Abstract
This study examines the production of five phenolic acids (chlorogenic acid, neochlorogenic acid, ferulic acid, caffeic acid and p-coumaric acid) following over-expression of AtPAP1 transcription factor by four transgenic root clones of Leonurus sibiricus after Agrobacterium rhizogenes transformation. The AtPAP1 expression level was estimated by quantitative real-time PCR. High levels of phenolic acids were found in the transgenic roots of L. sibiricus and were determined by high-performance liquid chromatography–mass spectrometry analysis. Additionally, transgenic roots showed antimicrobial potential and cytotoxic activity on glioma cells in IV grade. Our results suggest that L. sibiricus transformed roots with AtPAP1 gene over-expression may represent a potential source of phenolic acids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are known to produce a number of compounds of commercial value including medicines, fuels and industrial materials [1]. However, although the metabolites themselves are well characterized, little is known of the genes involved in their production. The use of transgenic manipulation may allow the biosynthesis of these compounds to be enhanced for industrial exploitation and evaluation [2]. One exhaustively studied factor is AtPAP1, otherwise known as Arabidopsis Production of Anthocyanin Pigment 1, a gene which encodes an MYB transcription factor from Arabidopsis. It is an important factor involved in the biosynthesis of anthocyanins [3,4,5]. Over-expression of AtPAP1 can effectively induce the accumulation of some phenylpropanoid derivatives such as anthocyanin in tobacco, hops, rose, Salvia miltiorrhiza and canola by regulating the related pathway genes, such as phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), anthocyanidin synthase (ANS) and flavonol synthase (FLS) [6,7,8]. As AtPAP1 is known to induce the biosynthesis of phenylpropanoids, the present study investigates their potential in metabolic engineering.
Leonurus sibiricus L., a member of the Lamiaceae, is a herbaceous plant grown in crop fields in many countries in Asia. The genus Leonurus contains about 20 species [9]. This plant is an annual, biannual or perennial, aromatic plant which is commonly used as a medical and culinary herb, but the literature reports that it shows analgesic, anti-inflammatory, antioxidant and anti-atherogenic potential, as well as antihemorrhagic, anti-diabetic, antibacterial, anti-tumour and allelopathic potential [10,11,12,13].
Our earlier studies showed that the normal and transformed roots of L. sibiricus contained phenolic acids such as chlorogenic acid, ferulic acid, caffeic acid, p-coumaric, ellagic acid, syringic acid and vanillic acid [14, 15]. Transformed roots are induced around the site of Agrobacterium rhizogenes infection, and their fast growth and phenotypic stability make them potential candidates for large-scale secondary metabolite production. The present study uses AtPAP1 to enhance phenylpropanoid production in L. sibiricus transgenic roots following A. rhizogenes-mediated co-transformation: this transcription factor is known to greatly increase the levels of phenolic acids in transgenic roots. The transgenic root extract of L. sibiricus was also found to possess antimicrobial and cytotoxic effect.
The data obtained in this study support the development of the metabolic engineering of high-value products in medicinal plants, and the tested system may well be ideally suited for studying the biosynthesis of secondary plant compounds of medicinal and economic value.
Materials and Methods
Construction of the Plant Expression Vector Harbouring the AtPAP1 Gene
The plant expression vector was constructed based on the pCAMBIA 1305.1 binary vector. The coding sequence of AtPAP1 was synthesized by Biomatik Co. (Canada) and cloned between specific sequences recognizable by NcoI and Eco72I restriction enzymes into the pUC57-AtPAP1 vector. After double enzyme digestion of both vectors, the AtPAP1 gene was cloned into pCAMBIA1305.1, replacing the GUSPlus gene and generating a pCAMBIA1305.1-AtPAP1 vector for plant transformation. Figure 1 shows a schematic representation of T-DNA in the recombinant pCAMBIA1305.1-AtPAP1 vector. A schematic map was generated using SnapGene software (from GSL Biotech; available at snapgene.com).
