 and SNAP-25 in the large SNARE complexes compared to non-treated controls (Fig. 5a (III versus I)). In rat TGNs that were sequentially exposed to BoNT/C1 and /D, the proportions of large (104–288 k) and small (38–104 k) complexes containing both the residual SNAP-25 and SNAP-25C1 were dramatically reduced, compared to those in cells treated with /C1 only (Figs. 5a (IV versus II) and S5b). The same observations were made with mouse TGNs (Fig. S4). In contrast to the treatment of rat TGNs with BoNT/A alone (cf. Fig. 4a (II)), in which intact and cleaved SNAP-25 assembled into the large complexes, exposure to /A followed by /C1 resulted in the appearance of small complexes containing SNAP-25A and VAMP 1 (Fig. 5a (V)), presumably due to the additional cleavage of syntaxin (∼80 %) by /C1 (Fig. 5a (V)). Full-length SNAP-25 remained in the large oligomers (Fig. 5a (V)), similar to that seen with /C1 treatment alone (Fig. 5a (II)). This deduction is supported by the profile remaining unchanged when /C1 was applied before /A (Fig. 5a (VI)). Notably, large and small complexes were nearly abolished by exposure to /D following /A and /C1 (Fig. 5a (VII)). It is concluded that when syntaxin 1 is truncated, the cleavage of VAMP 1 by /D abrogates the presence of intact and truncated SNAP-25 in the SDS-resistant complexes.
Large Rather than Small SNARE Complexes in Sensory Neurons Support CGRP Exocytosis Triggered by Higher [Ca2+]i
/C1 Cleavage of Syntaxin 1 Inhibits Capsaicin-Elicited CGRP Release Despite Being Resistant to BoNT/A, Consistent with the Essential Presence of Syntaxin 1 in Large Functional SNARE Complexes
Semi-quantification of the relative proportions of large and small SDS-resistant SNARE complexes detected in rat TGNs under various treatments (Figs. 4, 5 and S4) revealed somewhat different profiles for the toxins’ effects on the two predominant sizes present. Notably, toxins or combinations that cleave VAMP 1 or syntaxin 1 (BoNT/D alone, /A followed by /D or /C1, or /C1 on its own, /C1 followed by /D or /A or sequentially applied /A, /C1 and /D) significantly reduced the formation of large SDS-resistant SNARE complexes (Fig. 6a). All the VAMP 1-cleaving toxins (BoNT/D or combinations with others) also decreased the assembly of small complexes, whereas the latter were not reduced by the syntaxin 1-cleaving /C1 or combinations with /A (Fig. 6b).
Large but not small SNARE complexes support capsaicin-induced CGRP release. The combined intensities of syntaxin 1, intact SNAP-25 and truncated forms (shown in Figs. 4, 5 and S5) for the large (a) or small (b) complexes from toxin-treated cells were related to the respective levels in toxin-free controls. c Capsaicin stimulation overcomes the inhibition of CGRP release by BoNT/A, but not other serotypes. Note that BoNT/B was unable to inhibit exocytosis because its target VAMP 1 in rat is not susceptible to cleavage. Data plotted are means ± SEM.; n = 3. d Immunoblots showing the patterns of cleavage of SNAP-25, VAMP 1 and syntaxin 1 by various BoNTs alone or in combination
The above-noted patterns of SNARE complexes were related to CGRP release from rat TGNs elicited by capsaicin, an agonist of TRPV1 present on these sensory neurons: its release was not affected by the removal of nine C-terminal amino acids from SNAP-25 by BoNT/A (Fig. 6c) presumably due to SNAP-25A forming large complexes. On the other hand, /C1-induced cleavage of syntaxin 1 by ∼80 % (Fig. 6d) gave ∼85 % inhibition of capsaicin-elicited CGRP release (Fig. 6c). Although /C1 also cleaves off eight C-terminal residues from SNAP-25 (Fig. 6d), it can be assumed that this would not affect the capsaicin-evoked exocytosis. Thus, intact syntaxin 1 is necessary for the latter. Despite BoNT/A cleaving ∼80 % of SNAP-25 (Fig. 6d) and failing to decrease capsaicin-elicited release (Fig. 6c), sequential application of /A followed by /C1 gave ∼85 % cleavage of syntaxin 1 and SNAP-25 and inhibited (∼90 %) the capsaicin-elicited CGRP release (Fig. 6c, d). This accords with SNAP-25A becoming inserted into smaller complexes due to the cleavage of syntaxin 1 (cf. Fig. 5a (V)). Notably, the greater inhibition by /C1 or sequential addition of /C1 and /D or /A and /C1 than either /A or /D alone (Fig. 6c) suggests that truncation of two or more SNAREs (Fig. 6d) exerts compounded inhibition of capsaicin-elicited CGRP release. Sequential treatment with /A followed by /B failed to inhibit capsaicin-elicited CGRP release (Fig. 6c) due to rat VAMP 1 being non-susceptible to/B; in stark contrast, /A followed by /D or /C1 and then /D blocked ≥70 % of the release (Fig. 6c). Interestingly, in the case of /C1 followed by /A, though theoretically SNAP-25C1 contains a cleavage site for /A (Fig. 5b), Western blotting did not detect any SNAP-25A in rat TGNs only SNAP-25C1 (Fig. 6d); accordingly, this treatment reduced by ∼90 % capsaicin-evoked CGRP release due to cleavage of syntaxin 1 (Fig. 6c, d). One possible explanation is that the substrate site for /A in the SNAP-25C1 is not recognised.
