Abstract
Dihydrolipohyl dehydrogenase (DLD) is a FAD-dependent enzyme that catalyzes the reversible oxidation of dihydrolipoamide. Herein, we report medium optimization for the production of a recombinant DLD with NADH-dependent diaphorase activity from a strain of Bacillus sphaericus PAD-91. The DLD gene that consisted of 1413 bp was expressed in Escherichia coli BL21 (DE3), and its enzymatic properties were studied. The composition of production medium was optimized using one-variable-at-a-time method followed by response surface methodology (RSM). B. sphaericus DLD catalyzed the reduction of lipoamide by NAD+ and exhibited diaphorase activity. The molecular weight of enzyme was about 50 kDa and determined to be a monomeric protein. Recombinant diaphorase showed its optimal activity at temperature of 30 °C and pH 8.5. K m and V max values with NADH were estimated to be 0.025 mM and 275.8 U/mL, respectively. Recombinant enzyme was optimally produced in fermentation medium containing 10 g/L sucrose, 25 g/L yeast extract, 5 g/L NaCl and 0.25 g/L MgSO4. At these concentrations, the actual diaphorase activity was calculated to be 345.0 ± 4.1 U/mL. By scaling up fermentation from flask to bioreactor, enzyme activity was increased to 486.3 ± 5.5 U/mL. Briefly, a DLD with diaphorase activity from a newly isolated B. sphaericus PAD-91 was characterized and the production of recombinant enzyme was optimized using RSM technique.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dihydrolipohyl dehydrogenase (DLD; EC: 1.8.1.4), also known as lipoamide dehydrogenase, is a FAD-dependent enzyme that catalyzes the reversible oxidation of dihydrolipoamide to lipoamide in the presence of NAD+. DLD is a component of the large multienzyme complexes, pyruvate dehydrogenase and 2-oxo-acid dehydrogenase and plays a role in energy metabolism in eukaryotes [1, 2]. This flavoenzyme contains a reactive disulfide bridge and a FAD cofactor per subunit. It has been extensively isolated from a variety of organisms belonging to prokaryotes, eukaryotes and archaeobacteria [3, 4]. DLD enzyme is also termed diaphorase for its NAD(P)H dehydrogenase activity [5]. Diaphorases are a group of flavin-bound enzymes that catalyze the pyridine nucleotide-dependent reduction of electron-accepting molecules [6, 7]. The first such enzyme has been purified from pig heart muscle [8]. Other enzymes have been identified from various sources such as Clostridium kluyveri, Bacillus stearothermophilus, Thermus aquaticus and Thermus thermophlius [9, 10]. Diaphorases have potential application for the coupled colorimetric measurement of dehydrogenases, nitric oxide and ethanol. They have been used in biotransformation reactions [11], biosensor design [10] and in vitro diagnostic tests such as phenylketonuria (PKU) [12], maple syrup urine diseases (MSUD) and galactosemia as well [13]. Due to industrial applications of diaphorases, these enzymes have attracted researchers’ attention for screening of novel sources and cost-benefit production.
The optimization of medium constituents is the primary step for cost-effective production of enzymes. The conventional optimization technique is one-variable-at-a-time method. Response surface methodology (RSM) is a statistical approach that has been used to optimize production medium of different enzymes [14]. Compared with classical methods, RSM can evaluate interaction from the effects of individual components and their influence on the production. This offers suitable information for executing optimization processes while simultaneously incorporating multiple responses [15]. In this communication, we report on the identification of a strain of B. sphaericus which produces a DLD enzyme with NADH-dependent diaphorase activity. To enhance the production of the recombinant enzyme using RSM, a central composition design method was employed for medium optimization.
Materials and Methods
Chemicals and Media
Reagents for cultivation were purchased from Merck (Darmstadt, Germany). Lipoamide and NAD+ were obtained from Sigma (St. Louis, MO, USA) and NADH was from Roche (Germany). Escherichia coli BL21 (DE3) and pET-28b (+) were purchased from Novagen (Philadelphia, PA). The media, Luria–Bertani (LB), Terrific broth (TB), M9 Minimal salt (M9), SOB and SOC were prepared according to the literature [16].