Agrobacterium rhizogenes Transformation
Agrobacterium rhizogenes strain A4 was transformed by the freeze–thaw transformation method [16].
Agrobacterium rhizogenes-Mediated Plant Material Transformation
The A. rhizogenes A4 strain carrying pCAMBIA1305.1-AtPAP1 vector was grown for 2 days at 28 °C on 100 mL YEP medium supplemented with 50 mg/L kanamycin (for plasmid selection) in a 200-mL Erlenmeyer flask (rotary shaker 120 rmp). A. rhizogenes cells were collected by centrifugation at 4000 rmp for 20 min and diluted to OD600 = 0.7 with Murashige and Skoog (MS) medium.
The A. rhizogenes transformation was performed in 5-week-old shoots cultured in vitro on Murashige and Skoog medium with 3% sucrose and 0.8% agar. The shoots were cut at the nodes and immersed in bacterial cell suspension on a Petri dish for 1 min. The explants were dried using sterile filter paper to remove excess bacterial culture and incubated in the dark at 24 °C for 5 weeks on MS medium. Control explants were transformed identically in sterile YEB medium without A. rhizogenes. After 2 weeks, the hairy roots started to appear at the wound sites of the transformed explants.
This procedure was repeated three times using 20 plant explants per repetition. The hairy roots (1–2 cm in length) formed on plant explants were individually transferred into MS solid (0.8% agar) medium containing 250 mg/L cefotaxime and 25 mg/L hygromycin B and incubated at 26 °C in the dark. The growing transformed roots were separated into individual Petri dishes (each clone) and subcultured for 2 weeks. Following this, each transformed root clone was transferred into 50 mL (in 250-mL Erlenmeyer flask) of Schenk and Hildebrandt (SH) [17] liquid medium with 3% sucrose containing 250 mg/L cefotaxime and 25 mg/L hygromycin B. The roots were transferred into a fresh liquid medium five times every week. After 5 weeks, the concentration of cefotaxime and hygromycin B was reduced to 100 and 10 mg/L, respectively, and the hairy roots were subcultured every week for the following month. After this time, the antibiotics were eliminated from the medium and four clones (AtPAP1 TR clones 1–4 and next name as the transgenic root clones) exhibiting the best growth rate were chosen for further culture and analysis. These clones were cultured in 80 mL of SH liquid medium in a 300-mL Erlenmeyer flask on a rotary shaker at 80 rpm; the culture was performed in the dark at 26 °C for 5 weeks. After this time, the fresh weight and the dry weight of the culture were measured.
Confirmation of Transgenic L. sibiricus Roots
Total genomic DNA was isolated from hairy roots using the cetyltrimethylammonium bromide (CTAB) method [18]. PCR was used to confirm the presence of the transgenic fragment using hptII-specific primers (F: 5′- CTATTTCTTTGCCCTCGGAC-3, R: 5′ATGAAAAAGCCTGAACTCACC-3′) (1026 pz). The genomic DNA of hairy roots induced by Agrobacterium without pCAMBIA-AtPAP1 vector was used as a negative control. The PCR procedure was performed in a BIOMETRA UNO thermal cycler; the reaction consisted of 0.2 mM of dNTP, 0.5 uM of each primer, 1U of Taq DNA polymerase (Thermo Scientific), 2 mM MgCl2 with 200 ng of genomic DNA in 1×Taq buffer in 20 μL reaction volume. The PCR used the following stages: initial denaturation at 95 °C for 2 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 60 s. pCAMBIA-AtPAP1 vector isolated from A. rhizogenes was used as a positive control.
Establishment and Confirmation of Transformation L. sibiricus Roots
Five-week-old L. sibiricus shoots were transformed in vitro by infection with A. rhizogenes strain A4. The procedure was performed according to Sitarek et al. [14]. The roots were cultured in the dark in 300-mL Erlenmeyer flasks with 80 mL of SH medium using a rotary shaker (80 rpm). Successful transformation of the rolB and rolC genes to T-DNA was confirmed by PCR according to Skała et al. [19]. We obtained five transformed root lines, but for further studies we selected only one clone which showed the highest biomass and phenolic metabolite production (data not shown).