Diminishing the Large SNARE Complexes by BoNT/C1 Overcame the Ionomycin-Induced Ca2+ Reversibility of BoNT/A-Induced Inhibition of Exocytosis and Blocked CGRP Release in Response to All Stimuli
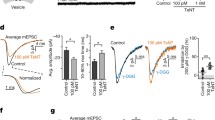
It became pertinent to investigate whether the Ca2+ reversibility of CGRP release inhibited by /A would be influenced by the presence of SNAP-25A in the large SNARE complexes, unlike SNAP-25C1 residing in mainly small forms. To test this, the release of CGRP induced by depolarisation, ionomycin or both from rat TGNs was quantified after pre-incubation with the different toxins or a combination. Notably, BoNT/A, /C1 or sequentially applied /A and /C1 resulted in 70–90 % inhibition of K+ depolarization-elicited CGRP release (Fig. 7a). Ionomycin (5 μM) overcame the inhibition by /A of K+-stimulated exocytosis (to ∼85 % of control), whereas the blockade by/C1, or /A followed by/C1, showed no significant reversal. In the case of capsaicin-treated cells, ionomycin failed to elicit any further CGRP release from cells treated with or without BoNT/A (Fig. 7b), suggestive of capsaicin achieving maximal release. In contrast to BoNT/A, /C1 or /A followed by /C1 reduced the release of CGRP evoked by capsaicin to ∼15 %, and this inhibition was unaffected by adding ionomycin (Fig. 7b). Thus, TRPV1-mediated Ca2+ reversibility of BoNT/A-induced inhibition could be overcome by BoNT/C1.
Effect of BoNTs on CGRP release from rat TGNs elicited by different stimuli. a Ionomycin (5 μM)-induced reversibility of the inhibition by BoNT/A of K+-elicited CGRP release is not seen with /C1 or /A followed by /C1. b Ionomycin did not cause any elevation of capsaicin-elicited CGRP release from cells treated with either /A, /C1 or /A followed by /C1. c Ionomycin (12.5 μM)-induced CGRP release was not blocked by /A, unlike /C1. d Both /A and /C1 inhibited CGRP release triggered by thapsigargin (10 μM). Vehicles for capsaicin (0.1 % ethanol) and ionomycin/thapsigargin (0.05 % DMSO) were included. Data plotted are means ± SEM; n ≥ 3; n.s., not significant, ***p < 0.001
To address whether Ca2+ entry pathways could affect BoNT/C1 inhibition of CGRP release, two reagents with distinct abilities to elevate [Ca2+]i were used to induce CGRP release. Notably, ionomycin-elicited CGRP release was not significantly affected by BoNT/A, whereas ∼75 % inhibition resulted from exposure to /C1 (Fig. 7c). Thus, BoNT/A is ineffective in inhibiting ionomycin-induced Ca2+-regulated CGRP release, as observed for capsaicin-evoked release. Next, the ability of /C1 was tested to inhibit CGRP release induced by internally stored Ca2+ using thapsigargin, an inhibitor of the ubiquitous sarco-endoplasmic reticulum Ca2+ ATPases. Incubation of TGNs with 15 μM thapsigargin elevated CGRP release up to ∼3 times the basal level (data not shown); importantly, BoNT/A or /C1 inhibited this augmented release (Fig. 7d). These collective findings demonstrated that diminishing the large SNARE complexes by BoNT/C1 blocks CGRP release independent of the stimulus used.
