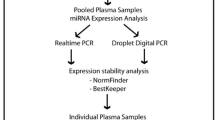
Abstract
Circulating microRNAs (miRNAs) are promising biomarkers for many diseases. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is a gold standard for miRNA expression profiling that requires proper data normalization. Since there is no universal normalizer, it is recommended to evaluate normalizers under every experimental condition. This study describes the identification of suitable endogenous normalizer(s) (ENs) for plasma miRNA expression in essential hypertension. Expression levels of 5 candidate ENs and 2 plasma quality markers were determined by qRT-PCR in plasma samples from 18 hypertensive patients and 10 healthy controls. NormFinder, GeNorm, and DataAssist software programs were used to select the best EN(s). Expression levels of the 5 candidate ENs were also analyzed in urine samples from hypertensive patients and compared to the plasma samples of the hypertensive patients. Among the analyzed candidates, hsa-miR-92a-3p was identified as the best EN, and hsa-miR-21-5p and hsa-miR-16-5p as the next best. Moreover, hsa-miR-92a-3p showed the most consistent expression between plasma and urine. In conclusion, this study showed that hsa-miR-92a-3p, hsa-miR-21-5p, and hsa-miR-16-5p may be used as normalizers for plasma miRNA expression data in essential hypertension studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypertension is among the most common chronic diseases, affecting more than one billion persons worldwide. The chronically elevated blood pressure is a common cause of chronic renal failure, myocardial infarction, stroke, heart failure, and even death [1]. Approximately 90–95 % of hypertension is the essential hypertension type, whose exact cause is unknown but has been recognized to result from a combination of genetic and environmental factors [2, 3]. Understanding causes of hypertension and identifying approaches to ensure appropriate management are top research priorities [4].
MicroRNAs (miRNAs) are small (22–26 nucleotides) non-coding RNA molecules that provide post-transcriptional regulation for many human genes [5, 6]. MiRNAs play important roles in many processes such as early development, cellular differentiation, and apoptosis [7]. Dysregulation of miRNAs has been observed in cancer, cardiovascular diseases, and other diseases [8, 9]. The discovery of miRNAs circulating in body fluids, such as serum and plasma, with high stability, has generated much interest in the assessment of such extracellular miRNAs as minimally invasive biomarkers for a variety of diseases, such as hypertension [10, 11].
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is a widely used method for profiling miRNA expression patterns with high sensitivity, specificity, speed, and reproducibility [12–14]. To obtain valid qRT-PCR data, accurate normalization of miRNA expression is essential [15]. One of the most common strategies to normalize miRNA qRT-PCR data is the “endogenous control method”, which involves standardization to endogenously expressed normalizer(s) using relative quantification [16]. The validity of this method depends on the endogenous normalizer used; the use of improper normalizer can lead to an incorrect conclusion [17]. Stable expression in different disease conditions, along with experimental variables, is one of the major characteristics of an ideal normalizer. However, a universal endogenous normalizer is unlikely to exist, so the stability of each candidate normalizer(s) has to be validated before conducting the experiment [15, 18]. This introduces a challenge, as the expression data of the tested candidate normalizer(s) need to be standardized. A number of software algorithms can be used to address this issue, including NormFinder [19], GeNorm [15], and DataAssist [20].
To the best of our knowledge, there are no published reports on the systematic evaluation of suitable endogenous normalizers for qRT-PCR data from circulating miRNA expression in hypertension. Therefore, the aim of this study was to select, from pre-selected candidates, the best endogenous normalizers for plasma miRNA expression qRT-PCR data in essential hypertension based on empirical evidence.