RNA Extraction and cDNA Synthesis
For transcript profiling, total RNA was taken from the roots of transgenic AtPAP1 TR clones 1–4 of L. sibiricus: it was isolated using Syngen Plant RNA MINI Kit reagent. Following this, a TranScriba kit (A&A Biotechnology, Poland) was used to synthesize first-strand cDNA in a 20 µL reaction mix according to the manufacturer’s instructions.
qRT-PCR Conditions and Analyses
Quantitative real-time PCR analysis was performed on the Agilent Technologies Stratagene Mx300SP working on MxPro software. The following primer sequences were used: AtPAP1 F-5′TGG AGG GTT CGT CCA AAG 3′ and R-5′CTT CTC CAT ACT TAT TAA TGC ACT GTC 3′. Briefly, each reaction was performed in a 10 µL mix containing 1 μL of cDNA, 04 μL of each primer and 5 μL of Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA) and distilled water. The elongation factor 1α (EF-1α) geneF-5′TGAGATGCACCACGAAGCTC-3′and R-5′CCAACATTGTCACCAGGAAGTG-3′ was chosen as an internal control for normalization. qRT-PCR conditions were 10 min of initial denaturation at 95 °C and then 40 cycles of 15 s at 95 °C 60 s at primers’ annealing temperature (Tm). Each sample was analysed in triplicate. In the melting curve analysis, the levels of the gene were normalized to that of the elongation factor 1α (EF-1α) gene used to test the specificity of amplification. The expression of the genes was calculated by the comparative Ct method [20].
Preparing Extracts from Transgenic Roots of L. sibiricus
The AtPAP1 TR clones 1–4 and TR without construct (10 g d.w. samples of lyophilized and powdered plant materials) were used as material. The extraction process was as follows: 15-min extraction in 500 mL 80% (v/v) aqueous methanol in a 35 °C ultrasonic bath, followed by two 15-min extractions with 300 mL of 80% (v/v) aqueous methanol. The product was then filtered and combined. The filtrate was then evaporated under reduced pressure, lyophilized to dryness and then kept in the dark for investigation. The extracts contained the following amounts of AtPAP1 (w/w) with regard to initial dry weight: 51.4% for TR clone 1, 49.7% for TR clone 2, 50.3% for clone 3, 51.8% for clone 4 and 51.7% for TR. These extracts were used for the HPLC analysis and the analysis of their biological properties.
HPLC Analyses
LC-MS/MS was used to identify the phenolic acids. The content of the acids was determined by HPLC [14].
Antimicrobial Activity
Microbial Strains and Growth Conditions
The following strains were tested: Staphylococcus aureus (ATCC 25923), Enterococcus faecalis (ATCC29212), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853) and the yeasts Candida albicans (ATCC 10231) and Saccharomyces cerevisiae (ATCC 2601). The growth conditions of all tested microorganisms were described previously by Sitarek el al [21].
Determining the Minimum Bactericidal Concentration (MBC), Minimum Fungicidal Concentration (MFC) and Minimum Inhibitory Concentration (MIC)
AtPAP1 clone 1 extract was used in this study. The minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC or MFC) of the tested extract were determined as detailed previously [22].
Cell Line
Normal human astrocytes (NHA) and human glioma cell lines (stage IV) were used in this study. The human glioblastoma primary cell line (stage IV) derived from a surgical specimen was established in the Department of Molecular Genetics. Cell culture was performed as described by Sitarek et al. [14, 15].