Discussion
Formation and functionality of SNARE complexes were examined in TGNs after treatment with BoNTs that cleave different SNAREs or at distinct bonds, by using 2-D gel electrophoresis and Western blotting, in conjunction with analysis of their inhibition of CGRP release induced by various stimuli. Though SNARE complexes were detected in BoNT-treated samples, only the larger form supported CGRP release elicited by elevating [Ca2+]i via distinct mechanisms. Considering the complexities of pain signalling, these findings might give insights into designing new generations of BoNT-based anti-nociceptives.
Intact VAMP 1 Is a Key Component for the Formation of SNARE Complexes in Peptidergic Sensory Neurons: Implications for Its Importance in Transmitter Release from Peripheral Nerves
It is already known that cleaving VAMPs 1, 2 and 3 by BoNT/D dramatically inhibits CGRP exocytosis from sensory neurons [26]; knockdown approach used herein specifically targeting VAMP 1 yielded direct evidence for involvement of this particular isoform. The effect of removing VAMP 1 in sensory nerves could not be compensated for by the other two isoforms present [26]. Clearly, VAMP 1 forms SNARE complexes independent of isoforms 2 and 3; also, stimulation augmented the amount in complexes which mediate activity-dependent exocytosis-coupled endocytosis (Fig. S2b). In contrast, fast Ca2+-triggered neurotransmitter release is virtually abolished by knocking out VAMP 2 in central neurons, and they could not be compensated by the other isoforms [39]. Attempts made to measure the stimulated release of glutamate, aspirate, GABA or glycine from small clear synaptic vesicle in TGNs proved to be difficult (data not shown); however, the possible contribution of VAMP 2 and 3 to mediating release of these classical small transmitter is not excluded, especially these isoforms found in the SDS-resistant complexes from sensory neurons. Further investigations on VAMP 1 are fully warranted because recent reports highlight its importance—rather than isoforms 2 and 3—for neurotransmitter release in the peripheral nervous system, not only in sensory but also motor neurons [26, 33, 40]. Moreover, VAMP 1 but not 2 or 3 is essential for substance P release and nociception in animal models; its proteolysis by BoNT/B in mouse, but not in rat where it is resistant, reduces intraplantar formalin-induced nocifensive behaviour as well as neuropathic pain in a model of spinal nerve ligation [41]. Cleavage of VAMP 1 by BoNT/D inhibits CGRP release from TGNs elicited by activation of TRPV1 that is non-susceptible to BoNT/A [29]. Furthermore, in nociceptive neurons, inflammation-induced insertion of functional transient receptor potential cation channel, subfamily A, member 1 into the surface membrane probably requires VAMP 1-dependent mechanisms [42]. Accordingly, analysis of the protein composition of neuropeptide-containing LDCVs from the dorsal horn of rat spinal cord, which contain afferent C- and Aδ-fibres of small neurons, confirmed that VAMP 1 is the only isoform associated with these vesicles [43].
Syntaxin 1 and VAMP 1 Preferably Bind Intact Rather Than /A- or /C1-Truncated SNAP-25 in Functional SDS-Resistant SNARE Complexes
It can be reasonably deduced from the data shown herein that syntaxin 1/intact SNAP-25 heterodimers can stabilise VAMP 1 in functional complexes to a greater extent than syntaxin 1/truncated-SNAP-25. This is consistent with a C-terminal domain of SNAP-25 being required for interaction with VAMP 1 but not for the binding of syntaxin 1 which occurs via its N-terminal domain [20]. In BoNT/A-treated TGNs, the large SDS-resistant SNARE complexes were found to contain only residual intact and four to fivefold more SNAP-25A, whereas BoNT/E cleaved SNAP-25E was absent from the complexes due to the removal of additional 17 C-terminal residues [29]. Although the absolute composition of these complexes (relative molecular mass (Mr) of 104–288 k) remains to be established, each SNARE should be equally represented; thus, it is unlikely that one complex could contain both truncated and full-length SNAP-25. Though VAMP 1 can bind syntaxin 1, SNAP-25 or SNAP-25A less avidly, after cleaving a majority of syntaxin 1 with/C1, both of the resultant /A- and /C-truncated SNAP-25 appeared in smaller complexes, whereas the residual intact SNAP-25 still showed up in the large forms. Our data suggest that cleavage of syntaxin 1 results in greater destabilisation of complexes containing /A- or /C-truncated-SNAP-25/VAMP 1 than those composed of intact SNAP-25/VAMP 1, although the same domain of syntaxin 1 is required for binding VAMP 1 and SNAP-25 [20]. As expected, cleaving syntaxin 1 followed by VAMP 1 led to a loss of intact and /A- or /C1-truncated SNAP-25 from both large and small complexes; this accords with the cleavage of residues 37–70 from VAMP 1 being known to prevent SDS resistance of SNARE complexes [20].