Screening and Isolation of Enzyme-Producing Strains
Screening of bacteria that produced DLD with diaphorase activity was carried out in medium containing dihydrolipoamide as a selective compound [3]. Twenty-five soil samples were collected from different locations of Tehran during spring and summer seasons of 2015. The samples were placed in sterile polyethylene bags and maintained at 4 °C until processed. One gram of each soil sample was suspended in selective liquid medium that contained (per liter): 0.5% dihydrolipoamide, 1 g NaCl, 2 g K2HPO4, 0.5 g MgSO4·7H2O, 5 g yeast extract, 5 g polypeptone in 1 L of tap water, pH 7.0, then incubated with shaking at 37 °C at 180 rpm for 24 h. Serial dilutions up to 10−4 were prepared. From each dilution, 0.05 mL was taken and placed on agar plates and incubated at 37 °C for 24 h [17]. The strains that grew under such conditions were selected for subsequent screening by enzyme production analysis. Production of DLD enzyme was done in the mentioned medium at 37 °C at 180 rpm for 24 h. After the completion of growth, the fermentation broth was centrifuged at 6500×g for 15 min at 4 °C. The harvested cells were washed twice with 0.9% NaCl solution, suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 7.4) and disrupted by ultrasonic oscillator for 10 min. Cells and insoluble materials were removed by centrifugation at 8000×g for 20 min at 4 °C. The supernatant solution was used as crude extract for DLD activity assay [18]. DLD activity was determined by the oxidation of dihydrolipoamide in the presence of NAD+ [18]. The reaction mixture (1 mL) contained 100 mM potassium phosphate buffer (pH 7.8), 1.0 mM EDTA, 0.4 mM dihydrolipoamide and 0.3 mM NAD+. A solution containing all assay components except dihydrolipoamide was used as the blank. The assay was monitored by the increase in absorbance at 340 nm for 3 min. The isolates that showed DLD activity were selected for diaphorase assay. Diaphorase activity was measured using an assay with thiazolyl blue tetrazolium bromide (MTT) as the terminal electron acceptor [19]. The standard reaction mixture contained 0.1 M potassium phosphate buffer (pH 7.6), 0.3 mM NADH, 0.4 mM MTT and the enzyme in a total volume of 0.7 mL. The increase in absorbance at 560 nm for 1 min was estimated and corrected for blank values lacking enzyme. One unit (U) of diaphorase activity was defined as the quantity of enzyme, which transfers electrons from 1 µmol of NADH to MTT per minute at 25 °C. The protein concentration was determined by Bradford method with bovine serum albumin (BSA) as the standard [20]. All assay experiments were done in triplicate, and the average results were used for data analysis.
Isolate Identification
Identification of the selected bacterium was done by 16S rRNA sequencing. Polymerase chain reaction (PCR) of 16S rRNA gene was performed with universal primer pair 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (AAGGAGGTGATCCAGCCGCA) [21]. The PCR program was as follows: 95 °C for 5 min, 30 cycles of 95 °C for 1 min, 45 °C for 1 min, and 72 °C for 2 min, and final extension at 72 °C for 10 min. The resultant PCR product was visualized by electrophoresis on 1% agarose gel, excised and gel-purified. DNA sequencing was performed by the commercial services of MacroGen Co. ltd. (Seoul, Korea). The 16S rRNA genes of the isolate and Bacillus type strains were aligned using the Molecular Evolution Genetic Analysis (MEGA) program, version 5.0 [22]. Phylogenic tree was constructed using the neighbor-joining algorithm using the MEGA version 5.0. The reliability of the internal branches of tree was statistically evaluated by bootstrap test with 1000 replications.