Materials and Methods
Study Samples
Sample sets included (1) 18 plasma samples from hypertensive patients (from the multicenter randomized clinical trial: Pharmacogenomic Evaluation of Antihypertensive Responses, PEAR [21]; (2) 9 urine samples from hypertensive patients (from the PEAR clinical trial); and (3) 10 plasma samples from healthy controls [22, 23]. The baseline demographics of study participants are presented in Table 1. All samples used in this study were from subjects who provided informed written consent and agreed to participate in the study and to the use of their samples for future research. The studies have been approved by the corresponding institutional review boards and have been performed in accordance with the ethical standards in the Declaration of Helsinki and its later amendments [21–23].
miRNA Isolation and cDNA Synthesis
Total RNA, including miRNA from both plasma and urine samples, was isolated using miRNeasy Serum/Plasma Kit (Qiagen, Valencia, CA, USA). The manufacturer’s instructions were followed except in the final step; the miRNA was eluted in 20 μL RNase-free water instead of 14 uL. Spike-In control and corresponding Ce_miR-39_1 primer assay (Qiagen, Valencia, CA, USA) were used, according to manufacturer’s instructions, to monitor miRNA purification and amplification. The cDNA was synthesized using miScript II RT Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The 20 μL reverse transcription reaction consisted of 10 μL RNA, 4 μL miScript HiSpec buffer, 2 μL 10× miScript Nucleics mix, 2 μL RNase-free water, and 2 μL miScript Reverse Transcriptase mix. Reactions were incubated for 60 min at 37 °C and then 5 min at 95 °C, using thermal cycler (Veriti® 96-Well Thermal Cycler, Applied Biosystems, CA, USA). The cDNA product was diluted by adding 200 μL RNase-free water to the 20 μL reverse transcription reaction. The synthesized cDNA was stored at −20 °C.
Selection of Candidate Endogenous Normalizers and Plasma Quality Indicators
As shown in Table 2, five candidate endogenous normalizers were included in the study. The candidates were selected based on the following criteria: (1) evidence of high expression in plasma, (2) evidence of being a normalizer or a candidate normalizer in at least one previous study, and (3) availability of a reliable quantification assay at the time of the study. Additionally, RNU6-2 and SNORD72 were selected as plasma quality indicators. These two targets were selected as they are highly expressed in different cell types, but not well expressed in serum/plasma. If the average Cycle Threshold (C t) value of these targets is below 32, this indicates that cellular contamination in serum/plasma is more than 0.1 % [24].
qRT-PCR for Profiling Mature miRNA Expression
The qRT-PCR was carried out using the miRNA-specific miScript miRNA PCR primer assays and the miScript SYBR Green Kit (Qiagen, Valencia, CA, USA). The qRT-PCR was performed in triplicate. Each 10 μL per well qRT-PCR reaction consisted of 1 μL cDNA, 2 μL RNase-free water, 1 μL 10× miScript primer assay, 1 μL 10× miScript universal primer, and 5 μL 2× QuantiTect SYBR Green PCR master mix. The cycling conditions for qRT-PCR started with initial HotStar Taq DNA Polymerase activation step at 95 °C for 15 min, then 40 cycles each of three steps (94 °C for 15 s, 55 °C for 30 s, and 70 °C for 30 s), and then the dissociation curve stage was added to verify specificity of the PCR products. The qRT-PCR was performed using QuantStudio™ 12 K Flex Real-Time PCR System (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA). The C t data were calculated using a manually set threshold for each target and an automatically set baseline, by QuantStudio™ 12 K Flex Software v1.2.2 (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA). Any C t > 37 was excluded from the analysis.
Stability Analysis of qRT-PCR Data
Three software programs were used, including GenEx version 6 (http://www.biomcc.com/genex-software.html), NormFinder version 0.953 (http://moma.dk/normfinder-software), and DataAssist™ version 3.01 (https://www.lifetechnologies.com/us/en/home/technical-resources/software-downloads/dataassist-software.html). These software programs are briefly described below.
NormFinder
NormFinder is a Microsoft Excel-based application that uses a model-based approach to assign a stability value to each candidate normalizer. It accounts for intra- and inter-group variation. The lower the stability value, the higher the stability of the candidate normalizer. The input data are supposed to be on a linear scale. When specifying the study groups, the output includes not only the best endogenous normalizers, but also the best combination of two endogenous normalizers [19]. Moreover, NormFinder implemented in GenEx software version 6 (will be called GenEx NormFinder) was used to assess the optimum number of endogenous normalizers required, through calculating the accumulated standard deviation (Acc.SD). The optimum number of endogenous normalizers is the one that achieves the lowest accumulated SD value [31].