MTT Cell Viability Assay
Cell viability was evaluated by MTT assay [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide] [23]. Briefly, human glioma cells and normal human astrocytes seeded onto 96-well microplates at 1 × 105 cells/100 μL per well were subjected to 24-h incubation with 0–1 mg/mL AtPAP1 TR clone 1 extract. The cells were then subjected to 4-h incubation with 100 μL MTT solution (5 mg/mL MTT in PBS). Formazan crystals were than dissolved in DMSO, the plates were read at 550 nm, and the IC50 value was determined according to a 0–1 mg/mL dose–response curve. At each concentration, all tests were performed in triplicate.
Statistical Analysis
Continuous data were presented as mean with standard deviation (SD). As the data were found to have a non-normal distribution, it was compared using the Mann–Whitney U test. p values < 0.05 were considered as statistically significant. Analyses were performed using STATISTICA software v.13 for Windows (StatSoft Inc).
Results
Establishment of L. sibiricus Transgenic Root Clones with Arabidopsis AtPAP1 Construct and TR
The AtPAP1 gene under the control of the pCAMBIA 1305.1 promoter was transferred into L. sibiricus by A. rhizogenes transformation. The first adventitious roots were observed 2 weeks after inoculation, and 68.5% of explants had responded after another 3 weeks (Fig. 2a1, a2). No roots developed on uninfected control explants. In addition, other 5-week-old shoots were transformed by A4 A. rhizogenes without the construct: a total of 40% of these shoots produced roots in the wound location. Four AtPAP1 clones (1 to 4) and one TR clone of L. sibiricus were obtained, and these were cultured for 5 weeks in SH liquid medium. After this time, the fresh weight of the transgenic roots clones increased 33–35 times from 2.5 to 83–87 g/L and the dry weight increased 29–31 times. The fresh weight of the TR showed lower increases in biomass (53 g/L of fresh weight and 9.2 g/L of dry weight). A culture time longer than 5 weeks resulted in the roots browning and dying. Of the four transgenic root clones of L. sibiricus, AtPAP1 clone 1 showed fastest growth in SH medium and was selected for further studies. Morphological observations found the transgenic roots with AtPAP1 transcript to be shorter and thicker with greater branches (Fig. 2b) than TR (Fig. 2c).
Molecular Characterization and Gene Expression of Transgenic Roots and TR
PCR analysis of AtPAP1 transgenic root clones 1–4 was performed with hptII-specific primers. Bands of expected sizes (1026 bp) were found in the corresponding transgenic samples, but not in the TR control root line without the construct (Fig. 3). To further verify gene expression in the tested root clones, transcripts of AtPAP1 were analysed by qRT-PCR using the EF-1α gene as a housekeeping reference gene. The results showed over-expression of the AtPAP1 gene in all transgenic root clones and lower expression in the TR control roots. Furthermore, the transcription levels varied among the transgenic clones. The highest relative expression was found in AtPAP1 clone 1 and the lowest in AtPAP1 clone 4 (Fig. 4).
Relative expression levels of AtPAP1 among control and transgenic clones. The expression levels were relative to EF-1α as described in methods. Each value represents mean ± SE (n = 3 replicates). *p < 0.05 compared AtPAP1 TR clone 1 with AtPAP1 TR clones 2, 3 and 4; **p < 0.05 compared AtPAP1 TR clone 4 with AtPAP1 TR clones 2 and 3
The Quantitative Analysis of Phenolic Acid Production in TR and Transgenic Roots Over-expressing AtPAP1
The results of HPLC analysis indicate that the accumulation of neochlorogenic acid, caffeic acid, ferulic acid, p-coumaric acid and chlorogenic acid was greater in AtPAP1 over-expressing transgenic roots clones 1–4 than in TR control roots without construct (Table 1). Our results showed that the most productive clone was AtPAP1 TR clone 1, which demonstrated greater production all of phenolic acids than TR control roots. The chlorogenic acid content in the AtPAP1 TR clone 1 root was 19.392 mg d.w., i.e. 4.72 times higher than in the TR control (4.104 mg/g of dry weight) (p < 0.05) (Table 1). Next, caffeic acid was 11.380 mg/g of dry weight in the AtPAP1 TR clone 1 root, i.e. 2.72 times higher than that in the TR control root (4.176 mg/g of dry weight). The accumulation of neochlorogenic acid, p-coumaric and ferulic acid was found to be 2.25 times, 1.73 times and 1.77 times greater in the AtPAP1 TR clone 1 root than the TR control root. In turn, for AtPAP1 clones 2 and 3, chlorogenic acid increased by 18.016 mg/g of dry weight and 9.128 mg/g of dry weight, respectively, and caffeic acid by 8.792 mg/g of dry weight and 6.634 mg/g of dry weight, respectively, compared with the TR control root. AtPAP1 clone 4 did not demonstrate any significant increase in the production of phenolic acids compared with TR control root (4.04 mg/g dry of weight and 4.176 mg/g dry of weight) (p < 0.05) (Table 1). AtPAP1 clone 1 showed the highest amount of phenolic acids and was therefore chosen for further biological studies.