Expression and Purification of Recombinant B. sphaericus DLD
To amplify the putative DLD gene, forward and reverse primers were designed based on the consensus DLD sequences among Bacillus species using DNASIS MAX software (DNASIS version 3.0, Hitachi Software Engineering Co., Ltd., Tokyo, Japan). The primers used were DLDFw (5′-CCCGGATCCATGGTAGTAGGAGA-3′) and DLDRev (5′-CCCGTCGACTTTTACAATGTG-3′), which contained the restriction sites for BamHI and SalI, respectively (underlined). PCR conditions were as follows: initial denaturation at 95 °C for 5 min, 30 cycles at 95 °C for 1 min, 60 °C for 1 min, 72 °C for 2 min and a final extension step at 72 °C for 7 min. The PCR product was double-digested with BamHI and SalI, gel-purified and then ligated into pET28a (+) expression vector. The construct bearing the desired gene (pET28aDLD) was transformed into E. coli strain DH5α competent cells for screening purposes. The identity of the cloned gene was also confirmed by nucleotide sequencing, and no mutation was revealed. The isolated plasmid was then transformed into E. coli strain BL-21 (DE3) competent cells for expression. E. coli strain BL21(DE3) cells bearing pET28aDLD were cultivated overnight in 100 mL of LB medium containing 40 μg/mL of kanamycin at 37 °C and 180 rpm. One hundred mL of precultured medium was transferred into 1 L of LB medium in culture flasks and incubated at 37 °C and 250 rpm. When cell density reached an OD600 of 0.6–0.8, DLD enzyme was expressed by the addition of 1 mM sterile isopropyl-β-d-thiogalactopyranoside (IPTG). After 5 h induction at 37 °C, cells were harvested, washed twice with 0.9% NaCl solution and stored at −20 °C. Expression of recombinant DLD in different conditions was evaluated by activity assay and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The cell pellet was dissolved in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 7.4) and disrupted by sonication. To purify the recombinant protein, supernatant of cell lysis was applied to a Ni–NTA affinity column (Qiagen, Germany) according to the manufacture’s instruction. The column was washed with 3 column volumes of the 50 mM Tris–HCl buffer (pH 7.0) containing 50 mM imidazole, and then, recombinant enzyme was eluted with an elution buffer (50 mM Tris–HCl, 50 mM NaCl, 10 mM EDTA, 300 mM imidazole, pH 7.0). SDS-PAGE was performed using discontinuous gels (10 cm × 10 cm) with a 12% separating gel and a 6% stacking gel [16]. Protein bands were visualized by staining with 0.25% Coomassie Brilliant Blue R-250 in the mixture of 50% methanol and 10% acetate.
Enzyme Characterization
The total molecular mass was determined by native PAGE. The protocol for native PAGE was a modification of non-denaturing PAGE [16]. The main differences from the Laemmli protocol were the absence of SDS in electrophoresis and sample buffers, samples were not boiled and resolving gel was overlaid with purified water. The effect of temperature was analyzed by performing the diaphorase activity assay at various temperatures (30–60 °C). Reaction mixture was pre-incubated at the desired temperatures for 4 min. The effect of pH on the enzymatic reaction of diaphorase was assessed by measuring the activity in the following buffers: 0.1 M sodium acetate (pH 3.0–5.0), 0.1 M potassium phosphate (pH 6.0–7.5), 0.1 M Tris–HCl (pH 8.0–9.0), 0.1 M glycine-NaOH (pH 9.0–11.0) and 0.1 M sodium carbonate (pH 11.5–12.0). The kinetic constants were determined using GraphPad Prism version 7.00, GraphPad Software, La Jolla California USA.
Medium Optimization
The media, LB, 2× YT, M9, SOB and SOC were used for basic medium optimization. The initial pH of the medium was adjusted to 7.0 and was not controlled during the fermentation. For primary seed culture, 1.0 mL of frozen cells of recombinant E. coli was inoculated in 50 mL medium and grown at 37 °C and 200 rpm for 16 h. The medium (500 mL) was then inoculated with 10% (v/v) of seed culture and incubated at 37 °C and 250 rpm. After 1 h of inoculation, IPTG was added to final concentration of 1 mM and cultures were further grown at 37 °C and 250 rpm for 5 h. Cultures were harvested to activity assay of produced enzyme as above. A central composite face-centered design (CCFD) was used to optimize medium composition of recombinant enzyme using Design-Expert software (version 8.0.4, State-Ease, Inc., USA). All variables were studied at two levels (+1, −1). All experiments were done in triplicate, and average enzyme activity was taken as dependent variable or response. Analysis of variance (ANOVA) was used for the determination of significant variables. The statistical significance of the proposed model was evaluated by Fisher’s statistical test (F-test). The regression equations were also summated to the F-test to determine the coefficient of determination (R 2). The fitted polynomial equation was expressed as three-dimensional surface plots to visualize the relationship between the responses (dependent variables) and the experimental levels of each factor (independent variables). The optimized parameters were tested experimentally to confirm the model validity [22, 23].