GeNorm
This algorithm calculates a stability value (M) for each candidate endogenous normalizer, which is the mean pairwise variation of that candidate with all other chosen candidate(s). Using a step-wise pairwise variation, it excludes candidates with the lowest stability. In this study, GeNorm implemented in GenEx software version 6 (will be called GenEx GeNorm) was used. It repeats the procedure until the two most stable endogenous normalizers are left. Similar to NormFinder, the lower score (M value) signifies higher expression stability of the candidate endogenous normalizer [15].
DataAssist
Like GeNorm, it uses a pairwise variation approach to assign a score to each candidate endogenous normalizer. The lower the score, the more stable the endogenous normalizer. The score is calculated once, with no repeated exclusion of the candidates with the highest score. To perform step-wise exclusion of the least stable candidates, the assay type can be changed from candidate control to target and the software can then calculate the new scores for the remaining candidate controls [20].
Statistical Analysis
The Statistical Package for the Social Sciences (SPSS) software version 17.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Normality was checked by Kolmogorov–Smirnov and Shapiro–Wilk tests, as well as Q–Q plots. Significance of circulating miRNA differential expression was checked by Student t test. Tests with two-sided p values <0.05 were identified as statistically significant.
Results
Choosing a Suitable Endogenous Normalizer(s) for Relative Quantification of Plasma miRNAs in Essential Hypertension
The expression levels of 5 candidate endogenous normalizers in 18 hypertensive and 10 healthy control plasma samples were compared. As shown in Fig. 1, there was no statistically significant difference between hypertensive and healthy control samples for any of the tested candidate normalizers (all p values >0.05). Moreover, all 5 candidate normalizers (hsa-miR-92a-3p, hsa-miR-21-5p, hsa-miR-25-3p, hsa-miR-16-5p, and hsa-miR-223-3p) were relatively abundantly expressed in plasma of hypertensive patients and healthy controls (average C t value range 22.3–24.7).
Differential expression of the candidate endogenous normalizers between hypertensive patients and healthy controls. The expression levels of five candidate endogenous normalizers were compared between plasma of hypertensive patients and plasma of healthy controls, using “Quantitative Real-Time Polymerase Chain Reaction, qRT-PCR”. The middle line in each box represents the median and the box borders represent the inter-quartile range. The lower and upper whiskers represent the 10th and 90th percentiles, respectively. “HTN” represents the hypertension group and “HC” represents the healthy control group
NormFinder Output
First, the data were analyzed by considering the study groups: hypertensive group and healthy control group. Among the 5 candidate endogenous normalizers, the software determined hsa-miR-92a-3p as the best endogenous normalizer (Fig. 2), and hsa-miR-92a-3p and hsa-miR-21-5p as the best combination of two candidates. Then the groups were ignored and the data were reanalyzed, using GenEx NormFinder. As shown in Fig. 3a, hsa-miR-92a-3p was the best endogenous normalizer. Moreover, as shown in Fig. 3b, the lowest accumulated SD was achieved with 4 endogenous normalizers. However, the accumulated SD did not change greatly from using 3 endogenous normalizers to using 4 endogenous normalizers.
Ranking of the candidate endogenous normalizers using NormFinder. The expression stability of five candidate endogenous normalizers was evaluated by the stability values that NormFinder generated by comparing plasma samples from hypertensive patient group and healthy control group; the lower the stability value, the more stable the candidate endogenous normalizer
a Ranking of the candidate endogenous normalizers using GenEx NormFinder. The expression stability of five candidate endogenous normalizers was evaluated by the standard deviation (SD) that was generated by GenEx NormFinder by ignoring the grouping into hypertensive and healthy control groups. The lower the SD, the more stable the candidate endogenous normalizer. b Optimum number of endogenous normalizers required. The number of candidate endogenous normalizers that achieves the lowest accumulated standard deviation (Acc.SD) is recommended by GenEx NormFinder as the optimum number of normalizers required
GeNorm Output
Using GenEx GeNorm, it automatically performed the pairwise comparison of the 5 candidate endogenous normalizers and kept removing the candidate with the highest M value until the best pair was defined (hsa-miR-92a-3p and hsa-miR-16-5p) and then output the ranking of the 5 candidates as shown in Fig. 4. The 5 candidate endogenous normalizers showed M values <1.5, which indicates that all of them had good stability [15].