MTT Viability on Glioma Cells After Treatment with AtPAP1 TR Clone 1 Transgenic Roots Extract of L. sibiricus
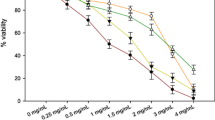
MTT assay was performed in grade IV glioma cells after treatment with AtPAP1 TR clone 1 transgenic roots extract of L. sibiricus after 24 h. The tested extract did not demonstrate any cytotoxic effect on normal cells in the concentration range 0–1 mg/mL after 24 h. The AtPAP1 TR clone 1 extract was found to have a cytotoxic effect on grade IV glioma cells at the IC50 concentration of 0.75 mg/mL (Fig. 5).
MTT assay of L. sibiricus TR and transgenic AtPAP1 TR clone 1 root extract in glioma cells (grade IV) and normal human astrocytes (NHA). Cells were treated with AtPAP1 TR clone 1 root extract at various concentrations 0.25, 0.50, 0.75 and 1 mg/mL for 24 h. The data represent the mean ± SD of three independent experiments. *p < 0.05 compared grade IV with NHA in appropriate time (24 h)
Antimicrobial Activity
The antimicrobial activities of the AtPAP1 TR clone 1 transgenic L. sibiricus root extract were quantitatively assessed by determining the MIC and MBC/MFC, as given in Table 2. The MIC and MBC/MFC values of AtPAP1 clone 1 transgenic L. sibiricus root extract were in the range of 125–2500 µg/mL for MICs and 500–5000 µg/mL for MBC/MFCs for all tested strains. Antibacterial activity was shown by the extract against all bacterial strains, with MIC values in the range of 125–250 μg/mL. The highest results were observed against S. aureus, P. aeruginosa and E. faecalis (125 μg/mL). L. sibiricus transgenic root extract was found to have substantial antifungal activity against S. cerevisiae and C. albicans with MIC values of 625 and 2500 μg/mL, according to the microdilution assay, and for MFC values 625 and 5000 μg/mL (Table 2).
Discussion
The genetic basis of the development of Arabidopsis thaliana, particularly regarding the identity of the floral organ, the regulation of the particular genes and the synthesis of the secondary compounds, has been extensively researched over the past 20 years [24, 25]. A number of methods have been used including in situ hybridization, the phenotypic analysis of mutants and transgenic plants, and DNA–protein interactions [26,27,28]. However, a key novel aspect of the present study is that it describes the first example of the successful engineering of the phenylpropanoid pathway by co-expression of the key transcription factor AtPAP1 in the transgenic roots of L. sibiricus.