Batch Fermentation in 5-L Bioreactor
To further optimize the production of recombinant diaphorase, additional cultural conditions were studied. Working volume was 3.0 L in 5-L stirred tank bioreactor (BioFlo 3000, New Brunswick Scientific, Edison, New JersEy, USA). The pH was controlled at 7.0 ± 0.2 by addition of 25% (v/v) ammonia and 20% (v/v) HCl. Pre-inoculum was prepared by transferring freshly grown cells into 500-mL flask containing 300 mL medium and incubating at 37 °C and 250 rpm for 12 h. The initial batch culture was started by addition of 10% (v/v) of primary seed culture into the bioreactor. After 1 h of cultivation in bioreactor (OD600 = 0.6–0.8), IPTG was added to a final concentration of 1 mM to induce expression and culture was further grown 37 °C and 400 rpm for 4 h. Culture was sampled at regular intervals for enzyme activity assay.
Results and Discussion
Isolation and Phylogenetic Analysis of Strain PAD-91
During the screening on selective medium containing dihydrolipoamide, 94 strains were isolated. In order to identify the enzyme-producing microorganisms, bacterial isolates were analyzed for DLD activity and NADH dehydrogenase ability. From these bacteria, only one strain, named as PAD-91, was found to have diaphorase activity. Strain identification was conducted by comparative sequence analysis of the 16S rDNA of isolate and other bacteria in GenBank database. The 16S rRNA nucleotide was analyzed with the BLAST program and showed 97% homology with B. sphaericus. According to the created phylogenetic tree (Fig. 1), it was inferred that the PAD-91 strain was closely related to B. sphaericus. To the best of our knowledge, there is no report about production of diaphorase by B. sphaericus.
Phylogenetic tree of the 16S rDNA sequences of strain PAD-91 associated with the other members of Bacillus species. Numbers at nodes are levels of bootstrap support (%) based on neighbor-joining (NJ) method of 1000 resampled datasets. The scale bar indicates 0.005 nucleotide substitution per position. Isolate is marked
Construction of Expression Plasmid for Recombinant Diaphorase
The DNA fragment containing putative DLD gene was obtained by PCR with primers designed on the basis of conserved sequences from other Bacillus strains and sequenced. The 1413-bp open reading frame (ORF) had a coding capacity of 470 amino acids (Fig. 2). Expression plasmid of DLD, pET28aDLD, was constructed and transformed in E. coli strain BL21 (DE3).
Recombinant Enzyme Properties
The molecular mass of the recombinant diaphorase enzyme was determined to be about 50 kDa by native PAGE (Fig. 3a). The subunit structure was examined by SDS-PAGE. SDS-PAGE gel analysis showed a single band indicating that the enzyme consists of one subunit (Fig. 3b). This finding was in agreement with the previous reports in which many diaphorases have been shown to be monomers with MW values ranging from 50 to 55 kDa [9]. The diaphorase reaction exhibited its maximal activity at 30 °C (Fig. 4a). As seen, a sharp decrease in enzyme activity was observed above 30 °C and was completely inactivated at 60 °C. The effect of various pH values on the enzymatic reaction of diaphorase was evaluated in pH range from 3.0 to 12.0 at 30 °C. The optimum pH of the diaphorase reaction was approximately 8.5, and it showed high activity in a pH range of 7.0–9.0 (Fig. 4b). The K m and V max values for the recombinant enzyme were determined to be 0.025 mM and 275.8 U/mL, respectively. The K m value for NADH (0.025 mM) was lower than other diaphorases such as B. stearothermophilus (0.07 mM) and C. kluyveri (0.038 mM) [9]. The calculated K m indicated high affinity for the NADH substrate. Moreover, the turnover number of the enzyme (108.9 s−1) was also higher than other previously purified diaphorases. Thus, it can be inferred that the important advantage of B. sphaericus diaphorase over previously studied enzymes was its kinetic features. Suitable kinetics parameters offer promise for our identified diaphorase to be a good candidate for biotechnological applications.