Ranking of the candidate endogenous normalizers using GenEx GeNorm. The expression stability of five candidate endogenous normalizers was evaluated by calculating the mean pairwise variation (M value) between each candidate and all the other candidates. Then, using step-wise approach, the software excludes the least stable candidate (with the highest M value) and M values are recalculated for the remaining candidates and so on till the most stable pair of candidates is reached
DataAssist Output
Using DataAssist, the 5 candidate endogenous normalizers were ranked according to pairwise comparison score. Then, the candidate with the highest score (lowest stability) was removed and the remaining 4 candidates were re-ranked, and so on till ending up with the pair with the lowest M value (highest stability): hsa-miR-92a-3p and hsa-miR-16-5p. The ranking of the 5 candidate endogenous normalizers from the least stable to the most stable was in the order of hsa-miR-223-3p, hsa-miR-21-5p, hsa-miR-25-3p, and hsa-miR-92a-3p/hsa-miR-16-5p.
Comprehensive Ranking
Putting the output of all the algorithms together by calculating the mean rank for each of the 5 candidate endogenous normalizers, hsa-miR-92a-3p followed by hsa-miR-16-5p and hsa-miR-21-5p was the best (Table 3).
Comparing the Expression of Different Candidate Normalizers in Plasma and Urine
The expression levels of the 5 candidate endogenous normalizers in 9 hypertensive urine samples were quantified using qRT-PCR and compared to the expression data of the 5 candidate endogenous normalizers in the 18 hypertensive plasma samples mentioned above. As shown in Fig. 5, all candidate normalizers were more abundant in plasma than urine, as indicated by the lower average C t value in plasma than urine in each respective candidate normalizer. Also, hsa-miR-92a-3p has the most consistent expression between plasma and urine, as indicated by the smallest difference in the median C t values between plasma and urine, when comparing to the other tested candidate normalizers.
Differential expression of the candidate endogenous normalizers between plasma and urine. The expression levels of five candidate endogenous normalizers were compared between plasma of hypertensive patients and urine of hypertensive patients using “Quantitative Real-Time Polymerase Chain Reaction, qRT-PCR”
Quality Measure of Plasma Using RNU6-2 and SNORD7
The expression levels for both RNU6-2 and SNORD72 were relatively low (average C t value >32), which indicates that the plasma samples were essentially free from cellular contamination [24].
Discussion
Circulating miRNAs are promising and minimally invasive biomarkers. The qRT-PCR is the gold standard for miRNA profiling. However, the validity of qRT-PCR analysis depends on proper normalization for the individual experimental conditions. In this study, we investigated 5 candidate endogenous normalizers to identify the best ones for normalizing the qRT-PCR data of essential hypertension studies in plasma.
The stability of the candidate endogenous normalizers was analyzed using NormFinder, GeNorm, and DataAssist. We applied NormFinder “requirements for validity of the results” while determining the number of candidate endogenous normalizers to be tested and the sample size required. It required having a minimum of 3 candidate normalizers and 2 samples per group and recommended using 5–10 candidates and 8 samples or more per group.
In this study, the ranking of the different candidate normalizers using NormFinder showed some differences as compared to that by DataAssist and GeNorm (Table 3). This was anticipated, due to the use of different approaches. However, GeNorm and DataAssist showed the same results, which was also anticipated because they use the same approach (see the “Stability Analysis of qRT-PCR Data” section of the “Materials and Methods” above). This highlights the importance of using software programs that are based on different approaches for better assessment of candidate endogenous normalizers.