The PAP1 gene from Arabidopsis encodes an MYB-type transcription factor: a key part of the MYBHLH-WD40 transcription factor complex; the complex is known to regulate the pathway of phenylpropanoid biosynthesis [29, 30]. Both the bHLH and MYB-type factors are widely used to investigate phenylpropanoid metabolism [31]. We suspect that AtPAP1 over-expression may induce the activation of phenylopropanoid pathway genes and was found to strongly influence the phenylpropanoids in tested transgenic root lines, resulting in high levels of phenolic acid production. In both tested lines, one of the major compounds was chlorogenic acid. The content of these compounds was about fivefold higher than that of the control transformed root without the construct. Howles et al. [32] showed that chlorogenic acid levels increased approximately threefold following over-expression of PAL in transgenic tobacco. Furthermore, some transcription factors induce widespread biosynthetic gene expression, and the over-expression of these genes may elevate the chlorogenic acid content in tomatoes by as much as 22 times [33]. Similarly, Anh Tuan et al. [34] found the introduction of AtPAP1 to be associated with elevated levels of mRNA for all the tested biosynthetic genes, together with a 9.89-fold increase in chlorogenic acid accumulation in Platycodon grandiflorum hairy roots.
However, Elomaa et al. [35] report that in Gerbera sp. (Asteraceae species) the over-expression of GMYB10 (a homologue of AtPAP1) results in the up-regulation of all early biosynthetic genes (PAL, C4H, CHI and F3H) and causes increased production of phenolic acids and flavonoids. In turn, Zhang et al. [8] reported that PAP1 expression in S. miltiorrhiza resulted in greater content of anthocyanins, phenolic acids and flavonoids. In contrast, Qiu et al. [36] showed that while PAP1 expression stimulates several pathway genes, this expression alone was not associated with any significant change in phenylpropanoid content in Sauseria involucrate.
Additionally, our study showed increased production of other phenolic acids such as caffeic acid (about threefold), ferulic acid (twofold), p-coumaric acid (2.5-fold) and neochlorogenic acid (about 1.80-fold). We suspect that many of the genes positioned early in the phenylpropanoid pathway are also activated when AtPAP1 is overexpressed in the L. sibiricus root, which can cause increased production of phenolic acids. However, given the current limitations in our knowledge of phenylpropanoid metabolism in L. sibiricus, further experimental work would need to be carried out to confirm this observation.
This is the first study to confirm that over-expression of AtPAP1 in this species induced high levels of phenolic acids accumulation. In addition, the expression profiles of transgenic clone 1 roots will enable a more fuller understanding of the regulation of the biosynthesis of phenylpropanoids at the transcriptional level in L. sibiricus roots.
This study is the first assessment of the antimicrobial potential of after the treatment by transgenic L. sibiricus root extracts against S. aureus, E. faecalis, E. coli, P. aeruginosa and the yeasts C. albicans and S. cerevisiae. MIC and MBC assay showed a broad spectrum of antibacterial activity against all the tested strains. Ahmed et al. [10] showed that different solvent extracts of aerial parts (carbon tetrachloride, chloroform, acetone and methanol) of L. sibiricus possess antibacterial activity. We hypothesized that the phenolic acids identified in tested extract may be responsible for these properties. Cetin-Karaca and Newman [37] note that phenolic acids (chlorogenic acid, coumaric acid, ellagic acid) showed antibacterial potential.
The study then evaluates the effect of transgenic root (AtPAP1 TR clone) activity on grade IV glioma cells by MTT assay. The transgenic roots (AtPAP1 TR clone) were found to have cytotoxic effects of L. sibiricus with IC50 = 0.75 mg/mL; however, normal cell viability remained unaffected. The TR extract of the roots without any construct is known to have a cytotoxic effect on grade IV glioma cells with IC50 = 2.4 mg/mL [14]. We suspect that the stronger cytotoxic effect is associated with the higher accumulation of phenolic compounds in transgenic roots expressing the AtPAP1 transcriptional factor, but more research is needed to confirm this hypothesis.