Native PAGE (a) and SDS-PAGE (b) analysis of recombinant enzyme. a Lane M standard protein molecular weight marker; lane 1 purified enzyme. b Lane M standard protein molecular weight marker; lane 1 whole cell lysate from a sample after medium optimization; lane 2 whole cell lysate from a sample before medium optimization (basal medium). The quantity of whole cell lysates and purified enzyme applied to SDS-PAGE and native PAGE gels were 40 and 10 μg, respectively
Effect of Basal Medium on Enzyme Production
In order to determine the optimal medium for recombinant strain E. coli BL21 (DE3) to grow and produce enzyme, the influence of liquid media of LB, 2× YT, SOB was investigated. The results indicated that 2× YT medium was the most favorable for cell growth. Under this condition, enzyme activity of 180 U/mL was achieved.
Effect of Carbon Source on Enzyme Production
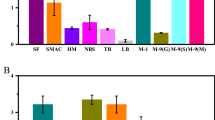
The influence of carbon source in 2× YT medium was studied by adding of maltose, glucose, lactose, sucrose, mannose and fructose at concentration of 10 g/L. As shown in Fig. 5, enzyme activity was the highest when sucrose was employed as carbon source. Low enzyme activity was seen in culture media containing glucose and maltose. The negative effect of glucose has been reported for medium optimization of other enzymes such as lipase [24, 25]. This phenomenon is in accordance with the suppressive role of this parameter, which is attributed to its catabolic repression influence [26]. In contrast, there are reports that glucose improves enzyme production [27].
Effect of Nitrogen Source on Enzyme Production
The nitrogen sources, yeast extract and tryptone in 2× TY medium were replaced by tryptone, yeast extract, beef extract, ammonium sulfate, ammonium nitrate and urea at concentration of 20 g/L. When yeast extract was used (Fig. 6), enzyme activity was the highest. So the yeast extract was found as the best nitrogen source. Yeast extract has been used in most fermentation studies as a supplier of vitamins and growth factor [28]. Similar observations have been reported in production of other enzymes such as validoxylamin glycosyltransferase [29], inulinase and invertase [30].
Effect of Inorganic Salts on Enzyme Production
The influence of inorganic salts in 2× YT medium was studied by adding of ZnSO4, FeSO4, MgSO4, MnSO4 and CaCl2 at concentration of 0.2 g/L. The results showed that MgSO4 was an important additive (Fig. 7). In contrast, other inorganic salts had negative effects on enzyme production. The reason of promoting function of Mg2+ may be that Mg2+ is highly necessary to maintain genomic stability and is an essential cofactor in almost enzymatic systems involved in DNA processing [15, 31]. However, in some cases Mg2+ has exhibited negative effects on the production of enzymes [32, 33].