This study also showed that hsa-miR-92a-3p was ranked the first with each of the three software programs used, which qualifies it to be the best normalizer among the tested candidates. On the other hand, hsa-miR-223-3p was ranked the last with each of the three software programs, which makes it the least stable normalizer, among the tested candidates, under these experimental conditions (essential hypertension studies using plasma for miRNA profiling). For each candidate normalizer, the mean of the rank reported by each of the 3 algorithms was calculated, which revealed that both hsa-miR-21-5p and hsa-miR-16-5p have the same comprehensive rank that is right after hsa-miR-92a-3p. Thus, hsa-miR-92a-3p is considered the single best EN and this as well as hsa-miR-21-5p and hsa-miR-16-5p are considered the top three.
Many studies have investigated pre-selected candidates to find suitable endogenous normalizers for the miRNA qRT-PCR expression data for specific tissue/body fluid and disease of interest. Davoren et al. examined the expression of 5 candidate miRNAs (let-7a, miR10b, miR-16, miR-21, and miR-26b) and 3 candidate small nucleolar RNAs (RNU19, RNU48, and Z30) to determine the most suitable endogenous normalizer(s) for miRNA qRT-PCR expression data in human breast cancer using breast cancer tissue. The study showed that the combination of miR-16 and let-7a was the best [32]. Song et al. examined 6 miRNA candidates (let-7a, miR-16, miR-93, miR-103, miR-192, and miR-451) and one small nucleolar RNA candidate, RNU6B, for suitability as endogenous normalizers for miRNA qRT-PCR data in gastric cancer studies using serum. miR-16 and miR-93 were shown to be the best [33].
Besides the endogenous normalizer method, the “global mean expression” is another commonly used method of qRT-PCR data normalization. As the name indicates, this method is based on using the mean expression level of all the miRNAs that were detected in the sample for normalizing the data. It is suitable for studies in which a large number of miRNAs are being analyzed, not those with a limited number of miRNAs [34].
It is worth mentioning that there was variation in the expression levels of the different candidate normalizers between subjects within the hypertensive group as well as the healthy control group. However, this is consistent with the literature where others have shown similar, or even larger, variation. For example, in the McDermott et al.’s study, the best 2 endogenous normalizers (miR-16 and miR-425) showed variation in the qRT-PCR C t values among the study groups. For miR-16, C t range is 13.565–18.765 in the cancer group and 13.585–17.812 in the control group. For miR-425, C t range is 18.100–24.206 in the cancer group and 17.459–23.395 in the control group [14]. Also, in Zhu et al.’s study, the most stable set of normalizers (miR-26a, miR-221, and miR-22*) showed variation in the qRT-PCR C t values among the study groups. For miR-26a, C t range is approximately 24–33 in the patient group and 24–31 in the control group. For miR-221, C t range is approximately 25–34 in the patient group and 26–33 in the control group. For miR-22*, C t range is approximately 29–37 in the patient group and 29–36 in the control group [35].
In order to check if any of the 5 candidate normalizers will show consistent expression in other body fluids, the expression levels of these candidates were compared between plasma and urine. Hsa-miR-92a showed another piece of evidence that it is the best endogenous normalizer among the tested candidates by having the most consistent expression between plasma and urine of hypertensive patients. The fact that other body fluids like urine, cerebrospinal fluid, and saliva have less miRNA than serum and plasma can explain the observed difference in the expression of all the candidates between plasma and urine. To overcome the low level of circulating miRNAs in urine, some studies recommend pre-amplification to be conducted for urine samples in miRNA expression analysis studies [36, 37]. NormFinder can be used not only to rank the candidate normalizers according to the expression stability, but also to determine the optimum number of endogenous normalizers required. The GenEx NormFinder results from this study showed that four (hsa-miR-92a-3p, hsa-miR-21-5p, hsa-miR-16-5p, and hsa-miR-25-3p) is the optimal number of endogenous normalizers that will provide the best normalization under these experimental conditions. However, using 4 endogenous normalizers may be time and resource consuming. We recommend using the best 3 endogenous normalizers (hsa-miR-92a-3p, hsa-miR-21-5p, and hsa-miR-16-5p) especially with the small difference in the accumulated SD between the 3 endogenous normalizers and the 4 endogenous normalizers.