Conclusion
This study represents the first successful example of the engineering of the phenylpropanoid pathway L. sibiricus roots by modification with selected transcription factors (AtPAP1) from Arabidopsis thaliana. Strong induction of phenolic acids including chlorogenic acid, neochlorogenic acid, p-coumaric acid, caffeic acid and ferulic acid was observed in transgenic L. sibiricus roots. Additionally, this novel cytotoxic and antimicrobial transgenic clone represents a sustainable source of high-quality L. sibiricus transgenic roots for medicinal and economic applications by the alteration of its production of bioactive compounds.
References
Dixon, R. A., & Strack, D. (2003). Phytochemistry meets genome analysis, and beyond. Phytochemistry, 62, 815–816.
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., & Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends in Plant Science, 15, 573.
Stracke, R., Werber, M., & Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology, 4, 447–456.
Du, H., Zhang, L., Liu, L., Tang, X.-F., Yang, W.-J., Wu, Y.-M., et al. (2009). Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry, 74, 1–11.
Qiu, J., Sun, S., Luo, S., et al. (2014). Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum. Plant Cell Reports, 33, 669. https://doi.org/10.1007/s00299-014-1585-8.
Gatica-Arias, A., Farag, M. A., Stanke, M., Matousek, J., Wessjohann, L., & Weber, G. (2012). Flavonoid production in transgenic hop (Humulus lupulus L.) altered by PAP1/MYB75 from Arabidopsis thaliana L. Plant Cell Reports, 31, 111–119.
Zvi, M. M., Shklarman, E., Masci, T., Kalev, H., Debener, T., Shafir, S., et al. (2012). PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytologist, 195, 335–345.
Zhang, Y., Yan, Y. P., & Wang, Z. Z. (2010). The Arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza. Journal of Agriculture and Food Chemistry, 58, 12168–12175.
Tasdemir, D., Wright, A. D., Sticher, O., Çalis, I., & Linden, A. (1995). Detailed 1Hand 13C-NMR investigations of some diterpenes isolated from Leonurus persicus. Journal of Natural Products, 58, 1543–1554.
Ahmed, F., Islam, M. A., & Rahman, M. M. (2006). Antibacterial activity of Leonurus sibiricus aerial parts. Fitoterapia, 77, 316–317.
Sayed, M. A., Haque, M. M., Roy, B., Hossain, S. M. J., & Das, S. R. (2012). Allelopathic effects of different extracts of honeyweed (Leonurus sibiricus) on seeds germination and seedlings growth of some selected vegetables. Journal of Natural Products, 5, 243–250.
Rahmatullah, M., Rahman, M. A., Haque, M. Z., Mollik, M. A. H., Miajee, Z. U. M., Begum, R., et al. (2010). A survey of medicinal plants used by folk medicinal practitioners of Station Purbo Para village of Jamalpur Sadar Upazila in Jamalpur district, Bangladesh. American-Eurasian Journal of Sustainable Agriculture, 4, 122–135.
Narukawa, Y., Niimura, A., Noguchi, H., Tamura, H., & Kiuchi, F. (2014). New diterpenoids with estrogen sulfotransferase inhibitory activity from Leonurus sibiricus L. Journal of Natural Medicines, 68, 125–131.
Sitarek, P., Skała, E., Toma, M., Wielanek, M., Szemraj, J., Nieborowska- Skorska, M., et al. (2016). A preliminary study of apoptosis induction in glioma cells via alteration of the Bax/Bcl-2-p53 axis by transformed and non-transformed root extracts of Leonurus sibiricus L. Tumour Biology, 37, 8753–8764.
Sitarek, P., Skała, E., Toma, M., Wielanek, M., Szemraj, J., Skorski, T., et al. (2016). Transformed root extract of Leonurus sibiricus induces apoptosis through intrinsic and extrinsic pathways in various grades of human glioma cells. Pathology & Oncology Research. https://doi.org/10.1007/s12253-016-0170-6.
Höfgen, R., & Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Research, 16(20), 9877.