Sequential Application of BoNTs Could Compound the Blockade of CGRP Exocytosis and Alter the Profile of SNARE Complexes Commensurate with the Inhibitory Patterns
Diminution of large SNARE complexes seems to correspond to the extent of inhibition by BoNTs of CGRP release (Fig. 6a, c); however, the changes are characteristic of the toxin serotypes used. Treatment of rat TGNs with BoNT/D cleaved VAMP 1 and decreased the amount of SDS-resistant SNARE complexes by up to ∼3-fold, including the large and small varieties (Fig. 6a, b). This is in great contrast to the effect of /B which cleaved VAMP 2 and 3 but did not reduce either complex type; in turn, neither K+- nor capsaicin-induced CGRP release was blocked. These results reaffirm that participation of VAMP 1 in SNARE complexes plays a predominant role in exocytosis from sensory neurons [41–43]. It is notable that inhibition of CGRP exocytosis by BoNTs was found to depend on the serotypes and stimulation used; for example, though SNAP-25A incorporates into complexes, only K+-evoked or thapsigargin-induced CGRP release gets inhibited (Fig. 7a, d). The contrasting inability of BoNT/A to inhibit capsaicin-induced release accords with SNAP-25A being capable of binding synaptotagmin [44] although displaying greatly reduced sensitivity to [Ca2+] [45] and allowing vesicle docking, fusion and CGRP release to occur when elicited by a prolonged large elevation of [Ca2+]i by capsaicin or ionomycin (Figs. 6c and 7a–c). Capsaicin-elicited release can be mediated by intact syntaxin 1 and VAMP 1 despite the presence of SNAP-25A in the large complexes; accordingly, BoNT/D reduced the functionally relevant large complexes and inhibited capsaicin-elicited CGRP release.
A different situation was seen with /C1-treated TGNs because syntaxin 1 is required to hold SNAP-25 and VAMP 1 in large complexes; only the lower size form occurred when syntaxin 1 was cleaved and these were non-functional in CGRP release, regardless of the stimuli used that involve distinct Ca2+ entry mechanisms. The inhibition by /C1 of CGRP exocytosis triggered by any stimulus (K+ depolarization, bradykinin, capsaicin, ionomycin or thapsigargin) makes it a universal inhibitor. These data reaffirm that formation of large (Mr of 104–288 k) SNARE complexes requires all three SNAREs, especially intact VAMP 1 and syntaxin 1 and these but not the small species (Mr of 38–104 k) support exocytosis. Possibly, the small complexes loosely dock synaptic vesicles onto the plasma membrane but cannot fuse [46]. Sequential application of BoNTs (/A-/C1, /A-/D) might give synergistic anti-nociceptive effects in rodents due to extensive disruption of functional SNARE complexes. Although BoNT/D has different activities between rodent and humans [47], the information gleaned herein might help future designing of a new generation of BoNT-based therapeutics.