To the best of our knowledge, this is the first study that tries to identify suitable endogenous normalizers for qRT-PCR analysis of plasma miRNA expression in hypertension studies. Future studies with larger sample size and number of candidate endogenous normalizers are recommended to further validate our findings.
Conclusion
The findings from this study suggest that, among the tested candidates, hsa-miR-92a-3p, hsa-miR-21-5p, and hsa-miR-16-5p may be used for normalizing plasma miRNA qRT-PCR expression data in essential hypertension studies.
References
World Health Organization. (2002). The World Health report: Reducing risks, promoting healthy life. Geneva: World Health Organization.
Carretero, O. A., & Oparil, S. (2000). Essential hypertension. Part I: Definition and etiology. Circulation, 101(3), 329–335.
Hamet, P., Pausova, Z., Adarichev, V., Adaricheva, K., & Tremblay, J. (1998). Hypertension: Genes and environment. Journal of Hypertension, 16(4), 397–418.
Synetos, A., Toutouzas, K., Stathogiannis, K., Latsios, G., Tsiamis, E., Tousoulis, D., & Stefanadis, C. (2013). MicroRNAs in arterial hypertension. Current Topics in Medicinal Chemistry, 13(13), 1527–1532.
Lewis, B. P., Burge, C. B., & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1), 15–20.
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., & Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science, 294(5543), 853–858.
Ambros, V. (2004). The functions of animal microRNAs. Nature, 431(7006), 350–355.
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A, 99(24), 15524–15529.
van Rooij, E., Sutherland, L. B., Liu, N., Williams, A. H., McAnally, J., Gerard, R. D., et al. (2006). A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A, 103(48), 18255–18260.
Weber, J. A., Baxter, D. H., Zhang, S., Huang, D. Y., Huang, K. H., Lee, M. J., et al. (2010). The microRNA spectrum in 12 body fluids. Clinical Chemistry, 56(11), 1733–1741.
Li, S., Zhu, J., Zhang, W., Chen, Y., Zhang, K., Popescu, L. M., et al. (2011). Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation, 124(2), 175–184.
Heid, C. A., Stevens, J., Livak, K. J., & Williams, P. M. (1996). Real time quantitative PCR. Genome Research, 6(10), 986–994.
Roberts, T. C., Coenen-Stass, A. M., & Wood, M. J. (2014). Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One, 9(2), e89237.
McDermott, A. M., Kerin, M. J., & Miller, N. (2013). Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS One, 8(12), e83718.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7), RESEARCH0034.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29(9), e45.
Bas, A., Forsberg, G., Hammarström, S., & Hammarström, M. L. (2004). Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scandinavian Journal of Immunology, 59(6), 566–573.
Schmittgen, T. D., & Zakrajsek, B. A. (2000). Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. Journal of Biochemical and Biophysical Methods, 46(1–2), 69–81.
Andersen, C. L., Jensen, J. L., & Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64(15), 5245–5250.
Xia, M., Sherlock, J., Hegerich, P., You, X., Lee, K.,Walworth, C., and Spier, E. (2010). DataAssist™—Data analysis software for TaqMan® real-time PCR data. In Proceedings of the International MultiConference of Engineers and Computer Scientists 2010, IMECS 2010 (Vol I) March 17–19, 2010, Hong Kong.
Johnson, J. A., Boerwinkle, E., Zineh, I., Chapman, A. B., Bailey, K., Cooper-DeHoff, R. M., et al. (2009). Pharmacogenomics of antihypertensive drugs: Rationale and design of the pharmacogenomic evaluation of antihypertensive responses (PEAR) study. American Heart Journal, 157(3), 442–449.
Welder, G. J., Wessel, T. R., Arant, C. B., Schofield, R. S., & Zineh, I. (2006). Complementary and alternative medicine use among individuals participating in research: implications for research and practice. Pharmacotherapy, 26(12), 1794–1801.