Optimization of Enzyme Production
The most effective factors including sucrose, yeast extract, NaCl and MgSO4 were selected based on the results of single-factor research. A four-factors and three-level experiment was designed. The coding levels of factors and experimental results are shown in Table 1. The property of polynomial model equation was expressed by R 2, and its statistical significance was determined by F test. The analysis of variance results are shown in Table 2. The ANOVA for response surface quadratic regression model showed that the model was highly significant (p value of 0.0005) with an F value of 6.02. In addition, the model R 2 was 0.97, indicating that the fitting degree of equation was good, and there was a high degree of correlation between the predicted and measured values. Thus, it could be applied to predict theoretical enzyme activity. An adequate precision measures the signal-to-noise ratio and a ratio greater than 4 is desirable. The adequate precision value of 9.865 indicated an adequate signal and suggested that the model could be used to navigate the design space. A low value of coefficient of variance (CV %) (6.77) indicated a high degree of precision and reliability of the experimental values. As found from Table 2, sucrose and yeast extract had significant effect on enzyme activity with p value <0.05, while NaCl and MgSO4 had insignificant effect. The relation between factors and enzyme activity was indicated by the surface plots (Fig. 8a–f). The response surface curve in Fig. 8a represents the interaction between yeast extract and sucrose, which showed that the maximum diaphorase activity was achieved with high levels of these variables. Figure 8b depicts the interaction between MgSO4 and sucrose. The results showed that enzyme activity was mostly affected by sucrose than MgSO4. The response surface curve for the interaction of yeast extract and MgSO4 is represented in Fig. 8c. The shape of response showed that diaphorase activity was mostly affected by yeast extract levels than MgSO4. The same trend was observed for the interaction of sucrose and NaCl (Fig. 8d) as well as yeast extract and NaCl (Fig. 8e) where the maximum response was mostly influenced with sucrose and yeast extract. Figure 8f shows the interaction of NaCl and MgSO4. As found, there was a less significant interaction between NaCl and MgSO4 and the optimum response was obtained with the intermediate levels of these two factors. There was no regular trend between concentration of these factors and enzyme activity. By analyzing the response surface plots, the optimum levels for sucrose, yeast extract, NaCl and MgSO4 were determined to be 10, 25, 5 and 0.25 g/L, respectively.
Experimental Validation of Optimized Medium
After repeated experiments employing the optimized medium, the average actual enzyme activity was 345.0 ± 4.1 U/mL, indicating a good agreement between the observed and predicted value. As shown in Fig. 3, the production level of target enzyme was higher compared with basal medium. Overall, these results confirmed the validity and precision of the model. The obtained activity for recombinant B. sphaericus diaphorase was significantly higher than reported ones in other literatures [9]. Therefore, produced enzyme has the potential to be used as a component of diagnostic tests such as phenylketonuria, maple syrup urine diseases and galactosemia [12].
Scale-up Production of Recombinant Diaphorase
Based on the shake flask results, target enzyme production in a batch fermenter was studied to obtain higher enzyme activity. The experiments with the initial pH of the production medium showed that amylase production was optimized at pH 7.0 (Fig. 9a). The effects of inoculum size (v/v) and agitation on diaphorase production were investigated. Maximum production was obtained with 10% inoculum. Further increase in inoculum size causes the reduction in diaphorase production (Fig. 9b). The decrease in enzyme production with the increase in the percentage of inoculum might be due to the initial rapid growth of bacterium and lack of sufficient nutrient available in the medium for higher volume of bacteria. Optimum diaphorase production was obtained at 90 rpm (Fig. 9c). Further increase in agitation that decreased diaphorase production might be due to cell lysis at higher agitation. The activity of recombinant enzyme under optimized conditions in bioreactor was higher (486.3 ± 5.5 U/mL) than that obtained in parallel sake flask culture (345.0 ± 4.1 U/mL) (Fig. 10).
Conclusion
In this article, we presented recombinant expression, characterization and medium optimization of a new DLD enzyme with diaphorase activity from a newly identified strain of B. sphaericus, named PAD-91. The DLD gene consisted of 1413 bp encoding a protein with MW of 50 kDa. Recombinant diaphorase produced in E. coli BL21(DE3) exhibited its optimal activity at temperature of 30 °C and pH 8.5. K m and V max values with NADH were determined to be 0.025 mM and 275.8 U/mL, respectively. The most appropriate medium for the production of recombinant enzyme was composed of sucrose 10 g/L, yeast extract 25 g/L, NaCl 5 g/L and MgSO4 0.25 g/L. At optimized condition, the actual recombinant diaphorase activity was calculated to be 345.0 ± 4.1 U/mL. Fermentation in 5-L bioreactor resulted in a further enhancement of the enzyme activity to 486.3 ± 5.5 U/mL. Collectively, optimization of culture medium and growth conditions reduced the cost of production and improved the feasibility of commercial production of the target enzyme. Our results make it valuable to do further investigations on diaphorase from B. sphaericus PAD-91.