Distinct BoNT Sensitivities of CGRP Release Induced by Ca2+ Entry via Dissimilar Routes
BoNT/A and /C1 equally inhibited CGRP release evoked by a Ca2+-ATPase blocker, thapsigargin, that triggers Ca2+ entry from internal stores; on the other hand, /C1 proved effective in blocking exocytosis induced by capsaicin or the Ca2+ ionophore, ionomycin. The level of blockade by BoNT/A of CGRP exocytosis elicited with K+ depolarization was diminished by ionomycin, whereas /C1 prevented relief of the inhibition by ionomycin, consistent with SNARE complexes containing SNAP-25C1 being mainly of small size. The distinct stimuli tested involve different Ca2+ entry pathways. For example, TRPV1 channels are well known to be critical for pain transduction; their agonist, capsaicin, caused little increase in [Ca2+] in the absence of extracellular Ca2+ indicating that the increase in [Ca2+]i is not derived from internal Ca2+ stores [48, 49]. Like capsaicin, ionomycin-evoked release relies on extracellular rather than internal Ca2+ stores [50]. K+ depolarization is known to elicit Ca2+ entry through voltage-activated channels and this, in turn, induces Ca2+ release from the internal store [51]. The association between alteration of Ca2+ channel activity and development of pathological pain conditions is attracting much attention. In fact, certain channels are implicated in inflammatory and neuropathic hypersensitivity, so selective targeting of Ca2+ channels has become a new strategy for pain management [52]. Injured neurons display an elevated dependence on store-operated Ca2+ [53, 54]. As diminishing the formation of large functional SNARE complexes by BoNT/C1, /D or certain combinations blocks CGRP release elicited by these distinct stimuli, our findings might allow specific targeting of a particular BoNT serotype or combination to combat pain initiated via various excitatory signals.
Abbreviations
- BoNT:
-
Botulinum neurotoxin
- CGNs:
-
Cerebellar granule neurons
- CGRP:
-
Calcitonin gene-related peptide
- DC:
-
Di-chain
- DMEM:
-
Dulbecco’s modified Eagle medium
- DRGs:
-
Dorsal root ganglionic neurons
- EIA:
-
Enzyme immunoassay
- HC:
-
Heavy chain
- LC:
-
Light chain
- LDCVs:
-
Large dense-core vesicles
- mAb:
-
Monoclonal antibody
- SC:
-
Single chain
- SNAP-25:
-
Synaptosome-associated protein of relative molecular mass 25 k
- SNARE complexes:
-
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor complexes
- SP:
-
Substance P
- TGNs:
-
Trigeminal ganglionic neurons
- TRPV1:
-
Transient receptor potential vanilloid type 1
- VAMP 1 and 2:
-
Vesicle-associated membrane protein isoforms 1 and 2
References
Sudhof TC, Rothman JE (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323(5913):474–477
Rothman JE, Warren G (1994) Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol 4(3):220–233
Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395(6700):347–353
Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90(3):523–535
Otto H, Hanson PI, Jahn R (1997) Assembly and disassembly of a ternary complex of synaptobrevin, syntaxin, and SNAP-25 in the membrane of synaptic vesicles. Proc Natl Acad Sci U S A 94(12):6197–6201
An SJ, Almers W (2004) Tracking SNARE complex formation in live endocrine cells. Science 306(5698):1042–1046
Lang T, Margittai M, Holzler H, Jahn R (2002) SNAREs in native plasma membranes are active and readily form core complexes with endogenous and exogenous SNAREs. J Cell Biol 158(4):751–760
van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS, Jahn R (2010) One SNARE complex is sufficient for membrane fusion. Nat Struct Mol Biol 17(3):358–364
Shi L, Shen QT, Kiel A, Wang J, Wang HW, Melia TJ, Rothman JE, Pincet F (2012) SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science 335(6074):1355–1359
Jahn R, Scheller RH (2006) SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol 7(9):631–643
Hua Y, Scheller RH (2001) Three SNARE complexes cooperate to mediate membrane fusion. Proc Natl Acad Sci U S A 98(14):8065–8070
Montecucco C, Schiavo G, Pantano S (2005) SNARE complexes and neuroexocytosis: how many, how close? Trends Biochem Sci 30(7):367–372