Schenk, R. U., & Hildebrandt, A. C. (1972). Medium and techniques for induction of growth of monocotyledonous and dicotyledonous plant cell cultures. Canadian Journal of Botany, 50, 199–204.
Murray, M. G., & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8(19), 4321–4325.
Skała, E., Kicel, A., Olszewska, M. A., Kiss, A. K., & Wysokińska, H. (2015). Establishment of hairy root cultures of Rhaponticum carthamoides (Willd.) Iljin for the production of biomass and caffeic acid derivatives. BioMed Research International, 181098, 1–11.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 3, 1101–1108.
Sitarek, P., Rijo, P., Garcia, C., Skała, E., Kalemba, D., Białas, A. J., et al. (2017). Chemical composition antibacterial, anti-inflammatory, antioxidant and antiproliferative properties of essential oils from hairy and normal roots of Leonurus sibiricus L. Oxidative Medicine and Cellular Longevity, 7384061, 1–12. https://doi.org/10.1155/2017/7384061.
Wayne, P. A. (2015). Performance standards for antimicrobial susceptibility testing: Twenty fifth international supplement M100-S25. Wayne: Clinical and Laboratory Standards Institute.
Mosmann, T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods, 65, 55–63.
Bowman, J. L., Smyth, D. R., & Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell, 1, 37–52.
Coen, E. S., & Meyerowitz, E. M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature, 353, 31–37.
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., & Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology, 139, 5–17.
Huggett, J., Dheda, K., Bustin, S., & Zumla, A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes and Immunity, 6, 279–284.
Chervoneva, I., Li, Y., Schulz, S., Croker, S., Wilson, C., et al. (2010). Selection of optimal reference genes for normalization in quantitative RT-PCR. BMC Bioinformatics, 11, 253.
Kirik, V., Kolle, K., Misera, S., & Baumlein, H. (1998). Two novel MYB homologues with changed expression in late embryogenesis-defective Arabidopsis mutants. The Plant Journal, 13, 729–742.
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60, 107–124.
Matus, J. T., Aquea, F., & Arce-Johnson, P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology, 8, 83.
Howles, P. A., Sewalt, V., Paiva, N. L., Elkind, Y., Bate, N. J., Lamb, C., et al. (1996). Overexpression of l-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiology, 112, 1617–1624.
Luo, J., Butelli, E., Hill, L., Parr, A., Niggeweg, R., Bailey, P., et al. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. The Plant Journal, 56, 316–326.
Anh Tuan, P., Yeon Kwon, D., Lee, S., Arasu, M. V., Al-Dhabi, N. A., Park, N., et al. (2014). Enhancement of chlorogenic acid production in hairy roots of Platycodon grandiflorum by over-expression of an Arabidopsis thaliana transcription factor AtPAP1. International Journal of Molecular Sciences, 15, 14743–14752. https://doi.org/10.3390/ijms150814743.
Elomaa, P., Uimari, A., Mehto, M., Albert, V. A., Laitinen, R. A., & Teeri, T. H. (2003). Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein–protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiology, 133, 1831–1842.
Qiu, J., Gao, F., Shen, G., Li, C., Han, X., Zhao, Q., et al. (2013). Metabolic engineering of the phenylpropanoid pathway enhances the antioxidant capacity of Saussurea involucrata. PLoS ONE, 8(8), e70665. https://doi.org/10.1371/journal.pone.0070665.
Cetin-Karaca, H., & Newman, M. C. (2015). Antimicrobial efficacy of natural phenolic compounds against gram positive foodborne pathogens. Journal of Food Research, 4(6), 14–27.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Rights and permissions
About this article
Cite this article
Sitarek, P., Kowalczyk, T., Rijo, P. et al. Over-Expression of AtPAP1 Transcriptional Factor Enhances Phenolic Acid Production in Transgenic Roots of Leonurus sibiricus L. and Their Biological Activities. Mol Biotechnol 60, 74–82 (2018). https://doi.org/10.1007/s12033-017-0048-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-017-0048-1