Zineh, I., Welder, G. J., DeBella, A. E., Arant, C. B., Wessel, T. R., & Schofield, R. S. (2006). Atorvastatin effect on circulating and leukocyte-produced CD40 ligand concentrations in people with normal cholesterol levels: A pilot study. Pharmacotherapy, 26(11), 1572–1577.
Shaffer, J., Schlumpberger, M., & Lader, E. (2012) miRNA profiling from blood—challenges and recommendations. SABiosciences Technical White Papers. Retrieved from http://www.sabiosciences.com/support_whitepapers.php.
Meyer, S. U., Pfaffl, M. W., & Ulbrich, S. E. (2010). Normalization strategies for microRNA profiling experiments: A ‘normal’ way to a hidden layer of complexity? Biotechnology Letters, 32(12), 1777–1788.
Kan, C. W., Hahn, M. A., Gard, G. B., Maidens, J., Huh, J. Y., Marsh, D. J., & Howell, V. M. (2012). Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer, 12, 627.
Viprey, V. F., Corrias, M. V., & Burchill, S. A. (2012). Identification of reference microRNAs and suitability of archived hemopoietic samples for robust microRNA expression profiling. Analytical Biochemistry, 421(2), 566–572.
Benson, E. A., & Skaar, T. C. (2013). Incubation of whole blood at room temperature does not alter the plasma concentrations of microRNA-16 and -223. Drug Metabolism and Disposition, 41(10), 1778–1781.
Kroh, E. M., Parkin, R. K., Mitchell, P. S., & Tewari, M. (2010). Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods, 50(4), 298–301.
Guo, L. J., & Zhang, Q. Y. (2012). Decreased serum miR-181a is a potential new tool for breast cancer screening. International Journal of Molecular Medicine, 30(3), 680–686.
Bergkvist, A., Forootan, A., Zoric, N., Strombom, L., Sjoback, R., & Kubista, M. (2008). Choosing a normalization strategy for RT-PCR: GenEx system aids in the selection of reference genes for standardizing mRNA measurements. Genetic Engineering Biotechnology News, 28, 13.
Davoren, P. A., McNeill, R. E., Lowery, A. J., Kerin, M. J., & Miller, N. (2008). Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Molecular Biology, 9, 76.
Song, J., Bai, Z., Han, W., Zhang, J., Meng, H., Bi, J., et al. (2012). Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Digestive Diseases and Sciences, 57(4), 897–904.
Mestdagh, P., Van Vlierberghe, P., De Weer, A., Muth, D., Westermann, F., Speleman, F., & Vandesompele, J. (2009). A novel and universal method for microRNA RT-qPCR data normalization. Genome Biology, 10(6), R64.
Zhu, H. T., Dong, Q. Z., Wang, G., Zhou, H. J., Ren, N., Jia, H. L., et al. (2012). Identification of suitable reference genes for qRT-PCR analysis of circulating microRNAs in hepatitis B virus-infected patients. Molecular Biotechnology, 50(1), 49–56.
Semmelmann, K., Lewis, M., Shaffer, J., & Lader, E. miRNA biomarker discovery—overcoming limiting sample material. SABiosciences Technical White Papers. Retrieved from http://www.sabiosciences.com/support_whitepapers.php.
Ramachandran, K., Saikumar, J., Bijol, V., Koyner, J. L., Qian, J., Betensky, R. A., et al. (2013). Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clinical Chemistry, 59(12), 1742–1752.
Acknowledgments
This study was ancillary to the PEAR study that was funded by the National Institutes of Health (U01 GM074492). Mohamed Hassan M. Solayman was funded by the Embassy of the Arab Republic of Egypt.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared no conflict of interest.
Ethical approval
All procedures were in accordance with the ethical standards of the corresponding institutional review boards and with the 1964 Helsinki declaration and its later amendments.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Solayman, M.H.M., Langaee, T., Patel, A. et al. Identification of Suitable Endogenous Normalizers for qRT-PCR Analysis of Plasma microRNA Expression in Essential Hypertension. Mol Biotechnol 58, 179–187 (2016). https://doi.org/10.1007/s12033-015-9912-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9912-z