Dolly JO, Wang J, Zurawski TH, Meng J (2011) Novel therapeutics based on recombinant botulinum neurotoxins to normalize the release of transmitters and pain mediators. FEBS J 278(23):4454–4466
Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Sudhof TC, Jahn R, Niemann H (1994) Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. J Biol Chem 269(3):1617–1620
Schiavo G, Shone CC, Bennett MK, Scheller RH, Montecucco C (1995) Botulinum neurotoxin type C cleaves a single Lys-Ala bond within the carboxyl-terminal region of syntaxins. J Biol Chem 270(18):10566–10570
Blasi J, Chapman ER, Yamasaki S, Binz T, Niemann H, Jahn R (1993) Botulinum neurotoxin C1 blocks neurotransmitter release by means of cleaving HPC-1/syntaxin. EMBO J 12(12):4821–4828
Schiavo G, Malizio C, Trimble WS, Polverino de Laureto P, Milan G, Sugiyama H, Johnson EA, Montecucco C (1994) Botulinum G neurotoxin cleaves VAMP/synaptobrevin at a single Ala-Ala peptide bond. J Biol Chem 269(32):20213–20216
Yamasaki S, Baumeister A, Binz T, Blasi J, Link E, Cornille F, Roques B, Fykse EM, Sudhof TC, Jahn R et al (1994) Cleavage of members of the synaptobrevin/VAMP family by types D and F botulinal neurotoxins and tetanus toxin. J Biol Chem 269(17):12764–12772
Yamasaki S, Binz T, Hayashi T, Szabo E, Yamasaki N, Eklund M, Jahn R, Niemann H (1994) Botulinum neurotoxin type G proteolyses the Ala81-Ala82 bond of rat synaptobrevin 2. Biochem Biophys Res Commun 200(2):829–835
Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof TC, Niemann H (1994) Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J 13(21):5051–5061
Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H (1995) Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. EMBO J 14(10):2317–2325
Skehel JJ, Wiley DC (1998) Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95(7):871–874
Chen YA, Scheller RH (2001) SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2(2):98–106
Wu Y, Gu Y, Morphew MK, Yao J, Yeh FL, Dong M, Chapman ER (2012) All three components of the neuronal SNARE complex contribute to secretory vesicle docking. J Cell Biol 198(3):323–330
Kubista H, Edelbauer H, Boehm S (2004) Evidence for structural and functional diversity among SDS-resistant SNARE complexes in neuroendocrine cells. J Cell Sci 117(Pt 6):955–966
Meng J, Wang J, Lawrence G, Dolly JO (2007) Synaptobrevin I mediates exocytosis of CGRP from sensory neurons and inhibition by botulinum toxins reflects their anti-nociceptive potential. J Cell Sci 120(Pt 16):2864–2874
Durham PL, Cady R (2011) Insights into the mechanism of onabotulinumtoxinA in chronic migraine. Headache 51(10):1573–1577
Doods H, Arndt K, Rudolf K, Just S (2007) CGRP antagonists: unravelling the role of CGRP in migraine. Trends Pharmacol Sci 28(11):580–587
Meng J, Ovsepian SV, Wang J, Pickering M, Sasse A, Aoki KR, Lawrence GW, Dolly JO (2009) Activation of TRPV1 mediates calcitonin gene-related peptide release, which excites trigeminal sensory neurons and is attenuated by a retargeted botulinum toxin with anti-nociceptive potential. J Neurosci 29(15):4981–4992
Wang J, Zurawski TH, Meng J, Lawrence G, Olango WM, Finn DP, Wheeler L, Dolly JO (2011) A dileucine in the protease of botulinum toxin A underlies its long-lived neuroparalysis: transfer of longevity to a novel potential therapeutic. J Biol Chem 286(8):6375–6385
Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41(1):207–234
Wang J, Meng J, Lawrence GW, Zurawski TH, Sasse A, Bodeker MO, Gilmore MA, Fernandez-Salas E, Francis J, Steward LE, Aoki KR, Dolly JO (2008) Novel chimeras of botulinum neurotoxins A and E unveil contributions from the binding, translocation, and protease domains to their functional characteristics. J Biol Chem 283(25):16993–17002
Wang J, Zurawski TH, Bodeker MO, Meng J, Boddul S, Aoki KR, Dolly JO (2012) Longer-acting and highly potent chimaeric inhibitors of excessive exocytosis created with domains from botulinum neurotoxin A and B. Biochem J 444(1):59–67
Malin SA, Davis BM, Molliver DC (2007) Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2(1):152–160