References
Liang-Jun, Y., Napporn, T., Nathalie, S., & Michael, J. F. (2013). Serum dihydrolipoamide dehydrogenase is a labile enzyme. Journal of Biochemical and Pharmacological Research, 1, 30–42.
Rachael, A. V., Pierre, R., & Grazia, I. (2011). Mutation in the dimer interface of dihydrolipoamide dehydrogenase promote site-specific oxidative damages in yeast and human cells. The Journal of Biological Chemistry, 286, 40232–40245.
Serrano, A. (1992). Purification, characterization and function of dihydrolipoamide dehydrogenase from cyanobacterium Anabaena sp. strain P.C.C.7119. Biochemistry, 288, 823–830.
Jacques, A. E. B., Willem, J. H. V. B., Walter, M. A. M. V. D., Franz, M., & Arie, D. K. (1989). Molecular cloning and sequence determination of the Ipd gene encoding lipoamide dehydrogenase from Pseudomonas flurescens. Journal of Generral Microbiology, 135, 1787–1797.
Madiagan, R. A., & Mayhew, S. G. (1993). Preparation of the apoenzyme of the FMN-dependent Clostridium kluyveri diaphorase by extraction with apofalvodoxin. Biochemical Society Transactions, 22, 578–585.
Tedeschi, G., Chen, S., & Massey, V. (1995). Active site studies of DT-diaphorase employing artificial flavins. The Journal of Biological Chemistry, 270, 2512–2516.
Melanie, A. A., & Zongchao, J. (2006). Modulator of drug activity B from Escherichia coli: Crystal structure of a prokaryotic homologue of DT-diaphorase. Journal of Molecular Biology, 359, 455–465.
Natalia, L. R., Valentina, A. S., Alexander, V. E., Seergey, V. K., Irina, G. G., Bruce, S. K., et al. (2006). pH-dependent substrate preference of pig heart lipoamide dehydrogenase varies with oligomeric state. The Journal of Biological Chemistry, 280, 16106–16114.
Cakraborty, S., Sakka, M., Kimura, T., & Sakka, K. (2008). Characterization of a dihydrolipoyl dehydrogenase having diaphorase activity of Clostridium kluyveri. Bioscience, Biotechnology, and Biochemistry, 72, 982–988.
Antiochia, R., Cass, A. E. G., & Palleschi, G. (1997). Purification and sensor applications of an oxygen insensitive thermophilic diaphorase. Analytica Chimica Acta, 345, 17–28.
Bhushan, H., & Spain, H. (2002). Diaphorase catalyzed biotransformation of RDX via N-denitration mechanism. Biochemical and Biophysical Research Communications, 296, 779–784.
Shahbazmohammadi, H., & Omidinia, E. (2011). New enzymatic colorimetric method for the quantitative determination of phenylalanine in dry-blood spots. Journal of Sciences, 22, 15–20.
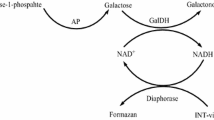
Kianmehr, A., Mahrooz, A., Ansari, J., Oladnabi, M., & Shahbazmohammadi, H. (2016). The rapid and sensitive quantitative determination of galactose by combined enzymatic and colorimetric method: Application in neonatal screening. Applied Biochemistry and Biotechnology, 179, 283–293.
Zhen, H., Liu, Q., & Shao, Y. W. (2009). Medium optimization for the production of a novel bioflocculant from Halomonas sp. V3a using response surface methodology. Bioresource Technology, 100, 5922–5927.
Zhang, H.-B., Mao, X.-Q., Wang, Y.-J., & Hu, X.-Q. (2009). Optimization of culture conditions for high-level expression of dextransucrase in Escherichia coli. The Journal of Food, Agriculture & Environment, 7, 75–78.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1994). Molecular cloning: A laboratory manual (2nd ed., pp. 1847–1857). Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
Shahbazmohammadi, H., & Omidinia, E. (2012). Isolation, purification and characterization of proline dehydrogenase from a newly isolated Pseudomonas putida POS-F84. Iranian Journal of Biotechnology, 10, 111–119.
Argyrou, A., Sun, G., Palfey, B. A., & Blanchard, J. S. (2003). Catalysis of diaphorase reactions by Mycobacterium tuberculosis lipoamide dehydrogenase occurs at the EH4 level. Biochemistry, 42, 2218–2228.
Boething, R. S., & Weaver, T. (1979). A new assay for diaphorase activity in reagent formulations, based on the reduction of thiazolyl blue. Clinical Chemistry, 25, 2040–2042.
Bradford, M. M. (1976). Rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principles of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Ki, J.-S., Zhang, W., & Qian, P.-Y. (2009). Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. Journal of Microbiological Methods, 77, 48–57.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
Shahbazmohammadi, H., Mostafavi, S. S., Soleimani, S., Bozorgian, S., Pooraskari, M., & Kianmehr, A. (2015). Response surface methodology to optimize partition and purification of two recombinant oxidoreductase enzymes, glucose dehydrogenase and D-galactose dehydrogenase in aqueous two-phase systems. Protein Expression and Purification, 108, 41–47.
Hasan-Beikdashti, M., Forootanfar, H., Safiarian, M. S., Ameri, A., Ghahremani, M. H., Khoshayand, M. R., et al. (2012). Optimization of culture conditions for production of lipase by a newly isolated bacterium Stenotrophomonas maltophilia. Journal of the Taiwan Institute of Chemical Engineers, 43, 670–677.
Rajendran, A., & Thangavelu, V. (2009). Statistical experimental design for evaluation of medium components for lipase production by Rhizopus arrhizus MTCC 2233 LWT-Food. Science Technology, 42, 985–992.
Gupta, N., Mehra, G., & Gupta, R. (2004). A glycerol-inducible lipase from Bacillus sp.: Medium optimization by a Plackett-Burman design and by response surface methodology. Canadian Journal of Microbiology, 50, 361–368.
Singh, J., Vohra, R. M., & Sahoo, D. K. (2004). Enhanced production of alkaline proteases by Bacillus sphaericus using fed-batch culture. Process Biochemistry, 39, 1093–1101.
Gupta, N., Sahai, V., & Gupta, R. (2007). Alkaline lipase from a novel strain Burkholderia multivorans: Statistical medium optimization and production in a bioreactor. Process Biochemistry, 42, 518–526.
Yongxian, F., Yang, Y., Xiaoqin, J., Xiaolong, C., & Yinchu, S. (2013). Cloning, expression and medium optimization of validamycin glycosyltransferase from Streptomyces hygroscopicus var. jinggangensis for the biotransformation of validoxylamine A to produce validamycin A using free resting cells. Bioresource Technology, 131, 13–20.
Dinarvand, M., Rezaee, M., Masomian, M., Jazayeri, S. D., Zareian, M., Abbasi, S., et al. (2013). Effect of C/N Ratio and media optimization through response surface methodology on simultaneous productions of intra and extracellular inulinase and invertase from Aspergillus niger ATCC 20611. BioMed Research International, 2013, 1–13.
Ruchi, G., Anshu, G., & Khare, S. K. (2008). Lipase from solvent tolerant Pseudomonas aeruginosa strain: Production optimization by response surface methodology and application. Bioresource Technology, 99, 4796–4802.
Singh, S., Moholkar, V. S., & Goyal, A. (2014). Optimization of carboxymethylcellulase production from Bacillus amyloliquefaciens SS35. 3 Biotech, 4, 411–424.
Sifour, M., Zaghloul, T. I., Saeed, H. M., Berekaa, M. M., & Abdel-Fattah, Y. R. (2010). Enhanced production of lipase by the thermophilic Geobacillus stearothermophilus strain-5 using statistical experimental designs. New Biotechnology, 27, 330–336.
Acknowledgments
Research finding was from Department of Biochemistry.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Statement
This article does not contain any studies with human participants performed by any of the authors.
Human and Animal Rights Statement
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Shahbazmohammadi, H., Omidinia, E. Medium Optimization for Improved Production of Dihydrolipohyl Dehydrogenase from Bacillus sphaericus PAD-91 in Escherichia coli . Mol Biotechnol 59, 260–270 (2017). https://doi.org/10.1007/s12033-017-0013-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-017-0013-z