Lawrence GW, Dolly JO (2002) Multiple forms of SNARE complexes in exocytosis from chromaffin cells: effects of Ca2+, MgATP and botulinum toxin type A. J Cell Sci 115(Pt 3):667–673
Skofitsch G, Jacobowitz DM (1985) Calcitonin gene-related peptide coexists with substance P in capsaicin sensitive neurons and sensory ganglia of the rat. Peptides 6(4):747–754
Meng J, Wang J, Lawrence GW, Dolly JO (2013) Molecular components required for resting and stimulated endocytosis of botulinum neurotoxins by glutamatergic and peptidergic neurons. FASEB J 27(8):3167–3180
Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER (2006) SV2 is the protein receptor for botulinum neurotoxin A. Science 312(5773):592–596
Schoch S, Deak F, Konigstorfer A, Mozhayeva M, Sara Y, Sudhof TC, Kavalali ET (2001) SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science 294(5544):1117–1122
Liu Y, Sugiura Y, Lin W (2011) The role of synaptobrevin1/VAMP1 in Ca2+-triggered neurotransmitter release at the mouse neuromuscular junction. J Physiol 589(Pt 7):1603–1618
Huang PP, Khan I, Suhail MS, Malkmus S, Yaksh TL (2011) Spinal botulinum neurotoxin B: effects on afferent transmitter release and nociceptive processing. PLoS One 6(4):e19126
Schmidt M, Dubin AE, Petrus MJ, Earley TJ, Patapoutian A (2009) Nociceptive signals induce trafficking of TRPA1 to the plasma membrane. Neuron 64(4):498–509
Zhao B, Wang HB, Lu YJ, Hu JW, Bao L, Zhang X (2011) Transport of receptors, receptor signaling complexes and ion channels via neuropeptide-secretory vesicles. Cell Res 21(5):741–753
Schiavo G, Stenbeck G, Rothman JE, Sollner TH (1997) Binding of the synaptic vesicle v-SNARE, synaptotagmin, to the plasma membrane t-SNARE, SNAP-25, can explain docked vesicles at neurotoxin-treated synapses. Proc Natl Acad Sci U S A 94(3):997–1001
Sakaba T, Stein A, Jahn R, Neher E (2005) Distinct kinetic changes in neurotransmitter release after SNARE protein cleavage. Science 309(5733):491–494
Popoff MR, Poulain B (2010) Bacterial toxins and the nervous system: neurotoxins and multipotential toxins interacting with neuronal cells. Toxins (Basel) 2(4):683–737
Eleopra R, Montecucco C, Devigili G, Lettieri C, Rinaldo S, Verriello L, Pirazzini M, Caccin P, Rossetto O (2013) Botulinum neurotoxin serotype D is poorly effective in humans: an in vivo electrophysiological study. Clin Neurophysiol 124(5):999–1004
Pecze L, Blum W, Schwaller B (2013) Mechanism of capsaicin receptor TRPV1-mediated toxicity in pain-sensing neurons focusing on the effects of Na+/Ca2+ fluxes and the Ca2+-binding protein calretinin. Biochim Biophys Acta 1833:1680–1691
Wisnoskey BJ, Sinkins WG, Schilling WP (2003) Activation of vanilloid receptor type I in the endoplasmic reticulum fails to activate store-operated Ca2+ entry. Biochem J 372(Pt 2):517–528
Keller JE, Neale EA (2001) The role of the synaptic protein snap-25 in the potency of botulinum neurotoxin type A. J Biol Chem 276(16):13476–13482
Usachev Y, Shmigol A, Pronchuk N, Kostyuk P, Verkhratsky A (1993) Caffeine-induced calcium release from internal stores in cultured rat sensory neurons. Neuroscience 57(3):845–859
Brittain JM, Duarte DB, Wilson SM, Zhu W, Ballard C, Johnson PL, Liu N, Xiong W, Ripsch MS, Wang Y, Fehrenbacher JC, Fitz SD, Khanna M, Park CK, Schmutzler BS, Cheon BM, Due MR, Brustovetsky T, Ashpole NM, Hudmon A, Meroueh SO, Hingtgen CM, Brustovetsky N, Ji RR, Hurley JH, Jin X, Shekhar A, Xu XM, Oxford GS, Vasko MR, White FA, Khanna R (2011) Suppression of inflammatory and neuropathic pain by uncoupling CRMP-2 from the presynaptic Ca2+ channel complex. Nat Med 17(7):822–829
Rigaud M, Gemes G, Weyker PD, Cruikshank JM, Kawano T, Wu HE, Hogan QH (2009) Axotomy depletes intracellular calcium stores in primary sensory neurons. Anesthesiology 111(2):381–392
Gemes G, Bangaru ML, Wu HE, Tang Q, Weihrauch D, Koopmeiners AS, Cruikshank JM, Kwok WM, Hogan QH (2011) Store-operated Ca2+ entry in sensory neurons: functional role and the effect of painful nerve injury. J Neurosci 31(10):3536–3549
Acknowledgments
This research is funded by a Principle Investigator Award (to J.O.D.) from Science Foundation Ireland which supports Dr. Meng and, in part, by Allergan Inc.
Conflict of Interest
We declare that this research is funded in part by Allergan Incorporated, P.O. Box 19534, Irvine, CA 92623, USA. The industrial sponsor had no role in study design, data collection and analysis or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplemental Fig. S1
BoNT/D but not /B inhibited K+-evoked CGRP release from rat cultured DRG neurons, whereas both blocked its release from mouse DRGs. The latter were dissected from postnatal day 5 rats (open bars) or mice (hatched bars), dissociated and cultured for 7 days before being exposed to 100 nM recombinant BoNT/D, /B or toxin-free medium for 24 h at 37 °C; K+-evoked Ca2+-dependent CGRP release was assayed using EIA, as before, and plotted as % of the value for the respective toxin-free control. Results are means ± SEM; n ≥ 3 (PDF 11 kb)
Supplemental Fig. S2
VAMP 1 is enriched in rat TGNs unlike central CGNs; VAMP 1 forms SNARE complexes with SNAP-25 and syntaxin 1 upon depolarisation. a Equal number of cells were lysed by SDS buffer for SDS-PAGE and Western blotting. Note that TGNs express more VAMP 1 than CGNs. b Upon stimulation of rat TGNs by HK, VAMP 1 was readily detectable in large (Mr of 104–288 k) SNARE complexes (PDF 60 kb)
Supplemental Fig. S3
The presence of VAMP 3 and a negligible amount of 2 in SDS resistant SNARE complexes. Rat TGNs were depolarized with high [K+] for 5 min at 37 °C before being lysed for 2-D electrophoresis. Antibodies against syntaxin 1, SNAP-25 and VAMP 2 were used for Western blotting. Note that VAMP 2 was hardly detected in SDS-resistant complex despite the presence of its free form (upper panel). Aliquots of the same samples were subjected to separate SDS-PAGE for probing VAMP 3 (lower panel). (PDF 91 kb)
Supplemental Fig. S4
In mouse TGNs, like rat, cleavage of syntaxin 1 leads SNAP-25C1 into small complexes. After cleaving syntaxin 1 and SNAP-25 with 100 nM/C1, cells were washed and stimulated with 60 mM K+ in the presence of 2.5 mM Ca2+ for 5 min, before lysis in SDS sample buffer for 2-D electrophoresis and immunoblot analysis. Note that the residual intact SNAP-25 resides predominantly in the large complexes whereas /C1-truncated SNAP-25 occurred in the small forms. Subsequent cleavage of VAMPs by /D reduced both complexes (PDF 178 kb)
Supplemental Fig. S5
Direct comparison of the levels of the two sizes of SNARE complexes in TGNs treated with BoNTs. After incubation with 100 nM toxins, the cells were depolarized with K+ for 5 min before being harvested in SDS buffer for 2-D electrophoresis. SNARE complexes of large (104–288 k) and small (38–104 k) sizes were analysed. a BoNT/D but not /B decreased the content of both sizes of SNARE complexes. b /C1 cleavage of syntaxin 1 resulted in the appearance of the majority of SNAP-25C1 in the small SNARE complexes; subsequent exposure to/D diminished the large and small SNARE complexes. c TGNs treated with BoNT/C1 followed by BoNT/A further reduced the intact SNAP-25 in the large complexes but did not preclude the presence of SNAP-25C1 in the smaller complexes; SNAP-25 cleaved by BoNT/A alone resided predominately in the large complexes. SNAP-25C1 occurs in the small rather than the large complexes after cleavage of syntaxin 1. d Treatment with /A followed by /D decreased both the large and small complexes (PDF 175 kb)
Rights and permissions
About this article
Cite this article
Meng, J., Dolly, J.O. & Wang, J. Selective Cleavage of SNAREs in Sensory Neurons Unveils Protein Complexes Mediating Peptide Exocytosis Triggered by Different Stimuli. Mol Neurobiol 50, 574–588 (2014). https://doi.org/10.1007/s12035-014-8665-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-8665-1